Abstract
Kaposi's sarcoma-associated herpes virus (KSHV)/human herpes virus 8 (HHV8) DNA sequences have been demonstrated in Kaposi's sarcoma (KS), as well as in some acquired immunodeficiency syndrome (AIDS)-related non-Hodgkin's lymphomas (NHL) and in multicentric Castleman's disease. Although KSHV DNA generally is abundant in KSHV-associated lymphomas, few copies of the virus are present in KS, a property that confounds detection by in situ methods. Previous in situ studies, which identified KSHV in lesions of KS, relied on the use of polymerase chain reaction (PCR) to amplify target DNA sequences before in situ hybridization (ISH) for localization or used ISH with radioactively-labeled probes to obtain adequate levels of detection sensitivity. In this study, a novel nonisotopic nucleic acid ISH method using catalyzed signal amplification and colorimetric detection without PCR-dependent target amplification was used to identify KSHV-specific sequences. The level of sensitivity was increased further by using a probe that detects viral cyclin D homolog transcripts, which are expressed at significant levels during latent viral infection. Thirty cutaneous lesions of KS (25 AIDS-related and five classical European type) were evaluated. AIDS-related NHL and cell lines derived from patients with AIDS-related NHL, all of which were known to harbor KSHV by Southern blot analysis, were used as positive controls. NHL and benign cutaneous vascular lesions not associated with AIDS were used as negative controls. For each of the 30 KS lesions studied, hybridization signals were detected in most of the spindle cells surrounding the atypical slit-like vascular channels and also were detected in some endothelial cells in well-formed blood vessels in the perilesional dermis. Plaque and nodular lesions generally contained more labeled cells than did early patch lesions. All AIDS-related NHL and cell lines contained KSHV-specific sequences; however, the non-AIDS–related NHLs and benign vascular lesions were negative. These results confirm the presence of KSHV sequences in cutaneous KS and provide in situ evidence of infection by this virus in early patch-stage lesions. This study also defines the in situ expression of the KSHV cyclin D homolog viral oncogene in cutaneous KS. The use of this sensitive nonisotopic ISH method should allow detection of other KSHV-specific gene products, further defining the pathobiology of this virus.
KAPOSI'S SARCOMA-ASSOCIATED herpes virus (KSHV) is a recently-discovered gammaherpes virus1 also called human herpes virus 8 (HHV8).2 KSHV DNA sequences have been demonstrated in Kaposi's sarcoma (KS),1 in an unusual form of acquired immunodeficiency syndrome (AIDS)-related non-Hodgkin's lymphoma (NHL), which usually presents as an effusion3-5 and in a significant percentage of multicentric Castleman's disease.6 Furthermore, KSHV has been identified in all forms of KS including AIDS-related (epidemic), classical European, endemic, and posttransplant (iatrogenic),1,7-13 suggesting a specific etiologic role for this virus in the pathogenesis of KS. To better define this role, the stage of tumor progression during which specific virally-encoded genes are expressed must be determined. For KS, histologic evidence of tumor progression is recognized as the evolution of early patch-stage lesions into larger plaques and ultimately into nodules and tumors.14 Several in situ hybridization (ISH) studies have demonstrated KSHV viral DNA sequences in lesions of KS15-17; however, these studies evaluated later plaque- and nodular/tumor-stage lesions exclusively and relied on labor-intensive procedures using the polymerase chain reaction (PCR) ISH15or ISH using radioactively labeled probes.16,17 These highly sensitive ISH techniques were necessary because very few copies of the virus are present in KS lesions,1 thus confounding detection by other less sensitive ISH methods. However, despite the enhanced sensitivity of detection conferred by radiolabeled probes, KSHV was not detected in all of the KS lesions in some of these ISH studies.16,17 Greater sensitivity was achieved by Boshoff et al15 who successfully identified KSHV in all of the plaque- and nodular/tumor-stage lesions studied by PCR-ISH and more recently, Staskus et al18 who used radioactively labeled probes to identify KSHV-specific sequences in all of their KS specimens including one very early patch-stage lesion.
