Abstract
Ribozymes are catalytic RNA molecules that recognize their target RNA in a highly sequence-specific manner. They can therefore be used to inhibit deleterious gene expression (by cleavage of the target mRNA) or even repair mutant cellular RNAs. Targets such as the mRNAs of oncogenes (resulting from base mutations or chromosome translocations, eg, ras or bcr-abl) and viral genomes and transcripts (human immunodeficiency virus–type 1 [HIV-1]) are ideal targets for such sequence-specific agents. The aim of this review is therefore to introduce the different classes of ribozymes, highlighting some of the chemistry of the reactions they catalyze, to address the specific inhibition of genes by ribozymes, the problems yet to be resolved, and how new developments in the field give hope to the future for ribozymes in the therapeutic field.
RIBOZYMES ARE ribonucleic acid (RNA) molecules with enzymatic activity that have a great potential as therapeutic entities because of their ability to either cleave deleterious RNAs or repair mutant cellular RNAs.1,2 They form basepair-specific complexes and catalyze the hydrolysis of specific phosphodiester bonds, causing RNA strand cleavage. Differences exist between ribozymes in size and structure and, although most naturally occurring ribozymes cleave intramolecularly in a cislinkage, the RNA component of RNase-P, which is involved in the processing of pre–t-RNA molecules, acts in trans, ie, intermolecularly.3
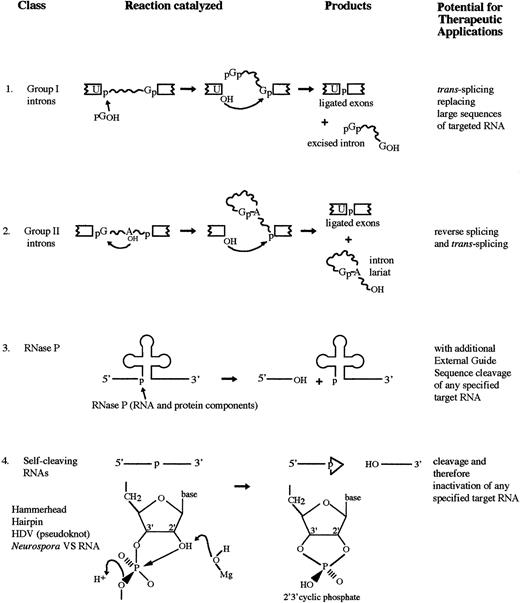
Like its protein counterpart, a catalytic RNA or ribozyme greatly accelerates the rate of a biochemical reaction and shows extraordinary specificity with respect to the substrates it acts upon and the products it produces. Ribozymes can cleave the normally unreactive bonds of a phosphodiester linkage in an RNA molecule resulting in a 3′ hydroxyl (3′OH) and 5′ phosphate (5′OH) and 3′ or 2′3′-cyclic phosphate.4 There are several different classes of ribozymes: the self-splicing group I and group II introns; RNase P; and several distinct catalytic motifs found in the small pathogenic RNAs (Fig 1).
RNA catalyzed reactions. (1) The two-step self-splicing reaction of group I introns. (2) The two-step self-splicing reaction of group II introns. An internal hydroxyl initiates the attack. (3) Cleavage of the 5′ leader sequence from pre-tRNA by RNase P. (4) Self-cleavage reaction of a number of small pathogenic RNAs and a few other RNAs.
RNA catalyzed reactions. (1) The two-step self-splicing reaction of group I introns. (2) The two-step self-splicing reaction of group II introns. An internal hydroxyl initiates the attack. (3) Cleavage of the 5′ leader sequence from pre-tRNA by RNase P. (4) Self-cleavage reaction of a number of small pathogenic RNAs and a few other RNAs.
SELF-SPLICING RNAs
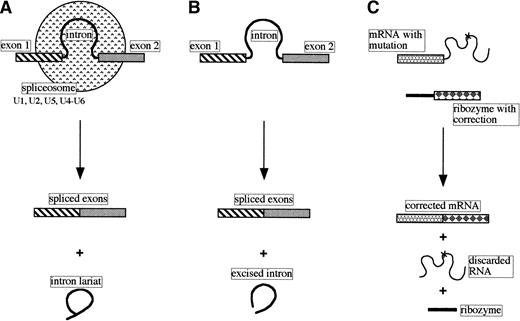
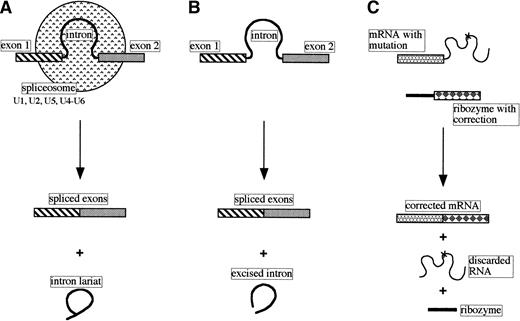
Many genes have their coding sequences (exons) interrupted by stretches of noncoding DNA called introns. Transcripts of such genes must undergo cleavage-ligation reactions to produce the mature functional RNA. The splicing of most nuclear pre-mRNAs involves a two-step process, generating an intron lariat and spliced exons, and has a requirement for a number of small nuclear ribonucleoprotein particles (snRNPs) and other proteins. Sequences in the RNA components of the snRNPs (U1, U2, and U5 small nuclear RNAs) recognize the 5′ splice site, branch point, and 3′ splice site, respectively, and, together with the U4 and U6 snRNPs, create the spliceosome where the intron is excised and the exons are ligated (Fig 2A; reviewed in Maniatis and Reed5).
