Abstract
The platelet membrane glycoprotein (GP)Ib-V-IX complex is the receptor for von Willebrand factor and is composed of four membrane-spanning polypeptides: GPIbα, GPIbβ, GPIX, and GPV. A qualitative or quantitative deficiency in the GPIb-V-IX complex on the platelet membrane is the cause of the congenital platelet disorder Bernard-Soulier syndrome (BSS). We describe the molecular basis of a novel variant BSS in a patient in which GPIbα was absent from the platelet surface but present in a soluble form in the plasma. DNA sequence analysis showed a homozygous dinucleotide deletion in the codon for Tyr 508 (TAT) in GPIbα. This mutation (GPIbαΔAT) causes a frame shift that alters the amino acid sequence of GPIbα within its transmembrane region. The hydrophobic nature of the predicted transmembrane region and the cytoplasmic tail at the COOH terminal are altered before reaching a new premature stop codon 38 amino acids short of the wild-type peptide. Although GPIbαΔAT was not detectable on the platelet surface, immunoprecipitation of plasma with specific monoclonal antibodies (MoAbs) identified circulating GPIbα. Transient expression of recombinant GPIbαΔAT in 293T cells also generated a soluble form of the protein. Moreover, when a plasmid encoding GPIbαΔAT was transiently transfected into Chinese hamster ovary (CHO) cells stably expressing the GPβ-IX complex, it failed to be expressed on the cell surface. Thus, a dinucleotide deletion in the codon for Tyr 508 causes a frameshift that alters the amino acid sequence of GPIbα starting within its transmembrane region, changes the hydrophobicity of the normal transmembrane region, and truncates the cytoplasmic domain affecting binding to the cytoskeleton and cytoplasmic proteins. This mutation affects anchoring of the GPIbα polypeptide in platelets and causes the observed BSS phenotype with circulating soluble GPIbα.
THE RARE CONGENITAL disorder of platelets initially described by Bernard and Soulier in 19481 and inherited in an autosomal recessive manner is characterized by a decreased number of large, morphologically abnormal platelets and a prolonged cutaneous bleeding time.2 Bernard-Soulier platelets are characterized by normal aggregation in response to agonists such as adenosine diphosphate and epinephrine and a failure to aggregate in the presence of ristocetin. Platelet aggregation induced by ristocetin is dependent on the interaction between von Willebrand factor (vWF) and the platelet glycoprotein (GP)Ib-V-IX complex.3 A critical functional role for the GPIb-V-IX complex was initially suggested by studies of Bernard-Soulier platelets, in which there was absent surface expression of GPIb, GPIX, and GPV.4
The platelet GPIb-V-IX complex consists of four type I membrane-spanning polypeptides that belong to the leucine-rich motif (LRM) family.5 GPIb consists of two subunits, α (140 kD) and β (27 kD), which are disulfide-linked.6,7 GPIX is noncovalently associated with GPIbα.8 GPV and GPIb-IX also form a noncovalent complex in the platelet membrane.9 The amino-terminus globular domain of the α subunit contains the binding site for vWF.10 The external portion of the α chain, termed glycocalicin (135 kD),11 can be cleaved from the platelet surface and found in circulation in normal plasma.12
The molecular basis of Bernard-Soulier syndrome (BSS) has been characterized in 15 published cases to date. The majority of the mutations characterized that result in BSS are within the GPIbα gene, five of which contain a nonsense mutation producing a truncated GPIbα protein,13-17 and an additional four mutations have been localized to the LRM of GPIbα.18-21 A single case having a mutation in GPIbα that changed a cysteine residue involved in disulfide bonding has also been described.22 Four mutations have been identified within GPIX, two of which resulted from an amino acid change in the LRM or the region flanking the LRM in GPIX,23,24 and two other point mutations that changed a cysteine to a tyrosine in GPIX25 and another that caused a nonsense codon.17 There has been a single report of a mutation within GPIbβ that resulted in BSS.26 Consistent with the description of these mutations in GPIbα, GPIbβ, and GPIX, data from transfection experiments have shown that all three polypeptides (Ibα, Ibβ, and IX) are needed for efficient surface expression of GPIbα and for efficient binding of vWF.6 27
In the present study, we report the molecular basis of a novel variant form of BSS. The patient described herein had a circulating protein recognized by monoclonal antibodies (MoAbs) to GPIbα despite the absence of GPIbα platelet surface expression. These results suggest that this truncated GPIbα was produced and secreted into plasma but failed to anchor to the platelet membrane, causing the observed phenotype.