In this study, cutaneous lesions of KS representing early patch-stage, plaque-stage, and late nodular/tumor-stage lesions were evaluated by ISH for the presence of KSHV-specific nucleic acid sequences. A novel ISH method based on colorimetric detection of a biotinylated KSHV-specific probe after catalyzed signal amplification19-21 was used. This ISH method is based on the catalyzed amplification of a detection system that does not require PCR amplification of nucleic acid target sequences or the use of radioactively labeled detection probes. Recently, this technique has been used to identify human papillomavirus-specific sequences in infected cell lines and in human tissues.22 Because it is likely that KSHV primarily exists in a latent state in KS lesional tissues,18,23,24 a latently-expressed gene was targeted in this ISH study to increase the sensitivity of viral detection. Large segments of the KSHV genome have been mapped and sequenced.2,25,26 Among the open reading frames (ORF) identified for KSHV is ORF72, a gene that encodes a protein with 257 amino acids and which shares homology with a number of human cellular cyclins including cyclin D125,27 and which is now designated v-cyc. Cyclins are a class of proteins, which are required for normal cellular proliferation,28 and cyclin D1 is now recognized as the PRAD1 oncogene implicated in a number of human malignancies.29-32. In vitro studies on cell lines harboring KSHV33 have shown that v-cyc is a latently-transcribed gene,34 which is expressed at significant levels in KS tissues.25 This gene encodes a protein, which is functional in its ability to phosphorylate and thus inactivate the retinoblastoma tumor suppressor protein.27 35 The in situ demonstration of KSHV cyclin D RNA sequences in early lesions of KS would, in addition to confirming the presence of the virus, provide further evidence linking KSHV to specific early steps in the pathogenesis of these lesions.
MATERIALS AND METHODS
Tissues and cell lines.
Formalin-fixed, paraffin-embedded tissues used in these experiments were obtained from the files of the Department of Pathology, The New York Hospital-Cornell Medical Center. Thirty lesions of cutaneous KS including 25 from patients with AIDS and five classical-type lesions from human immunodeficiency virus (HIV)-negative patients of European descent were included in the study (Table 1). The 30 specimens included 10 early patch-stage, 10 plaque-stage, and 10 nodular/tumor-stage lesions. As a positive control, three AIDS-related NHL, which had solid tissue involvement and which were known to contain KSHV sequences by Southern blot analysis, also were studied. As negative controls, five NHL not associated with AIDS, one cutaneous lymph node demonstrating reactive follicular hyperplasia, and five benign cutaneous hemangiomas were studied. All tissues were cut 5 μm thick on positively-charged glass slides (ProbeOn Plus, Fisher Scientific, Pittsburgh, PA).
Cell lines established from three patients with AIDS-related primary effusion lymphomas (PEL) and previously shown to harbor KSHV33 36 also were used as positive controls for ISH. These cell lines (BC-1, BC-2, and BC-3) were grown to a density of 0.3 to 0.5 × 106 cells/mL in RPMI 1640 supplemented with 2 mmol/L glutamine and 20% fetal bovine serum, 37°C in the presence of 5% CO2. The cells were harvested, washed in 1× phosphate-buffered saline (PBS), fixed in 10% phosphate buffered formalin for 1 hour, mixed with an equal volume of molten agar (final concentration, 2% wt/vol), allowed to solidify, and then routinely processed to produce a cell block from which paraffin sections were cut. In addition, a B-cell line (BJAB) known to lack KSHV, was used as a negative control for the mixing control described below.
High temperature target retrieval.
Before ISH, the tissue sections were deparaffinized, rehydrated through a graded series of alcohols, and incubated for 40 minutes in Target Retrieval Buffer (Dako Corp, Carpinteria, CA), which had been preheated and maintained at 95°C. The slides were allowed to cool at room temperature (RT) for 20 minutes before removal from the target retrieval buffer. The tissue sections next were incubated for 20 minutes in 0.2 N HCl, RT followed by a 3-minute wash in distilled H2O (dH2O). Endogenous peroxidase activity was blocked in the tissue sections by incubation in a 3% aqueous solution of H2O2 for 20 minutes, RT followed by a 3-minute wash in dH2O. After blocking endogenous peroxidase activity, tissue sections were used immediately for ISH.
Southern blot analyses.
For the three AIDS-related NHL and the three cell lines used in this study, fresh samples were available for DNA extraction and subsequent Southern blot analyses. Genomic DNA was extracted from cryopreserved tissue blocks or cell pellets using a salting-out procedure.37 Five-microgram aliquots of genomic DNA were digested with BamH I or Hind III restriction endonucleases according to the manufacturer's instructions (Boehringer-Mannheim, Indianapolis, IN). DNA fragments were separated by electrophoresis in 0.8% agarose gels, denatured with alkali, neutralized, and transferred to nitrocellulose filters according to the method of Southern.38 The filters were hybridized as previously described39 with 32P random primer-labeled (PrimeIt; Stratagene, La Jolla, CA) KS330Bam and KS631Bam probes25 to detect the presence of KSHV-specific sequences.
ISH probes.