Comparison of pre-mRNA nuclear splicing, self-splicing, and trans-splicing. (A) Nuclear pre-mRNA splicing with spliceosomes (small nuclear RNAs and proteins). (B) Self-splicing as performed by the self-splicing group I and group II introns. (C)Trans-splicing of a mutated mRNA with a modified group I catalytic intron.
Comparison of pre-mRNA nuclear splicing, self-splicing, and trans-splicing. (A) Nuclear pre-mRNA splicing with spliceosomes (small nuclear RNAs and proteins). (B) Self-splicing as performed by the self-splicing group I and group II introns. (C)Trans-splicing of a mutated mRNA with a modified group I catalytic intron.
Based on the nucleotide sequences and/or structures within and adjacent to the introns, introns have been classified into four classes: group I, group II, nuclear mRNA, and nuclear tRNA. Some examples of group I and II introns are capable of self-splicing in vitro in the absence of protein (Fig 2B).
Group I introns.
The self-splicing of group I introns, in the presence of a guanosine cofactor and magnesium, was first observed for the intron of the nuclear 26S rRNA gene in Tetrahymena thermophila.6 7 Self-splicing proceeds by two consecutive transesterification reactions, both initiated by nucleophilic attack. The excised intron, with a small deletion, can be converted into a true enzyme able to act in trans on specific substrates.
The RNA cleavage and ligation activities are intrinsic to the structure of the intron. By a number of basepaired regions, both the 3′ and 5′ splice sites are aligned for splicing by the internal guide sequence, close to the guanosine binding site.8-11 The catalytic domain of group I introns is formed by two structural domains, the crystal structures of which are now being determined.12 The interactions between the two domains and nucleotides important for cleavage activity are being elucidated.13 These studies are leading to understanding group I splicing: the mechanism of action and the roles played by the guanosine and metal cofactors.14-16
Despite not understanding fully the group I intron self-splicing mechanism of action these molecules are being manipulated to performtrans-splicing, ie, the intentional modification of the sequence of a targeted transcript in tissue culture cells.2 17 It can be seen in Fig 2C that a mutation in the RNA sequence can be corrected by replacing part of the mRNA with a new sequence using a suitably modified ribozyme. Although currently not very specific in choice of target RNA (the ribozyme recognizes andtrans-splices several mRNAs), it should be possible to use this approach to develop safe therapeutic ribozymes that can repair mutant RNAs associated with a variety of inherited diseases.
Group II introns.
Some group II introns are also able to undergo self-splicing. They differ from group I introns by the structure of their catalytic core and the products of the splicing: ligated exons and an excised intron-lariat18 (Fig 1). Again, the reaction consists of two transesterifications. The first step can be initiated by nucleophilic attack by an intronic 2′hydroxyl group on the phosphodiester linkage at the 5′ splice site, leading to the formation of the lariat structure. There is evidence for a second pathway, that involving attack by H2O or OH.19The second step is initiated by the 3′OH of the 5′ exon on the 3′ splice site. Basepairing interactions between sequences known as the exon binding site and the intron binding site hold the splice-sites in close proximity.20 This ability of group II introns to specifically bind the 5′ exon has been exploited to encourage the intron to catalyze reactions on exogenous substrates. Derivatives of group II introns reverse splicing by inserting themselves between ligated exons.21 This insertion and subsequent trans-splicing reactions can be used to shuffle sequences22 and therefore link any RNA molecule to any other RNA molecule, intentionally modifying the target sequence.
