Abstract
In hematopoietic cells, interleukin-2 receptor (IL-2R) γ chain (termed γc) is shown to be a component of the IL-4R system, whereas in nonhematopoietic cells, γc is absent and it is not a component of the IL-4R system. Here, we show that the IL-13R α′ chain (termed IL-13Rα′) but not the IL-13R α chain (termed IL-13Rα) can substitute for γc and, thus, IL-13Rα′ forms a novel component of the IL-4R system. This conclusion was drawn on the basis of chemical cross-linking, immunoprecipitation, the ability of IL-13Rα′ but not IL-13Rα to augment IL-4 binding affinity, and the requirement of IL-13Rα′ for IL-4–induced STAT6 activation in Chinese hamster ovary (CHO) cells transfected with various receptor subunits. Cotransfection of IL-4 receptor p140 (termed IL-4Rβ) with γc or IL-13Rα′ increased IL-4 binding affinity and allowed for STAT6 activation in response to IL-4. However, cotransfection of all three chains did not further increase IL-4 binding or alter the extent of STAT6 activation suggesting that all three chains together do not seem to participate in IL-4 function. Instead, IL-4Rβ heterodimerizes with γc or IL-13Rα′ and mediates STAT6 activation. Cotransfection of IL-4Rβ with IL-13Rα neither increased IL-4 binding affinity nor allowed for STAT6 activation in response to IL-4 indicating that IL-13Rα does not convert binding affinity nor transmit signals for IL-4. Because IL-4 phosphorylates JAK1 and JAK2 tyrosine kinases in nonhematopoietic cells, we investigated whether JAK1 and JAK2 are required for IL-4–induced STAT6 activation in various transfectants. Cotransfection experiments with different chains of IL-4R and kinase-deficient JAK1 and JAK2 mutants in CHO cells showed that JAK1 and JAK2 are required for optimal activation of STAT6 in the α′β transfectant but only partially in the βγc transfectant. Taken together, our results show that IL-13Rα′ is a novel functional component of the IL-4R system and that JAK1 and JAK2 mediate IL-4–induced optimal activation of STAT6 in nonhematopoietic cells.
INTERLEUKIN-4 (IL-4) is a growth and differentiation factor for human B- and T-lymphocytes.1 In contrast to its growth stimulatory effects on lymphocytes, IL-4 has growth inhibitory effects on many human carcinoma cell lines.2,3 The receptors for IL-4 have been shown to be expressed on a variety of cell types,4-6 and the effects of IL-4 involve IL-4 signaling through its receptors.
The structure of IL-4 receptor (IL-4R) has been extensively investigated; however, the exact structure is still unknown.1 The primary subunit of IL-4R was identified as a 140-kD protein, originally termed IL-4Rα.7 However, based on similarities in the extracellular domain (WSXWS motif and four cysteine residues at the fixed location) and the long intracellular domain between IL-4Rα and β chains of receptors for IL-3, IL-5, and granulocyte-macrophage colony-stimulating factor (GM-CSF), we have recently proposed to rename this chain IL-4Rβ.8,9 This recommendation was also based on the basis of its similarity with the IL-2Rβ chain, which, like IL-4Rp140, binds IL-2 but does not transmit signal on its own.10 The second subunit of the IL-4R system was shown to be the IL-2Rγ chain (termed γc),10,11 and recently we8,12 and other groups13 have proposed that the 60- to 70-kD protein form of IL-13R may also participate in mediating IL-4 effects and, thus, may constitute the third subunit of the IL-4R system (termed IL-4Rα).8
Recently, two different types of human IL-13 receptor chains were cloned. The IL-13R chain cloned from the human RCC cell line, Caki-1, is an approximately 70-kD protein and has a 50% homology to IL-5Rα on DNA level (termed here, IL-13Rα).14 On the other hand, Aman et al15 have cloned another type of IL-13R from the human T-cell leukemia virus-1 (HTLV-1)–infected MT-2 cell line by using the sequence of the murine IL-13R cDNA (termed here, IL-13Rα′).16 Unlike Caki-1 IL-13R (IL-13Rα), MT-2 IL-13R (IL-13Rα′) has no homology with any other cytokine receptors similar to cloned mouse IL-13R.
To directly examine which IL-13R chains are required for functional IL-4R, we reconstituted the IL-4R systems by transfection of two different IL-13Rα chains (α and α′), IL-4Rβ, or γc chains into Chinese hamster ovary (CHO-K1) cells. We then investigated their binding characteristics to IL-4, subunit structure of IL-4R by cross-linking and immunoprecipitation, and signal transduction mechanisms in response to IL-4.
MATERIALS AND METHODS
Materials.
Recombinant human IL-4 was kindly provided by Schering Corporation (Kenilworth, NJ). Recombinant IL-13 was purified as described.30 Polyclonal antibodies against c-Myc and γc antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Antibody for IL-4Rβ chain (P7) was provided by Immunex Corp (Seattle, WA). Horseradish peroxidase (HRP)-conjugated antimouse or rabbit IgG antibodies and streptoavidine HRP were obtained from Amersham (Arlington Heights, IL).
Cells.
The CHO-K1 cell line was obtained from American Type Culture Collection (Rockville, MD). Cells were cultured in a modified Eagle's minimum essential medium (AMEM) with 10 mmol/L HEPES, antibiotics, and 10% fetal bovine serum (FBS).
Plasmids and genes.
Human IL-4Rp140 (β) chain cDNA7 was kindly provided by Dr M. Widmer of Immunex Corp. Human IL-13Rα′15 and IL-2Rγ10 cDNA were cloned into pME18S mammalian expression vector. To epitope-tag the IL-13Rα′ with a c-Myc tag, new BamH1 sites were created at the C-terminus of IL-13Rα′ cDNA and it was cloned into the BamH1 site of CS+MT plasmid (derived from CS2 plasmid with Myc tag).17 IL-13Rα′ with c-Myc tag cDNA was recloned into the EcoR1 site of PME18S. The plasmids containing wild type JAK1 or its mutant (pME18SJAK1 and pME18SJAK1 delta)18were provided by Dr S. Watanabe (University of Tokyo Medical Science, Tokyo, Japan).19 JAK2 expression vector, pBOSJAK2, and kinase-deficient JAK2 expression vector, pBOSJAK2 D VIII, were provided by Dr D.M. Wojchowski (Pennsylvania State University, University Park, PA).20
Transient transfection of DNA.