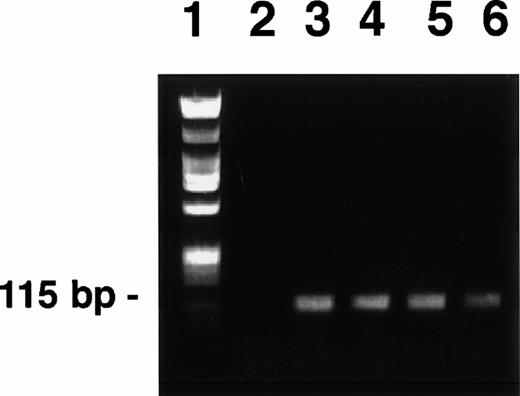
A 115-bp DNA fragment of the KSHV/HHV8 ORF 72 (cyclin D homolog) was amplified by PCR from DNA of the BC-3 cell line using previously published primers25 and PCR conditions.1 The amplification product was purified (QIAquick Spin, Qiagen Inc, Chatsworth, CA), quantified, and an aliquot was run on a 0.8% agarose gel (Fig 1) to verify fragment size and uniformity. The remaining PCR product was biotinylated (Fasttag kit, Vector Laboratories, Burlingame, CA) using the manufacturer's protocol. Biotinylated probes were purified by two cycles of gel filtration using Sephadex G50-packed columns (Pharmacia Biotech Inc, Piscataway, NJ) equilibrated with TNE buffer (100 mmol/L NaCl, 10 mmol/L Tris, pH 7.5, 1 mmol/L EDTA).
PCR-generated KSHV cyclin D probes derived from BC-3 (lanes 3 to 6). Each lane represents a PCR reaction product. Reaction products were pooled before biotinylation. Hind III-digested lambda phage DNA and Hae III-digested ◊X174 DNA mixture used as a molecular weight standard (lane 1). The Hae III-digested ◊X174 DNA was labeled and used as the irrelevant specificity probe mixture, as these fragments were of similar size to the KSHV probe. H2O control (lane 2). PCR-generated probes were prepared and labeled as described in the text.
PCR-generated KSHV cyclin D probes derived from BC-3 (lanes 3 to 6). Each lane represents a PCR reaction product. Reaction products were pooled before biotinylation. Hind III-digested lambda phage DNA and Hae III-digested ◊X174 DNA mixture used as a molecular weight standard (lane 1). The Hae III-digested ◊X174 DNA was labeled and used as the irrelevant specificity probe mixture, as these fragments were of similar size to the KSHV probe. H2O control (lane 2). PCR-generated probes were prepared and labeled as described in the text.
Irrelevant specificity probes were prepared by biotinylation of an aliquot of a cocktail of linearized Hae III-digested φX174 DNA routinely used as DNA molecular weight markers in agarose gels (Life Technologies, Gaithersburg, MD). This probe mixture was labeled and purified using the same protocols used for the KSHV cyclin D-specific probe.
ISH.
All tissues used for ISH were deparaffinized, rehydrated through a graded series of ethanol, submitted to high temperature target retrieval, and blocked of endogenous peroxidase activity as described above. A step using enzymatic digestion of the tissue before hybridization was not required in this protocol. Rehydrated tissue sections then were prehybridized with probe cocktail: final concentration 2× SSC (1× SSC = 150 mmol/L NaCl, 15 mmol/L sodium citrate); 10× Denhardt's solution40; 25% vol/vol ultrapure formamide (Life Technologies), 30% vol/vol low viscosity probe diluent41 (Research Genetics, Huntsville, AL), and 0.8 mg/mL sheared herring sperm DNA. For prehybridization, 30 μL of probe cocktail was placed directly onto the tissue section and a glass coverslip was applied to evenly distribute the cocktail making sure that bubbles were not introduced. The slides were heated for 5 minutes on the surface of a 95°C hotplate then incubated for 30 minutes, 37°C in a humidified chamber. After the prehybridization step, the coverslips were gently removed by immersion of the slides in Coplin jars containing Tris-buffered saline (TBST) (150 mmol/L NaCl, 10 mmol/L Tris, pH 7.5, 0.1% vol/vol Tween 20) for 5 minutes, RT. Excess buffer was carefully removed by wiping the slide around the tissue sections with absorbent paper. A 30-μL aliquot of probe cocktail containing the biotinylated KSHV Cyclin-specific probe (0.1 μg/mL) was then applied to the tissue sections and covered with a glass coverslip. The slides were heated for 5 minutes, 95°C on a hotplate to denature both probe and target sequences and then incubated for 1 hour, 37°C in a humidified chamber to allow hybridization. Coverslips were gently removed by soaking in TBST for 5 minutes, and the sections were washed twice for 5 minutes in 1× SSC, 55°C and then twice under conditions of greater stringency, 0.01× SSC, 55°C, 10 minutes. Washes and subsequent signal amplification and detection system reagents (see below) were applied to the tissue sections by capillary action.42
As a control to evaluate the specificity of hybridization, representative tissue sections were hybridized with a cocktail of biotinylated probes of irrelevant specificity. The irrelevant specificity probes were prepared as described above and were used at the same concentration as the KSHV-specific probe. In addition, selected tissues containing KSHV sequences as determined by ISH and/or Southern blot hybridizations were digested with DNAse-free Ribonuclease A (Boehringer Mannheim), 1 mg/mL, for 1 hour, 37°C before the high temperature target retrieval step. This control was performed to demonstrate that hybridizations were predominantly between the probe and cytoplasmic mRNA sequences in these tissues.