RNase P
Ribonuclease P (RNase P) is an ubiquitous endoribonuclease that processes the 5′ end of precursor tRNA molecules, producing 5′ phosphate and 3′OH termini.23 RNase P consists of both protein and RNA components and it was shown that the catalyst was the RNA moiety.24 As with the catalytic introns, a divalent cation is required as cofactor.25 RNase P can be directed to cleave any RNA when the target is in complex with a short, complementary oligonucleotide called an external guide sequence (EGS), thereby inactivating it.26-29
SELF-CLEAVING RNAs: SMALL PATHOGENIC RNAs
A number of small plant pathogenic RNAs (viroids, satellite RNAs, and virusoids), an RNA transcript from Neurospora mitochondrial DNA, and an animal virus, HDV, all undergo a self-cleavage reaction in vitro in the absence of protein (Table 1). The reaction, which requires magnesium and produces 2′3′-cyclic phosphate and 5′OH termini, results from several different catalytic motifs: the hammerhead, hairpin, and axehead or pseudoknot (Fig 3). It is generally thought that the self-cleavage reaction is an integral part of the small pathogenic RNAs' rolling circle method of replication.53 Circular monomeric plus and minus RNAs act as templates for the synthesis of longer-than-unit-length precursor RNAs. The production of monomeric forms from these concatamers requires specific cleavage thought to be performed by the RNA itself.
Small catalytic RNAs. (A) Hammerhead ribozyme split into substrate and catalyst,51 basepaired by means of two flanking arms (in bold). Numbering is that of Hertel et al.52 (B) Hairpin ribozyme split into substrate and catalyst.42 (C) Pseudoknot motif of the HDV ribozyme.45 Arrows show position of scissile bonds. IUB codes used throughout: N = A, C, G, U; R = A, G; Y = C, U; B = C, G, U; D = A, U, G; H = A, C, U; V = A, C, G.
Small catalytic RNAs. (A) Hammerhead ribozyme split into substrate and catalyst,51 basepaired by means of two flanking arms (in bold). Numbering is that of Hertel et al.52 (B) Hairpin ribozyme split into substrate and catalyst.42 (C) Pseudoknot motif of the HDV ribozyme.45 Arrows show position of scissile bonds. IUB codes used throughout: N = A, C, G, U; R = A, G; Y = C, U; B = C, G, U; D = A, U, G; H = A, C, U; V = A, C, G.
The reaction is a simple nonhydrolytic cleavage whereby the scissile bond undergoes a nucleophilic attack by the adjacent 2′OH group (Fig 1).54 The stereochemistry of cleavage and the role of Mg2+ in catalysis has been greatly studied.55,56 In the case of the hammerhead ribozyme, the crystal structure57,58 and fluorescence resonance energy transfer (FRET)59 measurements have shown the relative orientation of the core helices and have allowed the hammerhead catalytic core to be modelled.
Both of the crystallized hammerhead ribozymes contained modifications to prevent self-cleavage: the first57 contained an all DNA substrate analog; the second58, although all RNA, had the active site 2′ hydroxyl replaced with an inert 2′ methoxy group. As such, they may not represent the active structure. Scott et al60 have used time-resolved crystallography to determine the structure of an active hammerhead ribozyme in the absence of divalent cation or with Mg2+ but at a pH unsuitable for cleavage. A captured conformational intermediate showed that the most significant conformational changes were limited to the active site of the ribozyme.
It has been suggested that the function of the ribozyme catalytic core is twofold. First, it would destabilize the substrate strand to allow the scissile phosphodiester linkage to twist into a cleavable conformation. Second, the core would assist in positioning the divalent metal ion so as to facilitate catalysis.57 The more information gained about the active structure of the hammerhead ribozyme, the more rational the design of specific ribozymes can be. Once factors governing the specificity and mechanism of cleavage are elucidated, it may be possible to design new catalysts that efficiently cleave other sequences.
The hammerhead, hairpin, HDV, and Neurospora ribozymes have all been converted from the naturally occurring cis-active ribozymes to trans-active ribozymes by splitting the catalytic core from the substrate sequence. The Neurospora VS ribozyme has been converted once61 and is the least understood motif. More work has been performed on the trans-active HDV ribozyme. The development of this trans-active ribozyme is hampered by the lack of firm knowledge of the secondary and tertiary structure of the RNA and the alternative structures proposed.45,62,63 Recent studies are providing more detailed information.64,65 Despite this lack of understanding of the structure, trans-active HDV ribozymes have been developed.66 67
The development of the hammerhead and hairpin catalytic domains fortrans-cleavage are far more advanced. The structures and RNA folding are fairly well understood and the models for the design oftrans-acting hammerhead and hairpin ribozymes are well tried and tested.
The hammerhead (Fig 3A) consists of two flanking arms capable of basepairing with the substrate to form two helices (I and III) and a catalytic core with a helix (II) and several single-stranded regions. The ribozyme recognizes sequences either side of a substrate NUH site by means of the flanking arms. The catalytic core then cleaves the RNA 3′ of the NUH triplet. The hairpin ribozyme (Fig 3B) has also been split into substrate and catalytic core. Ribozymes are active in vitro in cell-free systems and in living cells. This activity and their sequence specificity makes them attractive as agents for the inhibition of gene expression.