MATERIALS AND METHODS
Case history.A 69-year-old white male whose mother and paternal grandfather immigrated to Wisconsin from Germany carried a long-standing diagnosis of BSS due to the presence of moderate thrombocytopenia (platelet count of 20,000 to 30,000/μL) with large platelets on peripheral blood smears, a profuse bleeding tendency requiring frequent transfusions, and an Ivy bleeding time of longer than 15 minutes. Previous studies in our laboratory have shown that his platelets do not bind to the MoAb AP-1, which recognizes GPIbα.28 His platelets failed to agglutinate in the presence of ristocetin, and no other hematologic abnormalities were noted. A brother and sister who were available for study were unaffected clinically.
MoAbs and glycocalicin.The anti-GPIbα antibody AP-1 blocks vWF binding to GPIbα.3 MBC 142.2 and MBC 142.6 are MoAbs raised against purified GPIbα. PECAM 1.3 is an anti-PECAM MoAb.29 MBC 123.1 and MBC 132.1 are MoAbs against GPIIIa and GPIIb, respectively. An anti-GPIX MoAb (FMC 25) was purchased from Harlan Bioproducts (Indianapolis, IN).
Purified glycocalicin was obtained from outdated platelets as previously described,30 and provided by Dr P.A. Kroner (The Blood Center of Southeastern Wisconsin).
Blood.Blood samples from the patient and the patient's family members who were available for study were collected into acid-citrate-dextrose. Platelet-rich plasma (PRP) from the siblings was prepared by centrifugation at 800g for 2 minutes. Platelets were isolated and washed three times by differential centrifugation and then resuspended in Phillips31 buffer (96.5 mmol/L NaCl, 85.7 mmol/L glucose, 1.1 mmol/L EDTA, and 8.5 mmol/L Tris) with 50 ng/mL prostaglandin E1 (PgE1 ; Sigma, St Louis, MO). Since BSS platelets are typically large and morphologically abnormal, a different technique was used to isolate platelets from the patient. Whole blood in 5-mL aliquots was layered onto 7.5 mL of a solution containing 1 part 32.8% (wt/vol) sodium metrizoate (Sigma) and 2 parts 4% (wt/vol) dextran (Sigma) in a 15-mL conical tube. The tube was then set at a 10° angle from the vertical position for 2 hours at room temperature to allow red blood cells to sediment. PRP was then gently aspirated, and aliquots were placed into 17 × 100-mm polypropylene tubes containing 50 ng/mL PgE1 and centrifuged at 1,000g for 10 minutes. The platelets were washed again in Phillips buffer containing PgE1 and centrifuged at 1,000g for 10 minutes.
Platelet lysates were prepared by resuspending the platelet pellet in 500 μL lysis buffer (96.5 mmol/L NaCl, 85.7 mmol/L glucose, 1.1 mmol/L EDTA, 8.5 mmol/L Tris, 5 mmol/L N-ethylmaleimide, 100 μg/mL leupeptin, 1 mmol/L phenylmethylsulfonyl fluoride, and 1% Triton X-100 [Pierce, Rockford, IL]). The lysate was vortexed for 3 minutes and then centrifuged at 4°C for 10 minutes at 16,000g. Aliquots of platelet lysate and platelet-poor plasma (PPP) were frozen at −80°C until analyzed.
Immunoblotting.Platelet lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on an 8% to 16% exponential gradient according to the method of Laemmli.32 The separated proteins were electroblotted onto a polyvinylidine difluoride membrane (Novex, San Diego, CA) as described by Towbin et al,33 blocked in phosphate-buffered saline (PBS) containing 5% powdered milk, and then incubated overnight with the anti-GPIbα MoAbs MBC 142.6 or MBC 142.11. The membrane was washed three times with PBS with 5% powdered milk, incubated with a goat anti-mouse IgG conjugated with horseradish peroxidase, washed again three times, and treated with SuperSignal Substrate (Pierce, Rockford, IL).