Individual plasmid DNA or a combination of multiple plasmid DNAs (6 μg/60-mm dish or 12 μg/100-mm dish) were transfected into semiconfluent CHO-K1 cells by Lipofectamine (GIBCO-BRL, Gaithersburg, MD) according to the manufacturer's instructions. Briefly, CHO-K1 cells (1 × 106/60-mm dish or 3 × 106/100-mm dish) were incubated with the DNA Lipofectamine mixture for 5 hours in Opti-MEM (GIBCO-BRL). The medium was then changed to AMEM with 10% FBS and incubated for 48 hours.
Radioreceptor binding assay.
Recombinant human IL-4 was labeled with 125I (Amersham Research Product, Arlington Heights, IL) by the IODO-GEN iodination reagent (Pierce, Rockford, IL) according to the manufacturer's instructions. The IL-4 equilibrium binding studies were performed by the method previously described.2,8 Briefly, 1 × 106 cells in 100 μL binding buffer (RPMI 1640 containing 0.2% human serum albumin and 10 mmol/L HEPES) were incubated for 2 hours with 100 to 200 pmol/L 125I–IL-4 with or without unlabeled IL-4 (50 nmol/L) or IL-13 (200 nmol/L) at 4°C. Cell-bound125I–IL-4 was separated from unbound125I–IL-4 by centrifugation through a cushion of phthalate oils. Pelleted cells were counted on a γ counter. To determine binding affinity, transfected cells were incubated with 100 pmol/L of125I–IL-4 with or without various concentrations of unlabeled IL-4. Scatchard data was analyzed by the LIGAND program (Provided by Dr P. Munson, National Institutes of Health).21
Affinity cross-linking of 125I–IL-4 to its receptor.
Transfected cells (5 × 106) were incubated with125I–IL-4 in the presence or absence of excess unlabeled IL-4 or IL-13 for 2 hours at 4°C. Bound 125I–IL-4 was cross-linked to IL-4R with disuccinimidyl suberate (DSS; Pierce) at a final concentration of 2 mmol/L for 20 minutes. The cells were then lysed at 4°C with modified radio immunoprotein assay (RIPA) buffer (1% NP-40, 300 mmol/L NaCl, 50 mmol/L Tris [pH 7.4], 1 mg/mL leupeptin, 1 mg/mL pepstatin A, 2 mg/mL aprotinin, 20 mmol/L phenylmethyl sulfonyl fluoride [PMSF], 1 mmol/L Na-vanadate, 25 mmol/L Na-F, 10 mmol/L Na-pyrophosphate, and 1 mmol/L EDTA). The resulting lysate was analyzed by electrophoresis through sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 7%). The gel was dried and exposed to radiograph film for 4 days at −80°C.
For immunoprecipitation, 125I–IL-4/IL-4R cross-linked complex was immunoprecipitated from the lysate for 2 hours at 4°C by incubating with protein A (G) sepharose beads, which had been preincubated with anti–IL-4Rβ (P7), γc, or c-Myc antibodies. The resulting conjugate was washed five times with lysing buffer, resuspended with reducing buffer, boiled for 5 minutes, and analyzed by 8% SDS-PAGE as described previously.
Electrophoretic mobility shift assay (EMSA).
After incubation with IL-4 (50 ng/mL) for 10 minutes, cells were washed with cold phosphate-buffered saline (PBS) and solubilized with cold whole cell extraction buffer (1 mmol/L MgCl2, 20 mmol/L HEPES pH 7.0, 10 mmol/L KCl, 300 mmol/L NaCl, 0.5 mmol/L dithiothreitol, 0.1% NP-40, 1 mmol/L PMSF, 1 mmol/L Na3VO4, and 20% glycerol). DNA protein interactions were assessed by EMSA by using the Bandshift kit from Pharmacia (Piscataway, NJ). Briefly, 50 μg of sample proteins were incubated for 20 minutes at room temperature with 1 ng of32P-labeled double stranded oligonucleotide probe SBE1 (signal transduction and activator of transcription [STAT]-binding element; 5′-gatcGCTCTTCTTCCCAGGAACTCAATG-3′;3′-CGAGAAGAAGGGTCCTTGAGTTACagct-5′) from the region flanking the transcription start site of the human sIL-1R antagonist gene that is necessary for response to IL-4 alone22 in binding buffer [10 mmol/L Tris-HCl (pH 7.5), 50 mmol/L NaCl, 0.5 mmol/L dithiothreitol (DTT), 10% glycerol, 0.05% NP-40, 0.05 mg/mL poly (dI-dC)2]. A 10× loading dye was added to samples that were then applied to a 4% nonreducing polyacrylamide gel and run at 150 V for 2 hours. Gels were dried for 2 hours and autoradiographed overnight four 4 days at −70°C. In some experiments, antimouse STAT6 (M-20) or antihuman STAT6 (S-20) rabbit polyclonal IgG (both from Santa Cruz Biotechnology, Santa Cruz, CA) were included in the reaction mixture for “supershift” assay.
RESULTS
125I–IL-4 binding assay on CHO-K1 cells transfected with IL-13Rα, IL-13α′, IL-4Rβ, and γc.