As described below, they have been used against oncogenes and viruses and have helped in the understanding of the role of genes in developmental processes. Many of the problems with ribozymes are similar to those outlined for antisense oligodeoxy-nucleotides (ODNs).68 Efficient entry into cells, ribozyme stability, and precise targeting to the substrate RNA sequences are receiving attention. Most, if not all, RNAs in vivo normally exist as ribonucleoprotein (RNP) complexes and not as free molecules. Interactions between ribozymes and substrates and these RNP proteins could greatly influence activity. Ribozyme binding could be inhibited by steric hindrance. Alternatively, proteins may enhance activity via annealing and strand-exchange activities. The intracellular environment, such as pH and the availability of divalent cations, must also be considered. For example, the intracellular Mg2+concentration is known to be approximately 0.8 mmol/L,69much lower than is commonly used for in vitro reactions (10 to 20 mmol/L). A primary aim is to show that the ribozyme functions in vivo in a catalytic manner producing two cleavage fragments.
Specific gene inhibition using ribozymes (Table2)
Specificity has, in many cases, been inferred from biological effects without measuring target mRNA or protein levels or looking for ribozyme cleavage products or for effects on unrelated mRNAs. Initial studies of ribozyme activity in cultured cells have come across difficulties in detecting the activity and the catalytic nature of the ribozyme. Several groups have resorted to using a reporter gene, such as chloramphenicol acetyltransferase (CAT),85 in which a reduction in the CAT activity is taken as demonstrating ribozyme activity, or neomycin phosphotransferase (npt).86 Ribozyme specificity has been demonstrated for the βAPP mRNA when this was shown to be reduced compared with control RNAs, α-actin, and G3PDH mRNA.87 However, in this last study, a mutant ribozyme (catalytically inactive) was also effective at reducing levels, suggesting that the ribozyme-mediated degradation of βAPP mRNA in COS-7 cells was not dependent on ribozyme cleavage.
Ribozyme cleavage has been demonstrated by an RNase protection assay, where the cleavage products protected the probe.86 But care has to be taken in interpreting such results. It was recently noted that a hammerhead ribozyme cleaved its substrate during RNA preparation and not while in the cell.82 88
Despite some of the problems already alluded to, many groups are actively investigating ribozymes' therapeutic applications.
OVERCOMING DRUG RESISTANCE
The reversal of drug resistance is a popular goal for ribozyme technology. Kobayashi et al70 showed they could reduce MOLT-3 cells from approximately 700-fold resistant to vincristine to only 20- to 30-fold resistant when an anti–MDR-1 ribozyme was transfected in and stably expressed (under the β-actin promoter). This increase in drug sensitivity, and reduction in MDR-1 mRNA levels, was shown to be proportional to the amount of ribozyme expression. They critically showed that, although they could not detect cleavage products, a disabled ribozyme incapable of cleavage had no effects on the drug resistance levels. They also tested their ribozyme in the non–drug-resistant parental cell line and found no effect: the ribozyme was not toxic. Kiehntopf et al71 found that liposome-mediated transfection of drug-resistant mesothelioma cell lines with either in vitro transcribed or chemically synthesized ribozymes significantly reduced expression of the MDR-1 gene. This restored sensitivity towards chemotherapeutic drugs. They were also able to demonstrate, using reverse transcription polymerase chain reaction, the reduction in full-length MDR-1 mRNA levels.
OVERCOMING THE TRANSFORMED PHENOTYPE
The mRNAs from oncogenes have also been targeted by ribozyme technology. H-ras is activated by a mutation at codon 12 (GGU → GUU), with the activated form being a substrate for hammerhead ribozyme cleavage. When transformed NIH3T3 cells that displayed a neoplastic phenotype in vitro and were tumorigenic in nude mice in vivo were transfected by a ribozyme designed to cleave the activated H-ras mRNA, the transformed phenotype was abrogated.72 A reduction in H-rasmRNA was observed. A mutant ribozyme resulted in cells with an intermediate phenotype, probably due to an antisense effect of the ribozyme hybridizing arms. This group also addressed the question of specificity by looking at the K-ras mRNA levels in control and ribozyme-treated cells and showed no cross-reactivity. Ohta et al73 demonstrated that a tissue-specific promoter for the expression of an anti–H-ras ribozyme produced better results than using a viral promoter. This result suggests that, depending on the target mRNA, the choice of viral promoter for ribozyme expression is not always the best option. The transfected ribozyme appeared to affect not only proliferation but also the differentiation process of the melanoma cells in vitro.89
BCR-ABL in chronic myeloid leukemia.