Polymerase chain reaction amplification of genomic DNA.Genomic DNA was isolated from peripheral blood lymphocytes as previously described.34 DNA was amplified by polymerase chain reaction (PCR) using primer pairs based on the published genomic sequence of GPIbα.35 For DNA sequence analysis, the full-length coding region for mature GPIbα was amplified with primers 162 to 181 (GGCCTGCATTTCCTCCTCACC) and 2653 to 2634 (AAGCTCCCGATGCTGCATGGG). The target sequences were amplified in a 50-μL reaction volume containing between 500 and 1,000 ng genomic DNA, 30 pmol of each primer, and 0.2 mmol of each dNTP in a reaction buffer consisting of 60 mmol/L Tris hydrochloride, pH 9.0, 15 mmol/L (NH4 )2SO4 , 2 mmol/L MgCl2 , and 1 U Taq polymerase (Perkin Elmer, Foster City, CA). PCR amplification was performed in a programable thermal cycler (model 9600; Perkin Elmer) for 35 cycles of denaturation for 45 seconds at 96°C, annealing for 1 minute at 60°C, and extension for 1 minute at 72°C. PCR products containing the entire coding region for GPIbα were cloned into the pCRII.1 cloning vector using the TA cloning kit (Invitrogen, San Diego, CA).
DNA sequencing.Direct sequence analysis of the entire coding region of PCR-amplified GPIbα and six independent clones from each of the patient, his brother, and sister was performed using the Prism Ready Reaction DyeDeoxy terminator cycle-sequencing kit and an Applied Biosystems (Foster City, CA) model 373A DNA sequencer. Sequencing primers were synthesized on an Applied Biosystems model 394 DNA synthesizer.
Transient expression.For transient expression studies, 293T cells provided by Dr D. Ginsburg (University of Michigan, Ann Arbor, MI) were used. The parent 293T cell line is a human renal epithelial cell transformed with SV40 large T antigen.36 293T cells were maintained at 37°C in a 5% CO2 , humidified chamber in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and 2 mmol/L L-Gln. A Pst I restriction fragment (nucleotides 1246 to 2206 and containing the two-base deletion) of the subcloned GPIbα PCR product (GPIbαΔAT) amplified from genomic DNA was subcloned into pGEM-7 containing the wild-type GPIbα cDNA, from which a PstI restriction fragment had been excised. A Xho I/Mlu I restriction fragment containing the entire coding region for GPIbα was then excised from this pGEM-7GPIbαΔAT and inserted into the mammalian expression vector pCI-NEO (Promega, Madison, WI), which carries the human cytomegalovirus promoter and the SV40 origin of replication. This expression vector (pCI-NEOGPIbαΔAT) now contained the entire coding region for GPIbα with a silent polymorphism Arg358 (A → G) present in the patient and the two-base deletion at codon Tyr 508 (TAT). Constructs containing both wild-type and mutant GPIbα were sequenced to ensure that no additional mutations had been introduced. Expression plasmids were introduced into 293T cells in the presence of lipofectamine (GIBCO-BRL, Grand Island, NY) following the protocol of Felgner et al.37 In brief, 4 × 106 cells were plated in 100-mm dishes and grown overnight; 1.6 mL OPTI-MEM-reduced serum media (GIBCO-BRL) containing 150 μg lipofectamine and 10 μg of the appropriate plasmid DNA was added, and the cells were incubated for 5 hours. Eight milliliters of culture medium was added, and incubation was continued at 37°C for 48 hours. Conditioned media were harvested, cleared by centrifugation at 10,000g for 10 minutes at 4°C, and then stored at −80°C until analyzed. Cells were detached from the plates with 3 mmol/L EDTA, and lysates were prepared using the same conditions as described for platelet lysate.
Immunoprecipitation.GPIbα MoAbs MBC 142.2 and AP-1 were coupled to cyanogen bromide–activated Sepharose 4B beads (Sigma). PPP, conditioned medium, or transfected cell lysate were precleared by incubation with uncoupled Sepharose CL-4B beads for 1 hour at room temperature. The beads were centrifuged at 1,000g, and the supernatant was added to the antibody-coupled beads and incubated for 2 hours at room temperature or overnight at 4°C. The beads were washed, and the immunoprecipitated complexes from PPP or conditioned medium eluted in a sample buffer containing 2% SDS. Cell lysate and platelet lysate were eluted with a sample buffer containing 5% SDS. All samples were boiled at 100°C for 3 minutes. The samples were then analyzed by SDS-PAGE on an 8% to 16% exponential gradient or 6% gels in the presence of 5% β-mercaptoethanol. Immunoblotting was performed in the same manner as already described using the antibodies MBC 142.6 and MBC 142.11 for detection.