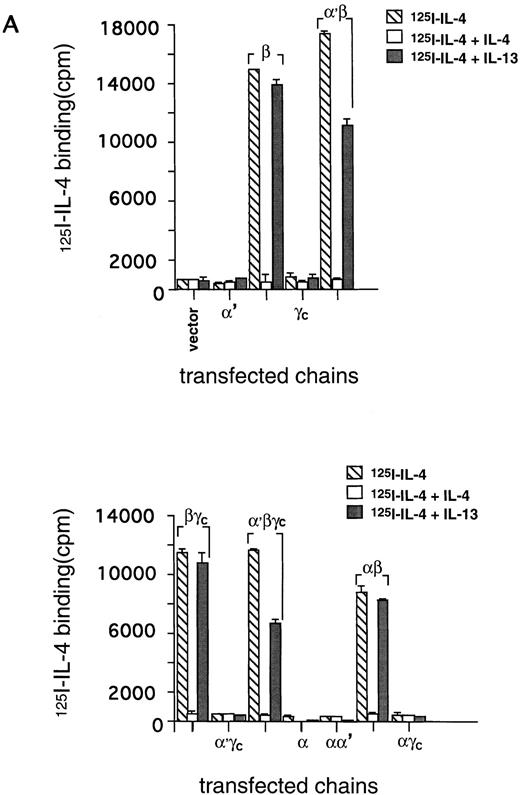
Previously, we proposed that IL-4R shares at least one chain of IL-13R in nonhematopoietic cancer cell lines.8,12 To examine directly the subunit structure of IL-4R, we introduced the cDNA of the IL-4Rβ chain into the CHO-K1 cell line along with recently cloned IL-13Rα chains (IL-13Rα and α′).14 15 As shown in Fig 1A, 125I–IL-4 bound to IL-4Rβ-transfected CHO-K1 cells, but it did not bind to cells transfected with IL-13Rα′ or γc alone. The IL-4 binding to IL-4Rβ–transfected cells was specifically inhibited by a 50-fold mol/L excess of unlabeled IL-4. However, IL-13 did not inhibit this binding. Interestingly, although IL-13Rα′- and γc-transfected cells did not bind to125I–IL-4, cotransfection of IL-4Rβ with these chains induced a high affinity binding (Fig 1B). In the case of α′β transfectants, 125I–IL-4 binding to its receptor was inhibited by a 50-fold mol/L excess of unlabeled IL-4 and also partially by IL-13 (Fig 1A, upper panel). On the other hand, in βγc-transfected cells, IL-4 bound with high-affinity125I–IL-4 binding was only inhibited by IL-4 and not by IL-13. In CHO-K1 cells transfected with all three α′βγc chains, 125I–IL-4 also bound with high affinity and this binding was completely blocked by unlabeled IL-4 and partially by IL-13 (Fig 1A, lower panel). These data suggest that IL-13Rα′ allows interaction of IL-13 with IL-4R, and when it is absent, such as in the case of the IL-4Rβ and γctransfectant, IL-13 does not interact with IL-4R.125I–IL-4 also did not bind to cells transfected with IL-13Rα alone. However, when it was coexpressed with the IL-4Rβ chain, the IL-4 binding was similar to that seen in cells transfected with β alone. Again, only IL-4 completely inhibited the binding of radiolabeled IL-4 whereas IL-13 did not inhibit this binding.
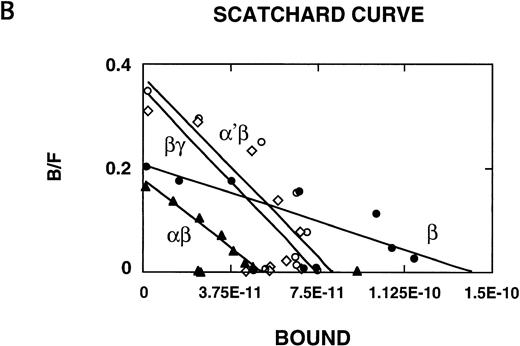
125I–IL-4 binding to CHO-K1 cells transfected with IL-13Rα′, IL-4Rβ, and γc. cDNA for various receptor chains (2 μg/chain) was transfected in CHO-K1 cells (1 × 106) by using Lipofectamine reagent for 48 hours. For IL-4 binding assay, 1 × 106 cells were incubated with 100 pmol/L of125I–IL-4 with or without a 200-fold molar excess of unlabeled IL-4 or IL-13. Binding assays were performed on two different occasions. Cell bound radioactivity was determined as described in Materials and Methods. (A) To determine binding affinity, transfected cells were incubated with 100 pmol/L of 125I–IL-4 with or without various concentrations of unlabeled IL-4. Scatchard data were analyzed by the LIGAND program (B). In four experiments, the binding data were fitted with only one site model.
125I–IL-4 binding to CHO-K1 cells transfected with IL-13Rα′, IL-4Rβ, and γc. cDNA for various receptor chains (2 μg/chain) was transfected in CHO-K1 cells (1 × 106) by using Lipofectamine reagent for 48 hours. For IL-4 binding assay, 1 × 106 cells were incubated with 100 pmol/L of125I–IL-4 with or without a 200-fold molar excess of unlabeled IL-4 or IL-13. Binding assays were performed on two different occasions. Cell bound radioactivity was determined as described in Materials and Methods. (A) To determine binding affinity, transfected cells were incubated with 100 pmol/L of 125I–IL-4 with or without various concentrations of unlabeled IL-4. Scatchard data were analyzed by the LIGAND program (B). In four experiments, the binding data were fitted with only one site model.
We also analyzed the binding affinity of IL-4R in CHO-K1 cells transfected with IL-4Rβ, IL-4Rβ plus IL-13Rα or α′, or γc by Scatchard analysis using the LIGAND program. This program allowed fitting of our data in only a one-site model. In four separate experiments, the data did not fit in a two-site model. As shown in Fig 1B, IL-4 bound to IL-4Rβ-transfected cells with high to intermediate affinity (Kd = 0.69 nmol/L); however, when this chain was cotransfected with IL-13Rα′ or γc, the affinity was approximately threefold higher (Kd = 0.21 nmol/L or 0.20 nmol/L, respectively) compared with β-transfected cells. In the cells transfected with α′, β, and γc chains (not shown), the affinity was similar to α′β- or βγc-transfected cells (Fig 1B). We also transfected the IL-13Rα chain14 into CHO cells; however, the IL-13Rα chain did not bind to 125I–IL-4 (Fig 1A), and cotransfection with IL-4Rβ did not appear to modulate IL-4R binding affinity (Fig 1B). These results suggest that IL-13Rα′ but not IL-13Rα is a novel component of the IL-4R system and it forms a high-affinity IL-4 receptor with IL-4Rβ as γc does.