Chromosome translocations and the resulting chimaeric genes are good targets for sequence-specific strategies. The hybrid mRNA will only be present in the cells with the translocation, and antisense ODNs or ribozymes targeted to the hybrid's junction should be specific for the hybrid and not affect the wild-type mRNA sequences. One such translocation results in the Philadelphia chromosome (Ph+) of chronic myeloid leukemia, and the bcr-abl oncogene. The expression of the bcr-abl protein tyrosine kinase is thought to be responsible for the malignant phenotype of Ph+ cells. Both the wild-type abl and bcr proteins are thought to be important for normal cell proliferation. This makes Ph+cells the ideal system to address the question of ribozyme sequence specificity. We have looked at the specificity of three different hammerhead ribozymes designed to cleave two splice variants of thebcr-abl mRNA90 in an attempt to clear up some of the contradictory results published.91-93 We showed the specific nature of one ribozyme (cleaved only bcr-abl RNA) but a lack of specificity of two others (cleaved both bcr-abl andabl RNAs). In a cell line system, the question of specificity has not really been addressed, but a decrease in cell proliferation,74 a decrease in bcr-abl mRNA and protein levels,75,76 and a decrease in bcr-ablkinase activity93 have been observed (Table 3). These studies all used slightly different hammerhead ribozymes: varying lengths of the flanking arms and various modifications to improve nuclease resistance or expression from vectors. Some groups observed quite substantial effects,75,76,93 whereas others have seen more modest91 or shown very few effects (our unpublished data). This is probably due to differences in the ribozymes' structures. Until the structure-function relationship is understood, such effects will not be predictable.
The hybrid gene AML1/MTG8 mRNA that results from a translocation between chromosomes 8 and 21 (associated with acute myeloid leukemia) is also being targeted by hammerhead ribozymes.94
OVERCOMING VIRAL DISEASE
Another clinical situation amenable to ribozyme targeting is viral disease, such as that associated with the human immunodeficiency virus (HIV). Two classes of ribozymes are being used to combat HIV: hammerhead79,81,95,96 and hairpin.97
Weerasinghe et al95 showed that MT4 cells transformed with vectors expressing an HIV-1 ribozyme were resistant to varying degrees of HIV-1 infection. The choice of constitutive versus inducible promoters for the expression of the anti–HIV-1 ribozymes had quite an influence on the degree of resistance to HIV-1 infection. A similar study was performed by Lo et al96 with a hammerhead ribozyme designed to cleave the HIV-1 tat RNA. Interestingly, although the anti-tat ribozyme-producing cells inhibited the replication of HIV-1, an antisense producing vector conferred a greater resistance to HIV-1 replication.
A third group have shown that T lymphocytes expressing anti–HIV-1 ribozymes showed resistance to HIV-1 replication.79 Cells transformed with a mutant ribozyme showed little resistance to viral replication. They argued that this demonstrated that the functional ribozymes were cleaving the HIV-1 RNA and were specific for this RNA. They have also looked at the effects of using different retroviral vectors to express the ribozymes: despite different ribozyme levels, there was a similar level of resistance to HIV-1 replication.98 This indicated that other factors had determining roles in the effectiveness of the ribozyme.
Bauer et al81 have made an important step by treating CD34(+) cells from individuals already infected with HIV-1. They transfected the cells with a retroviral vector containing a doubletat-rev ribozyme and showed up to a 1,000-fold inhibition of HIV-1 replication after further challenge with the virus.
A hairpin ribozyme has been shown to have similar effects to the hammerhead ribozyme: cells expressing the ribozyme were resistant to various strains of HIV-1. It was also demonstrated that the hairpin ribozyme significantly reduced the efficiency of the incoming virus to synthesize viral DNA.97 A hairpin ribozyme is in the preliminary stages of a phase I clinical trial against HIV-1.99 CD4(+) lymphocytes from HIV-1–infected donors were transduced with ribozyme or control vectors and viral replication was delayed by 2 to 3 weeks.80 The rapid progress of ribozymes to early clinical trials can be attributed to their specificity and ability to be delivered by viral vectors to the target cells. Another reason ribozymes could be successful against retroviruses is that they can potentially target several stages in the viral life cycle: the incoming genomic RNA, the mRNA transcribed from the integrated genome, and the progeny RNA genomes.