Flow cytometry.Chinese hamster ovary (CHO) βIX cells (kindly provided by Dr José A. López, Baylor College of Medicine, Houston, TX) are CHO cells that stably express human GPIbβ and GpIX at high levels.38 These cells were additionally transiently transfected with either the wild-type GPIbα or the construct containing the two-base deletion (GPIbαΔAT) or mock-transfected with the plasmid PMT2 alone. Transfected cells were detached from tissue culture plates with 0.5 mmol/L EDTA, suspended in PBS, centrifuged at 250g, and resuspended in Hanks balanced salt solution (HBSS) with 1% bovine serum albumin and 1% normal donkey serum. The cells (3 × 105) were transferred to each well of a 96-well V-bottom plate (Dynatech, Chantilly, VA) and incubated in HBSS with 1% bovine serum albumin containing the primary MoAbs for 60 minutes at room temperature. MBC 142.11 7 μg/mL and AP-1 5 μg/mL were used to detect the transiently expressed GPIbα. FMC 25 5 μg/mL was used to detect GPIX. The cells were then washed twice and incubated for an additional 30 minutes in a darkened room with a 1:100 dilution of phycoerythrin-conjugated affinity-purified F(ab′ )2 donkey anti-mouse IgG (Jackson Immunoresearch Laboratories, West Grove, PA). The cells were then washed twice, resuspended in 2% paraformaldehyde, and analyzed in a Becton Dickinson (San Jose, CA) FACScan flow cytometer.
RESULTS
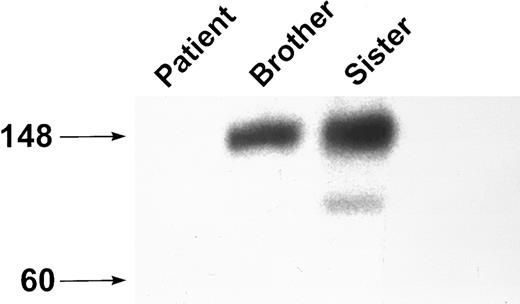
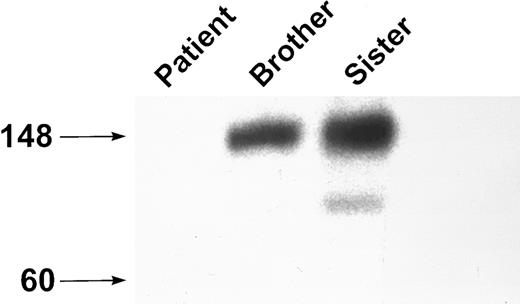
Analyses of platelet lysate.Previous studies have shown that the anti-GPIbα MoAb AP-1 fails to bind to this patient's platelets in a rapid whole-blood assay,28 whereas the MoAb AP-2, which recognizes GPIIb-IIIa, bound normally. Western blot analysis of platelet lysate with the MoAbs MBC 142.6 and MBC 142.11 showed no GPIbα in the patient's platelets. However, immunoblot analysis of platelets from the two siblings showed the presence of a band at approximately 140 kD, consistent with normal GPIbα (Fig 1). When this gel was subjected to an exposure 40 times longer than the control gel, a very faint band of about 140 kD was identified in the patient. Western blot analysis of platelet lysate with antibodies against the platelet membrane proteins GPIIb-IIIa and PECAM were normal in both the patient and the siblings (data not shown).
Western blot analysis of GPIbα. Platelet lysate was analyzed by SDS-PAGE on an 8% to 16% exponential gradient in the presence of β-mercaptoethanol and immunoblotted with the anti-GPIbα MoAb MBC 142.6. There is no GPIbα present in platelet lysate from the patient.
Western blot analysis of GPIbα. Platelet lysate was analyzed by SDS-PAGE on an 8% to 16% exponential gradient in the presence of β-mercaptoethanol and immunoblotted with the anti-GPIbα MoAb MBC 142.6. There is no GPIbα present in platelet lysate from the patient.