Affinity cross-linking of 125I–IL-4 to CHO-K1 cells transfected with IL-13α′, IL-4Rβ, and γc.
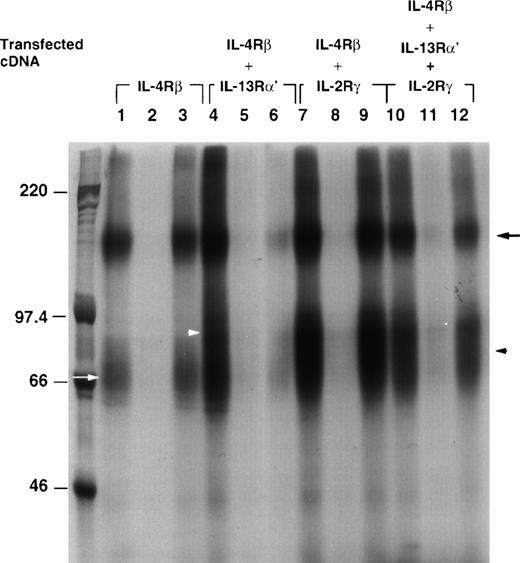
To visualize 125I–IL-4 binding to different chains, we cross-linked 125I–IL-4 to IL-4 receptors on CHO-K1 cells transfected with IL-4Rβ alone or cotransfected with IL-13α′, γc, or both. As shown in Fig2, 125I–IL-4 cross-linked to a major protein of approximately 155 kD in the IL-4Rβ chain transfectant (β; lane 1). A faint, broad, approximately 70-kD band was also detected (lane 1, white arrow). After the subtraction of the size of IL-4 (15 kD), these proteins were estimated to be 140 kD and approximately 55 kD. These data suggest that the 140-kD protein is a transcript of human IL-4Rβ cDNA and the approximately 55-kD band may be a degradation product of IL-4Rβ.23 These bands completely disappeared when the cells were coincubated with 200-fold mol/L excess of unlabeled IL-4 (lane 2), and this binding was not displaced by 200-fold mol/L excess of IL-13 (lane 3). No cross-linking of radiolabeled IL-4 was seen in cells transfected with vector alone (data not shown). In the IL-13Rα′ and the IL-4Rβ transfectant (α′β), an additional broad band was seen at 80 to 100 kD along with a 155-kD band (lane 4, white arrowhead). This band corresponded to the IL-13Rα′ chain (65-85 kD). These bands completely disappeared when cross-linking was performed in the presence of 200 mol/L excess of unlabeled IL-4 (lane 5). These bands also partially disappeared when cross-linking was performed in the presence of 200 mol/L excess of unlabeled IL-13 (lane 6). In the IL-4Rβ and γctransfectant (βγc), an additional, approximately 80-kD band (γc, approximately 65 kD, arrowhead), was observed along with a 155-kD band (lane 7). These bands completely disappeared when unlabeled IL-4 (lane 8) but not unlabeled IL-13 (lane 9) was added. In the triple chain transfectant (α′βγc), there were two bands at 155 kD and approximately 80 kD as in the βγc transfectants (lane 10). As observed with binding data, a 200-mol/L excess of IL-4 completely displaced 125I–IL-4 binding (lane 11); however, IL-13 only partially displaced 125I–IL-4 binding to these cells (lane 12).
Affinity cross-linking of IL-4R in transfected CHO-K1 cells. Two micrograms of cDNA for IL-4Rβ (lanes 1, 2, and 3), IL-4Rβ plus IL-13Rα′ (lanes 4, 5, and 6), IL-4Rβ plus IL-2Rγc (lanes 7, 8, and 9), and all three chains (α′, β, and γc; lanes 10, 11, and 12) were transfected to CHO-K1 cells. Transfected cells (5 × 106) were incubated with 125I–IL-4 in the absence (lanes 1, 4, 7, and 10) or presence of excess unlabeled IL-4 (lanes 2, 5, 8, and 11) or IL-13 (lanes 3, 6, 9, and 12) for 2 hours at 4°C. Bound125I–IL-4 was cross-linked to IL-4R with (DSS). The cells were then lysed at 4°C with modified RIPA buffer. The resulting lysate was analyzed by electrophoresis through an SDS-PAGE (7%) gel. The gel was dried and exposed to radiograph film for 4 days at −80°C. The molecular weight markers are shown on the left. The positions of different receptor chains are indicated (IL-4Rβ, black arrow; IL-13Rα′, white arrowhead; IL-2Rγc, black arrowhead; and approximately 55 kD, white arrow).
Affinity cross-linking of IL-4R in transfected CHO-K1 cells. Two micrograms of cDNA for IL-4Rβ (lanes 1, 2, and 3), IL-4Rβ plus IL-13Rα′ (lanes 4, 5, and 6), IL-4Rβ plus IL-2Rγc (lanes 7, 8, and 9), and all three chains (α′, β, and γc; lanes 10, 11, and 12) were transfected to CHO-K1 cells. Transfected cells (5 × 106) were incubated with 125I–IL-4 in the absence (lanes 1, 4, 7, and 10) or presence of excess unlabeled IL-4 (lanes 2, 5, 8, and 11) or IL-13 (lanes 3, 6, 9, and 12) for 2 hours at 4°C. Bound125I–IL-4 was cross-linked to IL-4R with (DSS). The cells were then lysed at 4°C with modified RIPA buffer. The resulting lysate was analyzed by electrophoresis through an SDS-PAGE (7%) gel. The gel was dried and exposed to radiograph film for 4 days at −80°C. The molecular weight markers are shown on the left. The positions of different receptor chains are indicated (IL-4Rβ, black arrow; IL-13Rα′, white arrowhead; IL-2Rγc, black arrowhead; and approximately 55 kD, white arrow).