HIV-1 is not the only virus being targeted. Beck and Nassal82 are targeting a hammerhead ribozyme to the encapsidation signal ε of Hepatitis-B virus, the causative agent of B-type hepatitis. Unfortunately, ribozyme cleavage has only been observed in vitro and in cell extracts, but not in intact cells. Even when the ribozyme was placed in cis with an artificial substrate, little ribozyme activity was observed (most of the activity could be attributed to antisense effects). The investigators speculated that there was something inhibiting cleavage within the cell, because efficient cleavage was observed upon RNA extraction. Hepatitis-C virus RNA is also the subject of selective targeting and destruction by ribozymes,83,100,101 as is the mRNA for influenza A virus.102
RIBOZYMES IN ANIMAL MODEL SYSTEMS
Some researchers are beginning to assay ribozyme effects in animal model systems.77,84,103 We have been investigating the action of an ex vivo purging of a murine cell line containing thebcr-abl oncogene with ribozymes against the bcr-ablmRNA before injection into SCID mice. Effectiveness of the ribozyme is demonstrated by the increased survival of the mice (when compared with mice injected with control-treated bcr-abl–containing cells).77
Flory et al84 have used a rabbit model of interleukin-1–induced arthritis to assess the localization, stability, and efficacy of exogenous anti-stromelysin hammerhead ribozymes. They observed that exogenously delivered ribozymes were taken up by cells in the synovial lining and synovial interleukin-1α–induced stromelysin mRNA levels were reduced. Catalytically inactive ribozymes were ineffective supporting a cleavage mechanism for ribozyme activity. Using nude mice, Czubayko et al78 have rather nicely demonstrated the relationship between the secreted growth factor pleiotrophin (PTN) from melanoma cells and the melanoma cells' metastasis to the lungs. Introducing ribozymes against PTN into a cell line, which reduced PTN mRNA and growth factor activity, concomitantly prevented the metastatic spread of the tumors. Larsson et al103 looked at hammerhead ribozymes against the β2-microglobulin (β2M) mRNA in both cell lines and transgenic mice. They observed ribozyme expression in lung, kidney and spleen with greatest reduction in the β2M mRNA levels in the lung. However, it should be noted that ribozyme levels and reduction in β2M mRNA varied widely between litter mates. Lieber and Kay104 have looked at ribozymes (against human growth hormone) expressed from adenovirus vectors after somatic gene transfer into transgenic mice. A reduction of up to 96% in growth hormone was observed in correlation with the mRNA levels. Using transgenic mice is obviously quite different from using a retroviral vector to express a ribozyme. Although inappropriate for therapeutic administration, transgenic animals can provide important information about ribozyme expression levels in different tissues and ribozyme efficacy, as well as information about the role of the targeted mRNA.
Very little has been done so far with respect to pharmacokinetic and pharmacodynamic data for ribozymes in animal model systems. However, one study of pharmacokinetic properties of synthetic, chemically modified hammerhead ribozymes after intravenous injections105 has shown the prolonged presence of a cytochrome P-450 ribozyme in the plasma (up to 48 hours postinjection) and perfusion into tissues other than the vascular system (kidney and liver). This group has also injected a similarly modified ribozyme to amelogenin mRNA, with no carrier to assist cellular uptake, and shown a surprisingly successful knock-out of the targeted gene's expression.106 The various works detailing transgenic animals with a gene construct for an antisense RNA or ribozyme sequence has recently been reviewed.107
OTHER USES OF RIBOZYMES
As well as their potential as therapeutic agents, ribozymes can be used to generate loss-of-function phenotypes to elucidate the roles of genes. This has been performed for the Fushi taragu gene in Drosophila,108 for c-fos,109 and for matrix metalloproteinase 2.110
Ribozymes have also been used in other systems to varying degrees of success: in plant protoplasts111,112 and in yeast (Saccharomyces cerevisiae113 and S pombe114). Transgenic plants with ribozymes to the lignin-forming peroxidase of tobacco have been created, with reduction in the peroxidase mRNA and protein levels observed.115
The design of ribozymes and problems to overcome
(For assistance in designing and applying ribozyme technology please refer to two recently published methods books: Ribozyme Protocols116 and Antisense and Ribozyme Methodology.117)
As detailed above, ribozymes have been successfully used to target specific mRNAs. To use a ribozyme strategy, several parameters must be decided. The first choice is the type of ribozyme to use. The hammerhead ribozyme is the best-studied type. The target sequence requirements for the hammerhead are slightly less stringent than for the hairpin, and, because the hammerhead is smaller than the hairpin, it is cheaper to synthesize and higher yields can be obtained. The next decision is the choice of target mRNA. It has been suggested that targeting the mutated gene sequence may not always be the most effective approach. Targeting an mRNA of a protein downstream of the mutated gene product may result in a more effective inhibition. However, this could lead to a decrease in specificity between normal and affected cells. The position of the NUH target site within the mRNA must then be selected. The efficiency of the ribozyme is dictated by the sequence (different NUH sequences can vary by >100-fold in the rates of their cleavage, and the sequence context of the site is also an important factor118) and accessibility of target site due to secondary and tertiary structures of the mRNA.119Obviously, before cleavage can occur, the ribozyme must bind to its substrate via the sequences either side of the target site. Experimental analysis such as nuclease mapping or chemical probing can be performed to monitor accessibility.120 Alternatively computer RNA folding programs can be used to help determine accessible sites121 122 with less expense but at a reduced reliability.