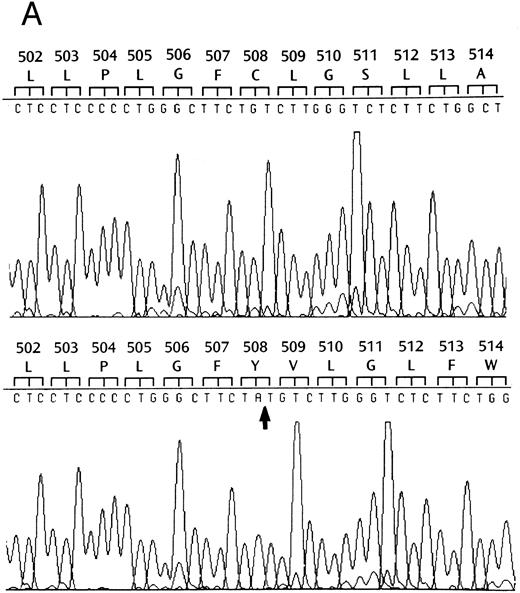
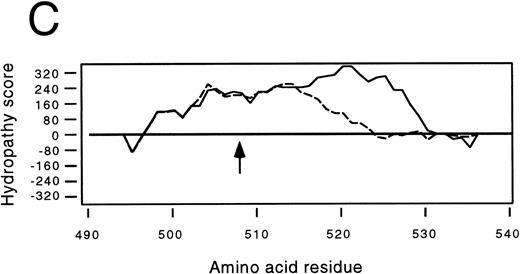
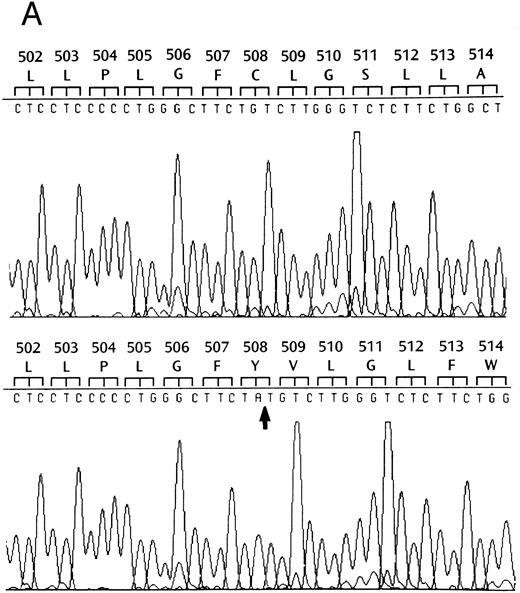
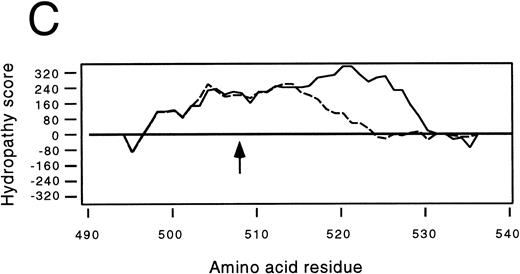
Identification of a two-base deletion in the GPIbα gene.To determine the genetic basis for the absence of GPIbα, we amplified the entire coding region of GPIbα from the patient's genomic DNA. The PCR-amplified DNAs of both the siblings and the patient were of the predicted size according to the published sequence.35 Sequence analysis of PCR-amplified DNA and six individual clones showed that nucleotides 2059 and 2060 were deleted (Fig 2A). This deletion causes a shift in the reading frame beginning at amino acid residue 508 within the transmembrane domain (Fig 2B), and predicts a premature stop codon after 80 altered amino acids. The regions from amino acid 495 to 535 in both the mutant and wild-type protein were analyzed by the method of Kyte and Doolittle,39 using a window of 10 residues with GeneWorks software (IntelliGenetics, Mountain View, CA). In the mutant protein, although the sequence following the frameshift is still hydrophobic, it is significantly less so than the wild-type protein, suggesting that the transmembrane region in the mutant protein may be truncated (Fig 2C) and not inserted in the platelet membrane. In addition to this deletion, there was a T → C polymorphism at nucleotide 532 within the 5′ untranslated region of GPIbα. An additional A → G polymorphism at nucleotide 1610 did not result in an amino acid change. The patient was homozygous for these polymorphisms40 and a previously described polymorphism having two repeats within the macroglycopeptide region of GPIbα.41 We followed the segregation of this deletion within the available family by amplification and direct sequencing of the entire coding region. In addition, the entire coding region was also cloned and sequenced in both siblings. We found that the patient is homozygous for this deletion, the sister inherited two wild-type alleles, and the brother is heterozygous for the deletion.
Effects of the dinucleotide deletion on the secondary structure of GPIbα. (A) DNA sequence analysis of GPIbα from a normal individual (bottom) and the patient (top). Nucleotides 2059 and 2060 are deleted in the patient; deleted nucleotides are shown with an arrow. (B) Alignment of the transmembrane region (doubly underlined) of the wild-type GPIbα and the mutant protein (GPIbαΔAT). The start of the alternate reading frame resulting from the AT deletion in the mutant coding sequence is shown in bold type. The charged amino acids following the transmembrane region are shown with a +. (C) Hydropathy plot using the method of Kyte and Doolittle39 performed on the transmembrane and surrounding amino acids of the wild-type GPIbα and GPIbαΔAT. The hydropathy plot for the mutant protein is shown as a broken line.