An additional control was performed to ensure that there was no diffusion of hybridization signals between cells. For this control, a B-cell line known to harbor KSHV (BC-3) was mixed in varying percentages with another B-cell line, which does not contain the virus (BJAB). Cell blocks and paraffin sections were prepared as described previously from the cell suspension mixures, followed by ISH with the KSHV cyclin probe. As a further test of labeling fidelity, the brown DAB colorimetric signal was further intensified (rendering it dark brown-to-black) by a 15-second incubation in NiCl2 (Vector Laboratories).
Catalyzed signal amplification.
A nonisotopic, colorimetric signal amplification system (GenPoint kit, Dako Corp) was used in these experiments to visualize specific hybridization signals. The procedure was followed according to the manufacturer's recommendations. For this procedure, after the stringent washes, tissue sections first were incubated with a streptavidin-horseradish peroxidase (SA-HRP) reagent for 15 minutes, RT and washed three times with TBST. After this step, the sections were incubated with a solution containing H2O2 and biotinyl tyramide19 20 for 15 minutes, RT and washed three times with TBST. This step results in the catalyzed deposition of additional biotins at the site of probe hybridization. The sections were then incubated a second time with SA-HRP for 15 minutes, RT and finally washed three times in TBST. Colorimetric signals were localized after incubation in 3,3'diaminobenzidine (DAB) for 5 minutes, RT. Signal development was stopped by placing the slides in dH2O. Hybridizations were identified as punctate brown colorimetric signals in the cellular cytoplasm and/or nucleus. Nuclear signals were considered to represent probe-target DNA hybridizations, whereas cytoplasmic signals were considered to represent primarily probe-target mRNA hybridizations. Tissue sections were counterstained with hematoxylin and permanently mounted. For KS lesions, sections also were counterstained with toluidine blue so that melanin in epidermal keratinocytes and dermal melanophages would turn green and be easily distinguishable from the brown DAB reaction product. Mounted tissue sections were viewed with a standard light microscope using bright field optics. All slides were interpreted by four pathologists (J.A.R., R.G.N., E.C., and D.M.K.).
RESULTS
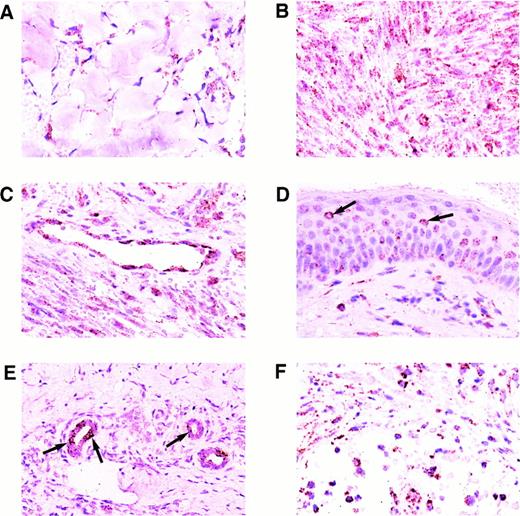
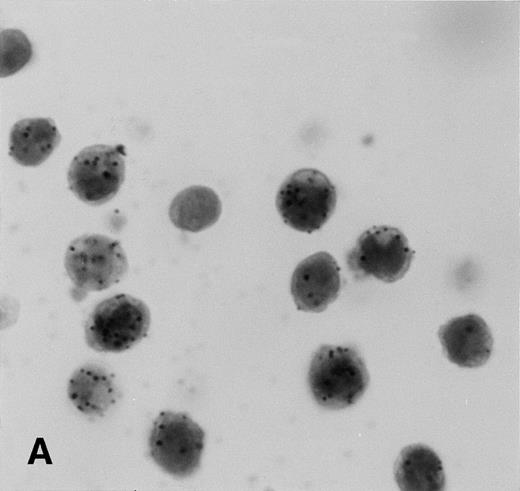
Results of the ISH for KS tissues are shown in Figs 2 and3. All 30 KS lesions contained colorimetric hybridization signals. Signals typically were located in the spindle cell component surrounding the characteristic slit-like vascular spaces of the lesions, in the endothelial cells of well-formed blood vessels in the surrounding dermis, and in small mononuclear cells scattered throughout the lesions. The lesional spindle cells usually contained many cytoplasmic hybridization signals, but only one or very few nuclear signals. Early patch-stage lesions generally had fewer signals than plaque-stage or nodular lesions, as these also had fewer spindle cells (Figs 2A and B and 3A). The intensity of labeling per cell did not differ significantly between patch-stage and later-stage lesions. Endothelial cells within well-formed vessels in the surrounding dermis typically contained abundant cytoplasmic and nuclear hybridization signals (Fig 2C). Of interest, not all of the blood vessels in the surrounding dermis contained labeled endothelial cells. In a few plaque-stage and nodular lesions, scattered epidermal keratinocytes or eccrine ductular epithelial cells contained hybridization signals as well. Only a few epidermal keratinocytes, limited to small foci within the epidermis, were labeled, and these usually contained only a few hybridization signals (Fig 2D). Some of the labeled keratinocytes were surrounded by nonlabeled cells indicating specific localization of the hybridization signal without diffusion. Rare eccrine ductular epithelial cells, which were labeled, typically had larger numbers of signals both in a nuclear and cytoplasmic distribution (Fig 2E), but often were surrounded by nonlabeled cells. The small labeled mononuclear cells scattered throughout the KS lesions contained abundant nuclear and cytoplasmic hybridization signals (Fig 2F).