Most computational algorithms for RNA structure prediction are based on calculation of the minimum free energy (ΔG). For a stretch of bases the program determines the most stable structure, the lowest ΔG. The computer then looks at the next stretch of bases (which may or may not overlap with the previous stretch) and calculate the next ΔG. In this way, local maxima and minima ΔG values are obtained that correspond to relatively unstable or stable regions of the RNA, respectively. The value of ΔG is dependent on which algorithm is used to determine the structure, and this is where the errors can be introduced. A minima in ΔG does not necessarily mean that a stable structure exists in vitro or in vivo. Current programs are also limited to the length of the sequence to be analyzed; distant interactions in a long mRNA cannot be predicted.
The length of the ribozyme's hybridizing arms (which basepair with the substrate) must strike a balance between providing specificity for the substrate while allowing product dissociation.123,124 It has been suggested that asymmetric arms may be more effective than symmetric arms.125 It is also important to realize that results suggesting an efficient arm length in a cell-free system may not be ideal in a cellular system.126,127 An increasing number of laboratories are applying in vitro selection, or in vitro evolution, techniques (SELEX) to develop more efficient ribozymes and ribozymes with new activities.128,129 The principal of the technique is based on the screening by selection of a pool of RNA molecules. Starting with either a totally or partially random pool of RNA molecules, a cycling of binding or activity, partitioning, and amplification steps results in a novel ribozyme or ribozymes with the desired binding or catalytic activity. Using this technique, and by chance, both RNA and antisense DNA molecules have been developed that are aptameric in activity, ie, they can interact directly with the target protein in a sequence-specific manner.130-132
Many groups are looking at ways to improve ribozyme catalytic activity. One way to do this is to use multiunit ribozymes (ie, transcripts with more than one catalytic domain) targeting different sites within the same mRNA. Chen et al133 developed a nona-ribozyme against HIV-1 and showed that this was more effective at cleaving substrate RNA in vitro and inhibiting HIV-1 replication in cell lines than mono-ribozymes. This multimeric ribozyme has a second advantage, ie, HIV-1 escape mutants are less likely. A multiunit anti–BCR-ABLribozyme (3 catalytic domains) was shown to be more effective than the three ribozyme domains added separately, both in vitro and in cell lines, by reducing the BCR/ABL mRNA.134
Efforts to increase exogenously delivered ribozymes' stability and hence activity have concentrated on chemical modifications to the ribozyme structure. 2′-fluoro or 2′-amino135and 2′-O-allyl and 2′-O-methyl106 136 are a few of the modifications being investigated.
Ribozymes can be delivered to cells in two ways: as preformed ribozymes (exogenous delivery) or as ribozyme genes, a method of endogenous delivery. The means of transfection (eg, lipofection or electroporation) is important for the former, whereas the choice of promoter (pol II, pol III, viral, etc) is important for the latter.137 The latter could be termed ribozyme gene therapy, whereby the introduced ribozymes downregulate the targeted gene expression or repair mutant mRNAs.138
Endogenous delivery has been achieved by inserting ribozyme sequences into the untranslated regions (UTRs) of genes transcribed by RNA polymerase II (pol II), such as the SV40 early promoter85or the actin gene.139 The RNA polymerase III (pol III) promoters from the U6 small nuclear RNA140 or from certain tRNAs have also been successfully used to express ribozymes and achieve effects.141 Tissue-specific promoters, such as the tyrosinase promoter, have also been investigated.73
Both retroviral-derived and more recently adeno-associated viral (AAV)-derived vectors are commonly used. Ribozyme genes can be expressed from the viral long terminal repeat (LTR) promoters or from introduced pol II or pol III promoters. Retroviral vectors are relatively efficient, safe, and capable of (randomly) integrating stably in the host genome of replicating cells. AAV is nonpathogenic and has the advantage of integrating into a defined region of the host genome without requiring cell division. Zhou et al98compared three types of promoters: Molony murine leukemia virus (MoMuLV) LTR, the human CMV promoter, and a human tRNAmetcassette for the transcriptional control of a pair of anti–HIV-1 ribozymes. The LTR promoter produced the highest expression levels of the ribozyme, but the ability of each to confer resistance to HIV-1 replication was very similar. The investigators concluded that other factors (antisense effects, protein influence, and localization) than the absolute levels of the ribozymes played a role in determining the effectiveness of the ribozymes. The recent work of Bertrand et al137 shows that colocalization is the most important factor. In this study, promoters used for the expression cassettes were derived from the human tRNAmeti (pol III), the human U1 snRNA (pol II), the human U6 snRNA (pol III), and the Rous sarcoma virus (RSV) LTR (pol II). Each cassette was introduced into a cell line with both AAV- and MoMuLV-based vectors. All of the cassettes produced ribozymes when in context of the AAV vector, whereas the tRNA and U6 cassettes were inactive with the viral vector. Anti-HIV ribozymes derived from the RSV cassette were expressed at the lowest levels and were cytoplasmic (consistant with being capped and polyadenylated). The other transcripts were predominantly nuclear and expressed at higher levels. When the cells were challenged with HIV, surprisingly only those containing the capped, poly-A RNAs that were cytoplasmically localized were able to suppress HIV replication.