Immunoprecipitation of IL-4Rβ, IL-13Rα′, and γc chains.
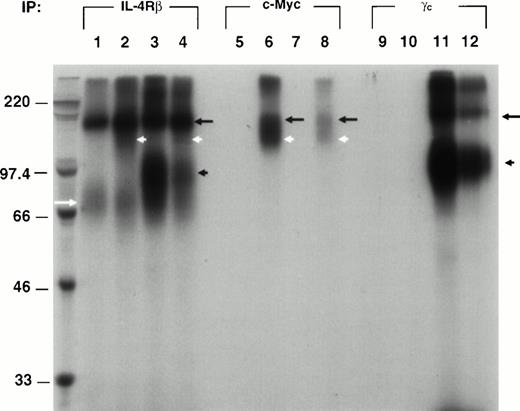
To unequivocally show whether IL-4Rβ interacts/associates with IL-13Rα′, we immunoprecipitated 125I–IL-4 cross-linked proteins from the four combinations of transfectants (β, α′β, βγc, and α′βγc) by using specific antibodies for appropriate chains. Because an antibody for IL-13Rα′ is not commercially available, we made an expression plasmid for the IL-13Rα′ chimera protein with a c-Myc epitope tag at C-terminus. The transfectants were cross-linked with 125I–IL-4 and lysed as described previously. The cell lysates were incubated with Protein A (G)-sepharose beads conjugated with anti–IL-4Rβ, –c-Myc, or -γc antibody, and immunoprecipitants were electrophoresed on 8% SDS-PAGE. As shown in Fig 3, when immunoprecipitated with the IL-4Rβ antibody (P7), a sharp 155-kD band and a diffuse and faint approximately 70-kD band were detected in β chain transfectants (lane 1). In α′β transfectants, an additional diffuse broad 110- to 130-kD band was detected (lane 2). Because this IL-13Rα′ chain has six repeats of c-Myc tag, the size of this protein is larger than that of IL-13Rα′ in affinity cross-linking data. In βγc transfectants, an additional dark, approximately 80-kD band was detected (lane 3), which corresponded to the γc chain. In α′βγc transfectants, a 155-kD band was detected along with additional, approximately 130-kD, 80-kD, and 70-kD bands, although additional bands were faint and diffuse (lane 4). When c-Myc antibody was used for IL-13Rα′ immunoprecipitation, a 155-kD and a 130-kD doublet were detected in only α′β and α′βγc transfectants (lane 6 and 8). On the other hand, no band was observed in β and βγc transfectants (lane 5 and 7). All four combinations of transfectants were also immunoprecipitated by anti-γcantibody to show γc interactions with IL-4Rβ. A 155-kD and an approximately 80-kD protein was detected in βγcand α′βγc transfectants (lane 11 and 12), whereas no band was detected in β and α′β transfectants (lane 9 and 10). These results indicate that IL-13Rα′ associates with IL-4Rβ as γc does. Moreover, in triple chain transfectants (α′βγc), two types of heterodimers (α′β and βγc) are probably induced, but there is no evidence for heterotrimer (α′βγc).
The IL-13Rα′ chain associates with IL-4β. cDNA for IL-4Rβ (lanes 1, 5, and 9), IL4Rβ plus IL-13Rα′ (lanes 2, 6, and 10), IL-4Rβ plus IL-2Rγc (lanes 3, 7, and 11), and all three chains (α′, β, and γc; lanes 4, 8, and 12) was transfected to CHO-K1 cells. Transfected cells were incubated with 1 nmol/L of 125I–IL-4.125I–IL-4/IL-4R cross-linked complex was immunoprecipitated from the cell lysate at 4°C by incubating with protein A (G) sepharose beads that had been preincubated with anti–IL-4Rβ (P7), γc, or c-Myc antibodies. The resulting complex was washed five times with lysing buffer, resuspended with reducing buffer, and analyzed by 8% SDS-PAGE as described previously. The molecular weight markers are shown on the left. The position of different receptor chains is indicated (IL-4Rβ, black arrow; IL-13Rα′, white arrowhead; IL-2Rγc, black arrowhead; and approximately 55 kD, white arrow).
The IL-13Rα′ chain associates with IL-4β. cDNA for IL-4Rβ (lanes 1, 5, and 9), IL4Rβ plus IL-13Rα′ (lanes 2, 6, and 10), IL-4Rβ plus IL-2Rγc (lanes 3, 7, and 11), and all three chains (α′, β, and γc; lanes 4, 8, and 12) was transfected to CHO-K1 cells. Transfected cells were incubated with 1 nmol/L of 125I–IL-4.125I–IL-4/IL-4R cross-linked complex was immunoprecipitated from the cell lysate at 4°C by incubating with protein A (G) sepharose beads that had been preincubated with anti–IL-4Rβ (P7), γc, or c-Myc antibodies. The resulting complex was washed five times with lysing buffer, resuspended with reducing buffer, and analyzed by 8% SDS-PAGE as described previously. The molecular weight markers are shown on the left. The position of different receptor chains is indicated (IL-4Rβ, black arrow; IL-13Rα′, white arrowhead; IL-2Rγc, black arrowhead; and approximately 55 kD, white arrow).
Activation of STAT6 in response to IL-4 in IL-4R chain transfectants.