Expression of the KSHV cyclin D gene in cutaneous KS lesions. (A) Early patch-stage lesion. Note hybridization signals in spindle cells forming the narrow vascular spaces in the dermis. (B) Nodular/tumor-stage lesion showing large numbers of hybridization signals in lesional spindle cells. (C) Well-formed blood vessel in the dermis adjacent to nodular lesion of KS. Note abundant hybridization signals in the vascular endothelial cells. (D) Plaque-stage lesion of KS with scattered keratinocytes in the overlying epidermis containing a few hybridization signals. Some of the labeled keratinocytes (arrows) are isolated and are surrounded by nonlabeled cells indicating that diffusion of label between cells did not occur. (E) Eccrine ductular epithelial cells containing hybridization signals. Only some of the eccrine epithelial cells are labeled in this plaque-stage KS lesion. Some of the labeled cells (arrows) are isolated and are surrounded by nonlabeled epithelial cells. (F) Nodular KS lesion containing scattered mononuclear cells, morphologically consistent with lymphocytes or histiocytes, which are labeled. All panels, ISH with catalyzed signal amplification, DAB chromogen, toluidine blue and hematoxylin counterstain. (A through F) Original magnification × 270.
Expression of the KSHV cyclin D gene in cutaneous KS lesions. (A) Early patch-stage lesion. Note hybridization signals in spindle cells forming the narrow vascular spaces in the dermis. (B) Nodular/tumor-stage lesion showing large numbers of hybridization signals in lesional spindle cells. (C) Well-formed blood vessel in the dermis adjacent to nodular lesion of KS. Note abundant hybridization signals in the vascular endothelial cells. (D) Plaque-stage lesion of KS with scattered keratinocytes in the overlying epidermis containing a few hybridization signals. Some of the labeled keratinocytes (arrows) are isolated and are surrounded by nonlabeled cells indicating that diffusion of label between cells did not occur. (E) Eccrine ductular epithelial cells containing hybridization signals. Only some of the eccrine epithelial cells are labeled in this plaque-stage KS lesion. Some of the labeled cells (arrows) are isolated and are surrounded by nonlabeled epithelial cells. (F) Nodular KS lesion containing scattered mononuclear cells, morphologically consistent with lymphocytes or histiocytes, which are labeled. All panels, ISH with catalyzed signal amplification, DAB chromogen, toluidine blue and hematoxylin counterstain. (A through F) Original magnification × 270.
Expression of KSHV cyclin D. (A) Higher magnification, same nodular-stage lesion of KS as shown in Fig 2B. Note abundant signal in the cytoplasm of lesional spindle cells; fewer nuclear signals are present. (B) Same nodular-stage KS tissue hybridized with the biotinylated probe of irrelevant specificity. Hybridization signals are not detected. (C) Nodular-stage lesion of KS hybridized with the KSHV-specific probe after RNAse digestion of the tissue. Only a few nuclear signals remain. (D) Benign cutaneous hemangioma, which did not contain any labeled cells. (E) BC-3 positive control cell line showing abundant cytoplasmic and nuclear hybridization signals. (F) Non-AIDS–related cutaneous NHL, which did not contain any labeled cells. All panels, ISH with catalyzed signal amplification, DAB chromogen, hematoxylin counterstain. (A, B, C, and E) Original magnification × 660. (D and F) Original magnification × 270.