Sullenger and Cech142 have successfully looked at using a viral packaging signal to direct their ribozymes to the same subcellular locations as the viral targets. It has also been observed that the 3′UTRs of some mRNAs carry the signal responsible for localization.143,144 These, and others yet to be discovered, could be attached to ribozyme sequences to aid their colocalization. In a hope to understand RNA trafficking in the cell, we have been following ribozyme distribution using fluorescently tagged ribozymes.145 Because the nucleus is thought by many to be the site of action for ribozymes, several groups are looking at ribozyme activity when expressed with a nuclear localization signal140 or in isolated nuclei.146 It can be concluded that the type of promoter and its context can determine the intracellular compartmentalization of the ribozyme and that colocalizing the ribozyme with the substrate is a primary determinant of ribozyme efficacy in vivo.
Measurements for assaying ribozyme activity can be made at the mRNA (RNase protection assays or reverse-ligation polymerase chain reaction147) or protein levels (Western blotting and antibody probing), function of the protein (eg, kinase activity) and effects on cell differentiation, or the onset of apoptosis. In general, ribozyme cleavage activity in the cell system or in vivo has not been empirically demonstrated, but instead has been inferred from the lack of effects by a catalytically inactive ribozyme if included as a control.
Stein and Krieg148 pointed out various considerations for the interpretation of data derived from the use of antisense ODNs. Charged ODNs are polyanions and as such can bind and sequester growth factors to the basement membrane. Phosphorothioate ODNs may exhibit nonantisense but sequence-dependent effects, eg, a G quartet149 or a TAT triplet at the 3′ end.150 These considerations, although not proven, are probably just as valid for ribozymes. To show specific ribozyme cleavage, the following controls are required: a catalytically inactive ribozyme (to show antisense effects), a conventional antisense ODN equivalent to the ribozyme arms (again, to show antisense effects), an active but unrelated ribozyme (to show specificity for target mRNA), a scrambled ODN, and finally the transfection agent or vector only (nonspecific toxicity effects).
Ribozymes and antibiotics.
It has been demonstrated that certain classes of antibiotics can interact with some ribozymes and, in some cases, influence their cleavage activity. Aminoglycoside antibiotics inhibit group I intron function but not group II introns.151 The aminoglycosides, in particular neomycin B, have been shown to inhibit hammerhead ribozyme activity,152 as have the tetracyclins.153 The HDV ribozyme is also inhibited by both aminoglycosides and tetracyclins.154 Contrary to this, theNeurospora VS ribozyme cleavage activity is actually enhanced by viomycin, a tuberactinomycin antibiotic.155 The modulation of ribozyme activity by other agents, in particular antibiotics, should be taken into consideration when designing experiments with cell cultures (often grown in the presence of antibiotics) and possibly in a clinical setting.
THE FUTURE FOR RIBOZYMES
For ribozymes to become realistic therapeutic agents several obstacles need first to be overcome. These obstacles are the efficient delivery to a high percentage of the cell population, efficient expression of the ribozyme from a vector or intracellular ribozyme concentration, colocalization of the ribozyme with the target, specificity of ribozyme for the desired mRNA, and an enhancement of ribozyme-mediated substrate turnover.
Despite these reservations, results with ribozymes so far look promising, particularly in the HIV-1 studies. As our knowledge of RNA structure, secondary and tertiary, increases, we will be able to target the RNA more rationally, which may help with the problems of specificity. At the same time, the understanding of the physical localization of RNA in cells and its tracking as it moves from the nucleus to cytoplasm will also help in ensuring colocalization of the ribozyme and target. Modifications of the ribozymes, eg, the 2′ ribose with allyl group, increases the stability to nucleases quite dramatically. Similarly, DNA sequences allied to the ribozymes in use in our laboratory increase the stability. Entry into cells with liposomes or via vectors are also looking hopeful. Catalytic activity is maintained but research is still ongoing at tackling the problem of increasing the number of ribozyme molecules in proportion to the substrate molecules—a requirement for success. These molecules must retain their catalytic potential, must reach an accessible site in the substrate, and eventually be synthesized from the appropriate vector chosen for clinical trials. Work in the antisense DNA field would also benefit from solutions to these problems.
Supported by Grant No. 9667 of the Leukaemia Research Fund, London, UK.
Address reprint requests to Helen A. James, PhD, School of Biological Sciences, University of East Anglia, Norwich, Norfolk, NR4 7TJ UK.