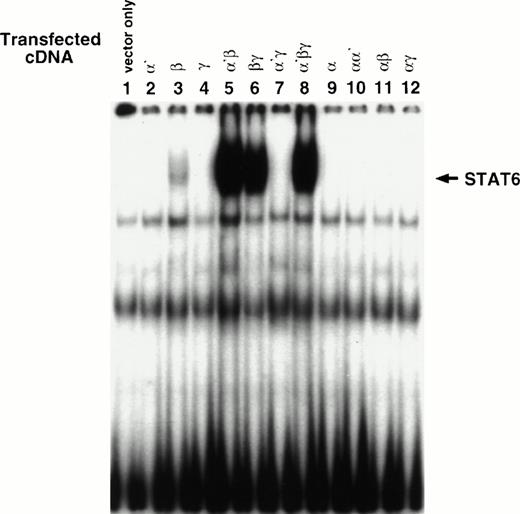
To determine whether α′β and αβ heterodimer complexes are biologically functional, we analyzed STAT activity in response to IL-4 in various transfectants. It has been shown that IL-4 activates only STAT6 protein in various cell types.8 24-26 As shown in Fig 4A, single chain transfectants of α′, γc, or vector alone did not activate STAT6-DNA binding activity (lanes 1, 2, and 4), whereas in some experiments, in β chain transfectants, STAT6 was activated at low levels (lane 3). The activation of STAT6 in response to IL-4 was optimal when the β chain was cotransfected with α′ (lane 5). As expected, STAT6 activation was also seen in βγc(lane 6) and α′βγc (lane 8) transfectants. We also investigated whether IL-13Rα can induce STAT6-DNA binding in response to IL-4. Any combination of the IL-13Rα chain failed to generate a STAT6-DNA binding complex (lane 9-12). These results suggest that α′β combination is sufficient to activate STAT6 protein in a comparable manner with the βγc or α′βγc combination.
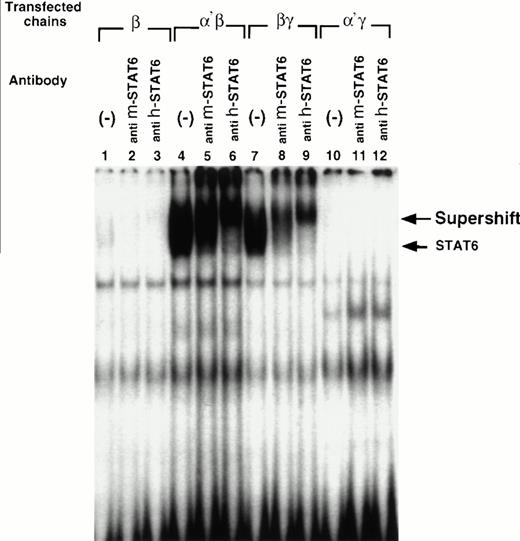
Cotransfection of the IL-13Rα′ but not the IL-13Rα chain with the IL-4Rβ chain is sufficient to reconstitute STAT activation in response to IL-4. CHO-K1 cells were transfected with various chains and then incubated with IL-4 (50 ng/mL) for 10 minutes, washed with cold PBS, and solubilized with cold whole cell extraction buffer. Fifty micrograms of sample proteins were incubated for 20 minutes at room temperature with 1 ng of 32P-labeled SBE1 probe in binding buffer. Then, samples were loaded on a 4% nonreducing polyacrylamide gel and run at 150 V for 2 hours (A). For supershift assay, antimouse STAT6 (anti m-STAT6) or antihuman STAT6 (anti h-STAT6) rabbit polyclonal IgG was included in the reaction mixture before electrophoresis. The gel was dried and analyzed by autoradiography (B).
Cotransfection of the IL-13Rα′ but not the IL-13Rα chain with the IL-4Rβ chain is sufficient to reconstitute STAT activation in response to IL-4. CHO-K1 cells were transfected with various chains and then incubated with IL-4 (50 ng/mL) for 10 minutes, washed with cold PBS, and solubilized with cold whole cell extraction buffer. Fifty micrograms of sample proteins were incubated for 20 minutes at room temperature with 1 ng of 32P-labeled SBE1 probe in binding buffer. Then, samples were loaded on a 4% nonreducing polyacrylamide gel and run at 150 V for 2 hours (A). For supershift assay, antimouse STAT6 (anti m-STAT6) or antihuman STAT6 (anti h-STAT6) rabbit polyclonal IgG was included in the reaction mixture before electrophoresis. The gel was dried and analyzed by autoradiography (B).
To confirm whether IL-4–induced SBE1-binding complex contains STAT6, an antibody supershift assay was performed. We used two different STAT6 antibodies because cross-reactivity of antibodies to CHO cell-derived STAT6 was not known. Antibody to mouse STAT6 caused a small shift, whereas antibody to human STAT6 caused a substantial shift in the electrophoretic mobility of the SBE1-binding activity (Fig 4B). These data confirm that the IL-4–induced SBE-1–binding complexes indeed contain STAT6 molecule.
JAK1 and JAK2 are required for the activation of STAT6 in response to IL-4 in α′β but not βγ chain transfectants.