Effects of the dinucleotide deletion on the secondary structure of GPIbα. (A) DNA sequence analysis of GPIbα from a normal individual (bottom) and the patient (top). Nucleotides 2059 and 2060 are deleted in the patient; deleted nucleotides are shown with an arrow. (B) Alignment of the transmembrane region (doubly underlined) of the wild-type GPIbα and the mutant protein (GPIbαΔAT). The start of the alternate reading frame resulting from the AT deletion in the mutant coding sequence is shown in bold type. The charged amino acids following the transmembrane region are shown with a +. (C) Hydropathy plot using the method of Kyte and Doolittle39 performed on the transmembrane and surrounding amino acids of the wild-type GPIbα and GPIbαΔAT. The hydropathy plot for the mutant protein is shown as a broken line.
Identification of soluble GPIbα in plasma and conditioned medium.Immunoprecipitation of plasma with MBC 142.2 and AP-1 and immunoblotting with MBC 142.6 demonstrated the presence of a protein of approximately 130 kD in both the proband and the siblings. However, this was significantly reduced in intensity in the proband versus the siblings (Fig 3). Platelet lysate from the patient that was subjected to the same conditions failed to demonstrate any GPIbα (data not shown).
Western blot analysis of plasma GPIbα. PPP was immunoprecipitated with MBC 142.2, analyzed by SDS-PAGE on an 8% to 16% exponential gradient in the presence of β-mercaptoethanol, and immunoblotted with MBC 142.6. WT is a normal non–family member wild-type sample. The patient's plasma contains a soluble GPIbα.
Western blot analysis of plasma GPIbα. PPP was immunoprecipitated with MBC 142.2, analyzed by SDS-PAGE on an 8% to 16% exponential gradient in the presence of β-mercaptoethanol, and immunoblotted with MBC 142.6. WT is a normal non–family member wild-type sample. The patient's plasma contains a soluble GPIbα.
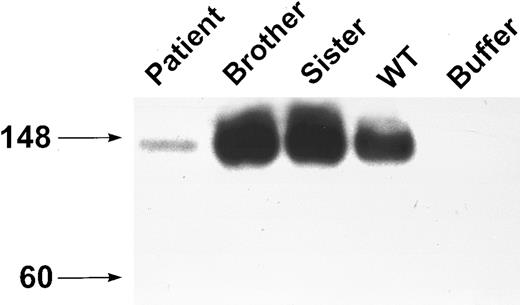
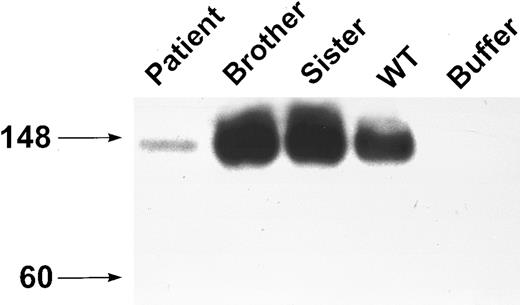
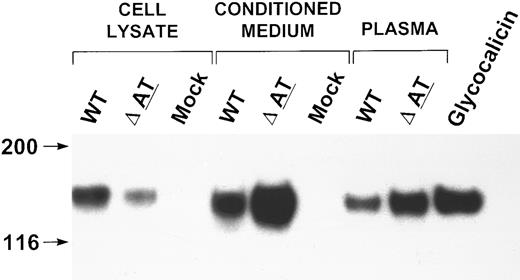
The effect of this mutation was evaluated using 293T cells transfected with either the wild-type GPIbα or the mutant gene (GPIbαΔAT). Conditioned medium was immunoprecipitated with MBC 142.2 and immunoblotted with MBC 142.6. A band of 130 kD was immunoprecipitated from the conditioned medium from both wild-type and GPIbαΔAT transfected cells (Fig 4). This band had a mobility similar to that of purified glycocalicin, and was identical in size to GPIbα-reactive material that was immunoprecipitated from the plasma of both the patient and his brother (Fig 4). However, considerably more of this product appeared in the conditioned medium of GPIbαΔAT-transfected cells versus medium from cells that had been transfected with wild-type GPIbα. A band of approximately 140 kD was identified in the lysate of cells that had been transfected with either the wild-type GPIbα or GPIbαΔAT (Fig 4). This product was slightly larger (∼10 kD) than the recombinant protein in the conditioned medium. A similar difference in size was noted in recombinant protein identified in cell lysates and conditioned medium when wild-type GPIbα was transfected into CHO cells.42 There was more of this product in the lysate of cells transfected with the wild-type GPIbα versus the mutant. These results suggest that GPIbαΔAT is more readily processed from the cell into the supernatant than wild-type GPIbα.