Expression of KSHV cyclin D. (A) Higher magnification, same nodular-stage lesion of KS as shown in Fig 2B. Note abundant signal in the cytoplasm of lesional spindle cells; fewer nuclear signals are present. (B) Same nodular-stage KS tissue hybridized with the biotinylated probe of irrelevant specificity. Hybridization signals are not detected. (C) Nodular-stage lesion of KS hybridized with the KSHV-specific probe after RNAse digestion of the tissue. Only a few nuclear signals remain. (D) Benign cutaneous hemangioma, which did not contain any labeled cells. (E) BC-3 positive control cell line showing abundant cytoplasmic and nuclear hybridization signals. (F) Non-AIDS–related cutaneous NHL, which did not contain any labeled cells. All panels, ISH with catalyzed signal amplification, DAB chromogen, hematoxylin counterstain. (A, B, C, and E) Original magnification × 660. (D and F) Original magnification × 270.
ISH performed on KS lesions with biotinylated probes of irrelevant specificity did not contain hybridization signals (Fig 3B) indicating that the hybridizations with the KSHV probe were specific. KS tissues, which had been digested with RNAse before ISH with the KSHV cyclin-specific probe, did not contain or contained significantly fewer cytoplasmic hybridization signals than did the matched tissue sections, which did not receive RNAse treatment (Fig 3A and C) indicating that the cytoplasmic hybridizations were RNA-specific. The five benign cutaneous hemangiomas evaluated in this study did not contain KSHV-specific sequences (Fig 3D).
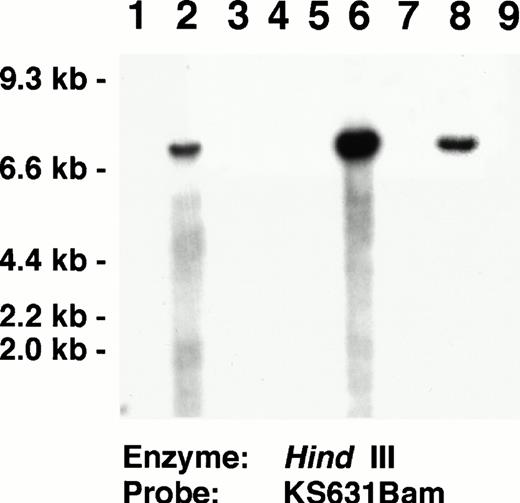
Each of the three AIDS-related NHL and three cell line positive controls studied contained KSHV-specific sequences (Fig 3E). The three positive control cell lines showed hybridization signals in virtually all of the cells with some cells having many nuclear signals. In the AIDS-related NHL tissues, lymphoma cells typically contained many nuclear and cytoplasmic signals. These tissues also contained scattered smaller mononuclear cells, which were labeled. Five primary cutaneous non-AIDS–related NHL and one cutaneous lymph node with reactive follicular hyperplasia did not contain KSHV-labeled cells (Fig 3F). All of these AIDS-related NHL and cell lines used as positive controls in the ISH experiments contained KSHV by Southern blot analyses (Fig 4).
Southern blot analysis identifying KSHV in AIDS-related NHL used as positive controls for the ISH experiments. Lanes 2, 6, 8:Hind III-digested DNA from AIDS-associated NHL containing KSHV-specific sequences identified by the KS631 probe. Lanes 3 to 5, 7, 9: AIDS-associated NHL lacking KSHV-specific sequences. Lane 1, Control DNA extracted from HL-60 promyelocytic leukemia cell line. Southern blots showing KSHV-specific sequences in the BC-1, BC-2, and BC-3 cell lines, which also were used as positive controls for ISH have been published previously.33 36
Southern blot analysis identifying KSHV in AIDS-related NHL used as positive controls for the ISH experiments. Lanes 2, 6, 8:Hind III-digested DNA from AIDS-associated NHL containing KSHV-specific sequences identified by the KS631 probe. Lanes 3 to 5, 7, 9: AIDS-associated NHL lacking KSHV-specific sequences. Lane 1, Control DNA extracted from HL-60 promyelocytic leukemia cell line. Southern blots showing KSHV-specific sequences in the BC-1, BC-2, and BC-3 cell lines, which also were used as positive controls for ISH have been published previously.33 36
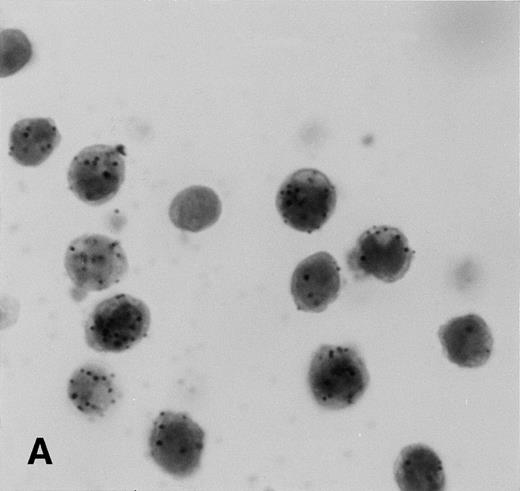
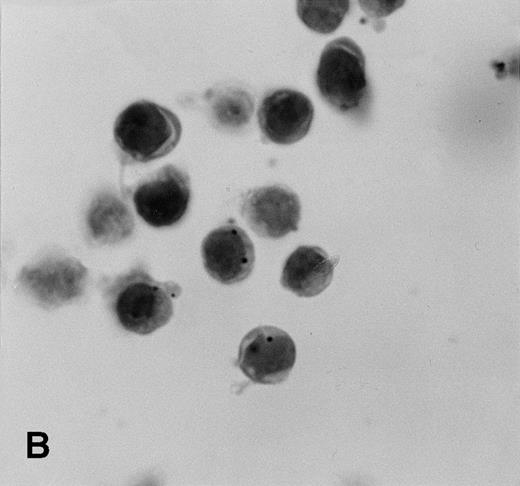
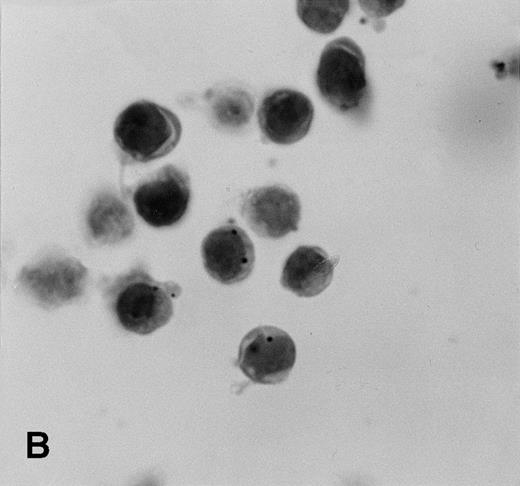