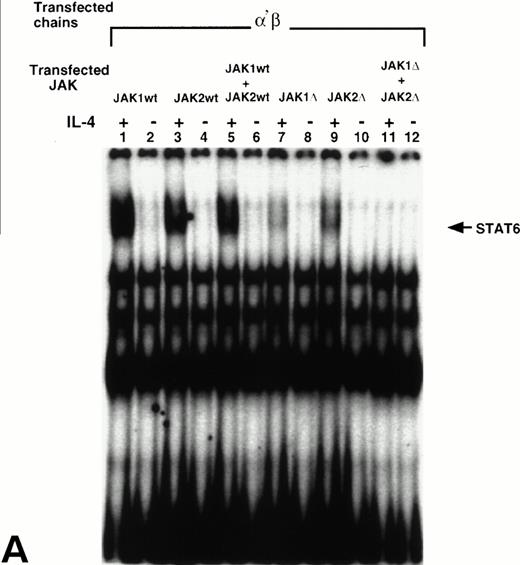
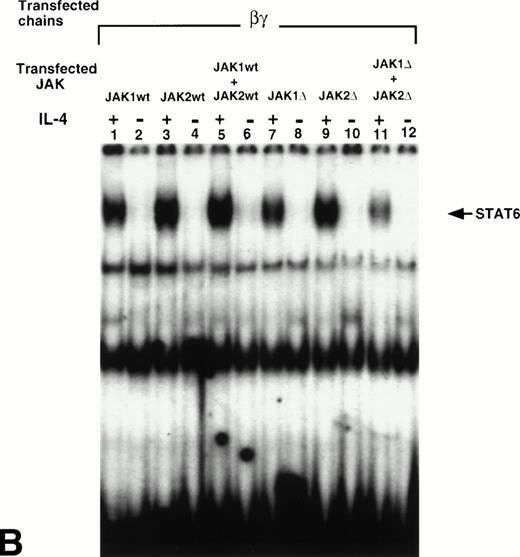
We have previously reported that JAK1 and JAK2 are phosphorylated and activated in response to IL-4 in nonhematopoietic cells that do not express γc, whereas JAK3 is phosphorylated in response to IL-4 in immune cells that do express γc.8,24 26 To show whether JAK1, JAK2, or both, are required for STAT6 activation in response to IL-4 in the cells that express α′β or βγc chains, we cotransfected wild type JAK1 (wtJAK1), JAK2 (wtJAK2), or both or kinase-deficient JAK1 (ΔJAK1), JAK2 (ΔJAK2), or both with α′β or βγc chains into CHO-K1 cells. STAT6 activation was then assessed by EMSA. In α′β transfectants, IL-4 induced activation of STAT6 when either wtJAK1, wtJAK2, or both were coexpressed (Fig 5A, lanes 1-6). When ΔJAK1 or ΔJAK2 were coexpressed with α′β chains, the activation of STAT6 in response to IL-4 was inhibited (Fig 5, lanes 7 and 9). However, when α′β chains were cotransfected with both ΔJAK1 and ΔJAK2 the STAT6 activation was completely blocked (lane 11). On the other hand, in cells transfected with βγcand ΔJAK1, ΔJAK2, or both, the activation of STAT6 in response to IL-4 was only partially inhibited (Fig 5B, lanes 7, 9, and 11). These results suggested that both JAK1 and JAK2 tyrosine kinases are required for optimal activation of STAT6 in response to IL-4 in the cells that express α′β chains of the IL-4R system. However, in cells that express βγc chains of the IL-4R system, JAK2 is not required whereas JAK1 is partially required for STAT6 activation by IL-4.
JAK1 and JAK2 are required for the activation of STAT6 in response to IL-4 in the cells that express IL-13Rα′ and IL-4Rβ chains of IL-4R. Wild type JAK1 (wtJAK1), JAK2 (wtJAK2), or both, and kinase-deficient JAK1 (▵JAK1), JAK2 (▵JAK2), or both, were cotransfected with (A) the IL-4Rβ chain and the IL-13Rα′ (α′β) or (B) the IL-4Rβ chain and γc (βγ) as described in Materials and Methods. After 48 hours, the cells were stimulated by IL-4 (50 ng/mL) and lysed in lysing buffer. Fifty micrograms of sample proteins were incubated for 20 minutes at room temperature with 0.5 to 1 ng of 32P-labeled SBE1 probe. The samples were loaded on a 4% nonreducing polyacrylamide gel and run at 150 V for 2 hours. The gel was dried and analyzed by autoradiography.
JAK1 and JAK2 are required for the activation of STAT6 in response to IL-4 in the cells that express IL-13Rα′ and IL-4Rβ chains of IL-4R. Wild type JAK1 (wtJAK1), JAK2 (wtJAK2), or both, and kinase-deficient JAK1 (▵JAK1), JAK2 (▵JAK2), or both, were cotransfected with (A) the IL-4Rβ chain and the IL-13Rα′ (α′β) or (B) the IL-4Rβ chain and γc (βγ) as described in Materials and Methods. After 48 hours, the cells were stimulated by IL-4 (50 ng/mL) and lysed in lysing buffer. Fifty micrograms of sample proteins were incubated for 20 minutes at room temperature with 0.5 to 1 ng of 32P-labeled SBE1 probe. The samples were loaded on a 4% nonreducing polyacrylamide gel and run at 150 V for 2 hours. The gel was dried and analyzed by autoradiography.
DISCUSSION
In this report, we show that the IL-4Rβ (β) and the IL-13Rα′ (α′) chains can induce high affinity binding to IL-4 as does the β + γc complex, although the α′ chain by itself does not bind IL-4. By using chemical cross-linking studies, we also show that the α′ chain is associated with the β chain in response to IL-4. Thus, the IL-4Rβ chain can form a complex with either the α′ chain or γc, but whether all three chains form a trimeric complex is not clear. Because antibody to anti–c-Myc–tagged IL-13Rα′ or γc did not immunoprecipitate all three chains, our data suggest that IL-4 does not seem to cross-link to all three chains simultaneously.
In contrast to the participation of IL-13Rα′, IL-13Rα did not seem to participate in the formation of the IL-4R complex. IL-13Rα chain transfectants neither bound 125I–IL-4 nor increased binding affinity to IL-4 when coexpressed with IL-4Rβ. These results suggest that IL-13Rα is not a component of the IL-4R system.
We have previously reported that in nonhematopoietic cancer cells, a high number of IL-4R are expressed.2,3,8,23 Although the γc chain is not expressed in these cells, IL-4R is still functional and STAT6 protein is activated in response to IL-4.8 IL-13R was also expressed in these cells and IL-13 inhibited 125I–IL-4 binding to its receptor.8 25 Thus, we hypothesized that part of IL-13R may be a functional component for the IL-4R system. Our current data confirm this hypothesis.