Western blot analysis of expressed WT and mutant GPIbα. Cell lysate, conditioned medium of 293T transfected cells, and plasma from the patient and his brother were immunoprecipitated with MBC 142.2, analyzed simultaneously by SDS-PAGE on a 6% gel, and immunoblotted with MBC 142.6. The immunoprecipitate of the brother's PPP was diluted 1:25, and the patient's PPP was diluted 1:2. Purified glycocalicin 0.38 ng was analyzed on this gel simultaneously.
Western blot analysis of expressed WT and mutant GPIbα. Cell lysate, conditioned medium of 293T transfected cells, and plasma from the patient and his brother were immunoprecipitated with MBC 142.2, analyzed simultaneously by SDS-PAGE on a 6% gel, and immunoblotted with MBC 142.6. The immunoprecipitate of the brother's PPP was diluted 1:25, and the patient's PPP was diluted 1:2. Purified glycocalicin 0.38 ng was analyzed on this gel simultaneously.
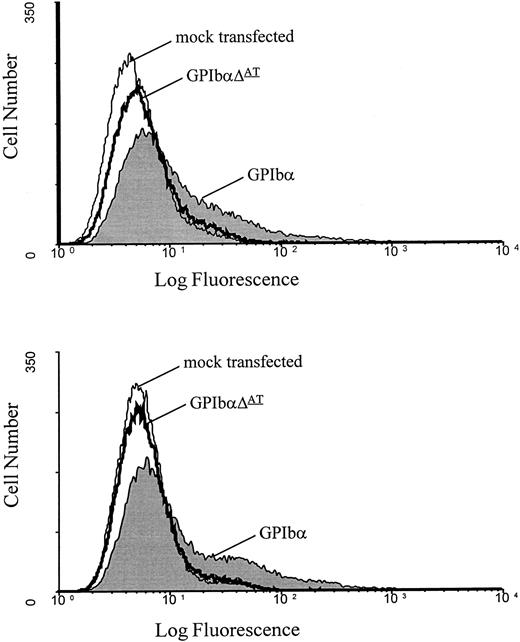
Transfection of DNA into CHOβ-IX cells.Transfected alone, GPIbα is not expressed on the cell surface in appreciable quantities.27 Studies examining the role of GPIbβ and GPIX have shown that all three subunits are required for efficient expression of the ligand-binding subunit on the surface of transfected cells.27 Therefore, to investigate the effect of this mutation on surface expression, both wild-type GPIbα and mutant GPIbαΔAT were transiently transfected into CHO cells that stably express both GPIbβ and GPIX.27 These cells were then incubated with the anti-GPIbα MoAbs MBC 142.11 and AP-1 followed by a phycoerythrin-conjugated donkey anti-mouse antibody, and then analyzed by flow cytometry. Figure 5 shows that a significant fraction of the cells transfected with the wild-type GPIbα construct exhibited an appreciable increase in surface fluorescence compared with the mock control. In contrast, there was no significant change in the surface fluorescence of cells transfected with the mutant GPIbαΔAT compared with the mock control (Fig 5).
FACS analysis of GPIbα and GPIbαΔAT-transfected CHO βIX cells. Analysis of transfected cells with the MoAbs AP-1 (top) and MBC 142.11 (bottom). There is no significant increase in fluorescence with the mutant protein.
FACS analysis of GPIbα and GPIbαΔAT-transfected CHO βIX cells. Analysis of transfected cells with the MoAbs AP-1 (top) and MBC 142.11 (bottom). There is no significant increase in fluorescence with the mutant protein.
DISCUSSION
This report characterizes a novel mutation responsible for BSS and the production of a soluble GPIbα. The BSS patient reported in this study has a homozygous two-base deletion of the last two base pairs of the codon for Tyr 508 (TAT) in GPIbα. This mutation causes a translational frameshift that alters the amino acid sequence of GPIbα within the transmembrane domain before reaching a new premature stop codon 38 amino acids short of the wild-type peptide at the cytoplasmic tail. No GPIbα was detected on the platelet surface with AP-1, but a protein similar to glycocalicin was immunoprecipitated from the patient's plasma using two different anti-GPIbα MoAbs, AP-1 and MBC 142.2. An identical product was identified in the conditioned medium of 293T cells transfected with GPIbαΔAT cDNA. In addition, when transiently transfected into CHO cells stably expressing GPIbβ and GPIX, there was no surface expression of GPIbαΔAT. These results suggest that this dinucleotide deletion produces a glycocalicin-like protein that circulates in plasma because the truncated protein fails to anchor in the platelet membrane.