Control sections prepared from mixtures of BC-3 (KSHV-positive) and BJAB (KSHV-negative) cell suspensions contained appropriate percentages of cells labeled for KSHVv-cyc (Fig5).
KSHV cyclin expression in BC-3 and BJAB cell suspension mixtures. (A) 80% BC-3 (KSHV-positive), 20% BJAB (KSHV-negative); (B) 20% BC-3, 80% BJAB. These results show appropriate percentages of cells with punctate labeling indicating that diffusion of the label between cells, a possible artifact of PCR-ISH-based assays, does not occur in this procedure.
KSHV cyclin expression in BC-3 and BJAB cell suspension mixtures. (A) 80% BC-3 (KSHV-positive), 20% BJAB (KSHV-negative); (B) 20% BC-3, 80% BJAB. These results show appropriate percentages of cells with punctate labeling indicating that diffusion of the label between cells, a possible artifact of PCR-ISH-based assays, does not occur in this procedure.
DISCUSSION
Based on epidemiological studies, it has been suggested that KS is an infectious, sexually-transmitted disease independent of HIV infection.43 In the US, KS is much more prevalent in homosexual and bisexual men than in other groups at high risk for HIV infection.44 For women who develop KS, there is a correlation with prior sexual contact with bisexual men.45Recently, DNA sequences from a novel human herpes virus KSHV/HHV8 were identified in lesions of KS.1 Since this discovery, many studies have confirmed the presence of KSHV in all subtypes of KS, strongly suggesting an etiologic role for this virus.7-13Previous ISH studies identified KSHV in plaque and nodular lesions of KS.15-18 In these tissues, KSHV DNA sequences were identified in the spindle cells surrounding the atypical vascular spaces. Many other PCR-based studies have identified the virus in early patch stage lesions of KS, but did not use in situ methods to define the specific cell types harboring the virus. Recently, however, Staskus et al18 showed three different KSHV-specific transcripts in lesions of KS; one of these transcripts was present in one very early patch-stage lesion included in their study. Another of these transcripts encodes the major capsid protein (MCP) of KSHV (ORF25).26 MCP-specific sequences were identified in low levels in lesional spindle cells in only a few later-stage lesions indicating that KSHV exists primarily in a latent state or undergoes an abortive replication cycle in these lesions.18 In the present study, KSHV-specific sequences were identified in each of the stages of progression recognized for KS. For all 10 early patch-stage lesions, KSHV-specific sequences consistently were identified in the fusiform, spindle-shaped cells surrounding the atypical vascular spaces, as well as in some endothelial cells of well-formed blood vessels in the adjacent dermis. This represents the first in situ demonstration of KSHV cyclin D mRNA in the earliest histopathologically-recognizable lesion of KS. In our study, a similar distribution of KSHV-infected cells was seen in later plaque-stage and nodular lesions, corroborating results of previous ISH-based studies, which defined the cellular distribution of KSHV.15-18 In addition to the previously described cellular distribution of KSHV, it also was observed that small mononuclear cells containing the virus and morphologically consistent with lymphocytes and/or histiocytes were present within the tissues. In addition, a few later-stage lesions contained keratinocytes in the epidermis and/or epithelial cells in eccrine structures harboring KSHV cyclin sequences. The in situ demonstration of transcriptional activity of KSHV v-cyc in early cutaneous lesions of KS provides further evidence for an etiologic role of this virus.