Two types of patterns were observed when excess of IL-4 or IL-13 were included to compete for 125I–IL-4 binding or cross-linking in various transfectants. In the first pattern, IL-4 completely blocked the binding of 125I–IL-4, whereas IL-13 did not inhibit125I–IL-4 binding in cells expressing β alone, αβ, or βγc. In the second pattern, IL-13 significantly inhibited binding and cross-linking of 125I–IL-4, although not completely, in cells expressing α′β or α′βγc. The mechanism of partial inhibition of125I–IL-4 binding by IL-13 is not completely clear. We believe that in the presence of all three chains, IL-4 has an option to form a complex with either IL-4Rβ and γc or IL-4Rβ and IL-13Rα′chains. Because IL-13Rα′ does not bind to IL-13 in the absence of IL-4Rβ9,12,15,16 and because IL-4Rβ is most likely sequestered in the complex with γc to form a high affinity IL-4 receptor, IL-13 is unable to block 125I–IL-4 binding completely. Similarly, in IL-4Rβ and IL-13Rα′ transfectant IL-4 will have higher affinity than IL-13. Thus, IL-4 will completely block but IL-13 will partially block 125I–IL-4 binding. Finally, the lack of competition of IL-4 binding by IL-13 in β, αβ, or βγc transfectants can be explained at least partly by the lack of binding of IL-13 to the β or βγc chain (Murata and Puri, unpublished data). These data agree with our previous report in which IL-13 did not compete for the binding of IL-4 in Raji and MLA144 cell lines.12
It is of interest to note that IL-4R heterodimerization is quite different from the IL-2R or IL-3R system. In these receptors, IL-2Rβ or IL-3Rβ has no binding affinity to IL-2 or IL-3 respectively; however, when complexed with the IL-2Rαγ or IL-3Rα chain it can form a high-affinity receptor. Moreover, β chains are important for their signal transduction. On the other hand, the IL-4Rβ chain has an intermediate affinity by itself and oligomerization with IL-13Rα′ or γc only causes a modest twofold to threefold increase in binding affinity to IL-4 (Fig 1B). Moreover, in α′βγc transfectants, receptor affinity was not significantly different from that of the α′β or βγc transfectants. These results suggest that the accessory molecules (α′ and γc) for IL-4R are not critical for increasing the IL-4 receptor affinity.
To show whether the IL-4Rβ and IL-13Rα′ chain complex was effective in STAT6 activation, we examined STAT6 activation in various transfectants. Our data indicate that the IL-4Rβ chain by itself can bind to IL-4 and when overexpressed alone can cause modest activation of STAT6 protein. These data are consistent with recent reports that showed that homodimerization of the IL-4Rβ chain can cause STAT6 activation.27,28 The activation of the STAT6 protein was robust when the β chain was cotransfected with the α′ chain and this activation level was similar to that caused by the βγc combination. Recently, we have observed that the α′ chain is always expressed along with the IL-4Rβ chain in cells that lack γc expression29 and STAT6 is activated in response to IL-4 without γc in some nonhematopoietic cancer cell lines.8 25 These results confirm that the IL-13Rα′ chain is a functional component of the IL-4R system and that the β chain requires either α′ or γc for STAT6 activation. Unlike the α′ chain, transfection of α chain along with the β chain did not cause activation of STAT6 protein indicating that IL-13Rα is not a functional component of the IL-4R complex.
It is still unknown why α′β oligomerization is used for the functional IL-4R system in nonhematopoietic cells. The signaling pattern by both combinations of chains (β+α′ or β+γc) in response to IL-4 is identical except for the JAK kinases used. In nonhematopoietic cells that lack γc, JAK1, JAK2, and Tyk2 are phosphorylated and activated in response to IL-4. On the other hand, JAK3 is phosphorylated instead of JAK2 in hematopoietic cells that express γc. We showed here that JAK1 and JAK2 are required for STAT6 activation in response to IL-4 in the cells that express α′β chains of IL-4R but are not required in the cells that express βγc chains (Fig 5). Interestingly, the substrates for these JAK kinases (STAT6 and IRS-1/IRS-2) are phosphorylated and activated in both cases. Thus, it is possible that STAT6 and IRS-1/IRS-2 pathways are common for the function of IL-4R in hematopoietic and nonhematopoietic cells. It is also possible that other unknown pathway(s) of signaling may contribute to specific functions of IL-4. Alternatively, the α′β heterodimer is used by both the IL-4 and the IL-13 receptor system and the βγc heterodimer is used by only the IL-4 receptor system as shown in T cells. In T cells, IL-13 does not compete for IL-4 binding, and these cells do not express IL-13R nor do they respond to IL-13. Additional investigations are required to show the role of the two different types of IL-4R system.
In summary, we have reconstituted a functional IL-4R in CHO-K1 cells by transfecting two different IL-13R chains (α and α′), IL-4Rβ, and γc chains. We provide experimental evidence for the first time that IL-13Rα′, but not IL-13Rα can associate with the IL-4Rβ chain, and α′β heterodimerization is sufficient to elicit STAT6 activation in response to IL-4. Moreover, both JAK1 and JAK2 are required for optimal activation of STAT6 in response to IL-4 in the cells that express the IL-13Rα′ chain along with IL-4Rβ but only partially in the cells that express βγc. These results suggest that the IL-13Rα′ chain is a functional component of the IL-4R system, and that may, in part, be responsible for the redundant biological response of IL-4 and IL-13.
ACKNOWLEDGMENT
We thank Dr G. Johnson for reading the manuscript, Dr N.I. Obiri for labeling IL-4 and reading the manuscript, Dr Waldemar Debrinski for IL-13, Dr T. Tomoda for providing PME18S and CS+MT plasmids, Dr W. Leonard for IL-13Rα′ and γc cDNA, Dr D.M. Wojchowski for pBOSJAK2 and pBOSJAK2ΔVIII, Dr S. Watanabe for plasmids for JAK1 and mutant JAK1, Dr Pasual Ferrara for IL-13Rα, Dr Ray Donnelly for SBE1 probe, Dr S.R. Husain for helpful comments on this manuscript and Ms P. Dover for excellent technical assistance.
Address reprint requests to Raj K. Puri, MD, PhD, Laboratory of Molecular Tumor Biology, Division of Cellular and Gene Therapies, Center for Biologic Evaluation and Research, FDA, NIH-Building 29B, Room 2NN10, 29 Lincoln Drive, MSC 4555, Bethesda, MD 20892-4555.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.