The molecular determinants involved in association of the subunits that comprise the GPIb-V-IX complex are not well understood. In cultured cells, full-length GPIbα associates with the other components of the GPIb-IX complex and is inserted into the membrane of these cultured cells.27,43 Deletion of amino acids Trp-586 to Ser-606 (numbering according to Wenger et al35 ) from the cytoplasmic domain of GPIbα abolished the interaction of the entire GPIb-IX complex with the cytoskeleton.44 However, truncation of the carboxy-terminal half of the cytoplasmic domain of GPIbα to a length significantly shorter than the mutation described in this report did not affect association of GPIbα with the GPIb-IX complex, insertion of the complex into the membrane, or binding of vWF to the extracellular domain of the complex.44 Thus, deletion of amino acids Trp-586 to Ser-606 of the cytoplasmic tail does not appear to be critical for coordinate expression of the complex on the surface of mammalian cells.
Transfection of mammalian cells with vectors containing the coding sequence for the entire human GPIbα have yielded variable results depending on the host cell used. When the cDNA containing the full-length GPIbα alone was transfected into COS cells, a 48/46-kD doublet was immunoprecipitated from the cell lysates with an anti-GPIbα MoAb.45 Transfection with full-length GPIbα cDNA and a GPIbα cDNA truncated at an internal Xba I into COS cells also demonstrated a 48/46-kD doublet in the culture medium.46 It has been suggested that the reason for truncation of the recombinant polypeptide is intracellular cleavage of the translation products of the full-length transcript.46 Transfection with GPIbα alone into COS cells failed to produce detectable GPIbα on the surface of these cells.47 These results are in accordance with those of López et al,27 who were also unable to identify GPIbα on the cell surface of CHO cells in the absence of GPIbβ and GPIX complex. In contrast, Meyer et al42 reported that when GPIbα alone is transfected into CHO cells and subjected to several rounds of gene amplification, a 110-kD recombinant protein similar to glycocalicin is found in the supernatant and GPIbα is detectable on the cell surface. The results of our transfection experiments in 293T cells are similar to the results reported for transfection into CHO cells.42 The difference in results between these mammalian cell lines may be due to active retention of GPIbα within the endoplasmic reticulum of COS cells,27 45-47 when GPIbα is transfected in the absence of GPIbβ and GPIX complex.
Whereas overexpression of GPIbα may lead to its appearance on the surface of cultured mammalian cells, it appears that the other subunits are required in vivo for expression of a functional complex, since mutations in both GPIX48 and GPIbβ26 have been described that cause BSS. Recently, Kunisihima et al16 described a patient with a homozygous single-nucleotide substitution causing a nonsense mutation at residue 444 of GPIbα, which generated a truncated molecule that lacked part of the extracellular region, the transmembrane region, and the entire cytoplasmic domain. There was no detectable GPIbα on the surface of the patient's platelets, but the patient had detectable glycocalicin both in plasma and within the platelets. These results are analogous to the results described in this investigation, in which there was no surface expression of GPIbα yet a protein similar to glycocalicin could be detected in the plasma. The results of our transfection experiments in 293T cells demonstrated a product that was similar to glycocalicin in both the wild-type and mutant transfections.
In summary, GPIbαΔAT changes the hydrophobicity of the transmembrane region and also truncates and changes the cytoplasmic sequence of this mutant polypeptide. GPIbαΔAT does not become localized to the plasma membrane in cells stably expressing GPIbβ and GPIX. These results suggest that the transmembrane region of GPIbα is critical in the assembly and association of the GPIb-V-IX complex.
ACKNOWLEDGMENT
The authors thank Dr Laurence Tempelis for providing clinical data, Dr Philip A. Kroner for providing glycocalicin, and Dr Chris Ward for assisting with graphics.
Supported by US Public Health Service Grants No. R29 HL56027 (D.K.) and PO1 HL44612 (R.R.M. and P.J.N.), a Grant-in-Aid from the National American Heart Association (D.K.), and Grant No. RR0344 (Clinical Research Center).
Address reprint requests to Dermot Kenny, MD, Blood Research Institute, The Blood Center of Southeastern Wisconsin, PO Box 2178, Milwaukee, WI 53201-2178.