It has been suggested that KSHV may progress from a latent phase to a lytic phase in some cell types.46,47 This is especially true for the B lymphocytes of AIDS-related PEL and the cell lines derived from them, both of which may evolve a lytic phase yielding assembled viral particles.3,36 Other studies have shown that the spindle cells of KS are likely latently infected with KSHV.18,23,24 Our observation that the nuclei of lesional spindle cells generally contain only few hybridization signals, but many cytoplasmic signals, is consistent with that conclusion. Furthermore, prior treatment of the spindle cells with RNAse eliminated or greatly reduced the amount of cytoplasmic signals generated after ISH, indicating that the majority of cytoplasmic labeling was due to hybridization of the probe to cytoplasmic mRNA and not to viral DNA sequences. Nodular/tumor-stage lesions of KS also showed a marked reduction in hybridization signal intensity with prior RNAse digestion. This would further support the argument advanced by Staskus et al18 that a lytic phase of viral replication does not occur or is abortive during the progression of KS lesions. Additional in situ experiments are in progress to identify other latently-expressed genes in early patch-stage lesions of KS.
Because some cells infected with KSHV (including B lymphocytes) can progress to a lytic phase of viral replication, it is possible that KS lesional tissues may contain spindle cells, which are latently infected together with other types of cells capable of productive viral replication. Thus, it seems possible that lesions of KS may arise in the setting of a lytic infection of B lymphocytes. It has been shown that some patients have detectable KSHV in the peripheral blood or KSHV-specific antibody titers before the development of KS lesions.48 49 Further elucidation of the pathobiology of the virus will rely on identifying specific viral genes, which are transcriptionally active at different stages of KS lesions, as well as in other cell populations from patients who subsequently develop KS.
In this study, a novel colorimetric ISH procedure using a catalyzed signal amplification system was used to identify KSHV-specific sequences. This amplification system was adapted for use from a similar system designed for solid-phase immunocytochemical assays such as Western blot procedures.19,20 Recently, Zehbe et al22 used a variation of this catalyzed signal amplification technique using streptavidin-nanogold with silver enhancement to identify with high specificity and sensitivity human papillomavirus nucleic acid sequences in infected cell lines and human tissues. The system used in the current study allowed the sensitive and specific identification of KSHV cyclin sequences in 100% of the routinely processed, formalin-fixed, paraffin-embedded archival tissue samples of KS studied. As such, the level of sensitivity of this method appears to be equal to that of previous studies that used PCR ISH15 or ISH with radiolabeled probes,18 both of which also identified KSHV in 100% of the KS lesions studied. Furthermore, this method allowed the sensitive detection of KSHV even in the earliest recognizable patch-stage lesions of KS, an observation that has been documented in only one specimen so far using radiolabeled probes.18 Other ISH methods using radioactively-labeled probes identified KSHV-specific sequences in a more variable 25% to 67% of the lesions evaluated.16 17 Because the procedures described here did not require use of radioactively labeled probes or the in situ amplification of specific target sequences by PCR before ISH, the problems inherent to those techniques such as the requirement for radioactivity precautions, long emulsion development times, or for avoidance of sample contamination by extraneous DNA were avoided. This ISH procedure allowed direct visualization of the specific cell types harboring KSHV with preservation of cellular morphology and without the diffusion of amplified DNA products, which may occur in PCR ISH-based assays. The complete ISH procedure required approximately 5 hours to perform, which is significantly less time that that required for detection of radioactively labeled probes after ISH and comparable to the time needed for PCR ISH. This procedure should be useful with other types of probes designed for specifically targeting low copy numbers of mRNA and DNA sequences. For KSHV, recognition of the specific sequence of transcriptional events and of the specific cells infected will further define the pathobiology of this virus.
Supported in part by US Public Health Service Grants No. CA73531 and CA68939 from the National Cancer Institute, National Institutes of Health.
Address reprint requests to Jon A. Reed, MD, Department of Pathology, The New York Hospital-Cornell Medical Center, 1300 York Ave, Room F-309, New York, NY 10021.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.