Abstract
The mechanisms that regulate the selective infiltration of eosinophils in certain allergic diseases are still poorly understood. The CC chemokine eotaxin is a potent chemoattractant, highly specific for eosinophils. Recent studies have implicated that eotaxin plays an important role in the recruitment of eosinophils in different inflammation processes. A number of other chemokines, cytokines, and chemoattractants also have chemotactic activities for eosinophils and some of them present high selectivity for eosinophils. To further study the role of eotaxin in inflammation, we generated mutant mice with the eotaxin gene disrupted and replaced by the Escherichia coliβ-galactosidase gene. These mice developed normally and had no histologic or hematopoietic abnormalities. Furthermore, our studies showed that the lack of eotaxin did not affect the recruitment of eosinophils in the inflammation models induced by Sephadex beads and thioglycollate, as well as in an experimental lung eosinophilia model induced by ovalbumin aerosol challenge, even at the onset of the inflammatory response. The replacement of the eotaxin gene by the β-galactosidase gene provided a useful marker to monitor the activity of the eotaxin promoter under normal conditions and after antigen challenges. Immunohistochemical staining suggested that endothelial cells were the major sources of eotaxin expression.
THE CHEMOKINES ARE a family of small secreted proteins that have chemotactic activities for leukocytes and play a key role in leukocyte trafficking.1-5 They are also important in processes such as hematopoiesis, anaphylaxis, angiogenesis, and malignancy.1,2,5 The importance of chemokines and their receptors in inflammatory and infectious diseases, especially in the infection by human immunodeficiency virus–type 1 (HIV-1), has recently been described.6,7 There are two major subfamilies of chemokines, designated as CXC (or α) and CC (or β), based on the conserved cysteine motif near the amino terminus of the protein.4,5 The CXC chemokines, which include interleukin-8 (IL-8), Gro-α, -β, and -γ, IP10, and SDF-1, attract neutrophils and T cells in general. The largest subfamily, the CC chemokines, attract mainly mononuclear cells and also lymphocytes, eosinophils, and basophils. These include monocyte chemotactic proteins (MCP-1, -2, -3, -4, and -5), macrophage inflammatory proteins (MIP-1α and MIP-1β), RANTES (regulated upon activation, normal T cell expressed and secreted), and eotaxin. There are also two single-member subfamilies, one of which is the C (or γ) subfamily with lymphotactin, which has only one conserved cysteine residue at the N-terminal and selectively attracts lymphocytes.8 The other subfamily, with the chemokine named fractalkine or neurotactin, contains a CX3C motif that exists in either membrane-bound or secreted forms.9,10 The receptors for the different chemokines are a family of seven-transmembrane G-protein–coupled receptors. So far, there are approximately 20 such receptors cloned and most of them have identified ligands.5,6 These include the CXCR receptors (CXCR1-4) for CXC chemokines and the CCR receptors (CCR1-8) for CC chemokines. The relationship between chemokines and receptors is complicated, because most chemokines can bind to more than one receptor and most receptors have more than one ligand.6
The CC chemokine eotaxin was originally discovered in the bronchoalveolar lavage (BAL) fluid of guinea pigs sensitized and challenged with ovalbumin aerosol.11 This protein, and its homolog found later in mice and humans,12-16 is a potent and specific chemoattractant for eosinophils. This is in contrast to most other CC chemokines, which have the capacity to attract more than one type of leukocyte.17 Eotaxin specifically binds to the CCR3 receptor, which is expressed selectively and at high levels on eosinophils.16,18,19 Other chemokines with potent chemotactic activities on eosinophils include eotaxin-2,20MCP-4,21,22 RANTES,23 MCP-3,24,25and, to a lesser extent, MIP-1α,23 MCP-2,25and, possibly, MCP-5.26 Most of these chemokines also use the CCR3 receptor when they are active on eosinophils.17,27One exception is MIP-1α, which uses another CC receptor also expressed on eosinophils, CCR1.19,28 Unlike eotaxin, most of these chemokines also bind to other receptors and are active on other leukocytes, such as monocytes, lymphocytes, and basophils. Some classical chemoattractants, such as C5a, leukotriene B4 (LTB4), and platelet-activating factor (PAF), have also been found to be active on eosinophils.17,29 30
The accumulation of eosinophils is characteristic of allergic diseases such as asthma and some parasitic infections.31,32 The mechanisms that regulate the infiltration of eosinophils during these processes are being elucidated. Recent studies indicate that eotaxin may play an important role in the recruitment of eosinophils in different inflammation models.12-14,33 Models of antigen-induced airway allergic inflammation and lung eosinophilia have been developed in both guinea pig and mouse.11,13,34 The eotaxin mRNA in lung is induced in response to antigen challenge and parasitic infection.13,34 Prechallenge treatment of mice with anti-CD3 monoclonal antibody (MoAb) inhibits the induced eotaxin mRNA expression and significantly reduces eosinophil infiltration in the lung.34 Administration of antibodies specific for eotaxin in vivo reduce the accumulation of eosinophils.35On the other hand, eotaxin expression is found constitutively in some tissues, such as thymus, lymph nodes, skin, and heart, where there is no eosinophil infiltration.12-14 It has been shown that in vitro eotaxin can inhibit the infection of certain HIV-1 strains that use CCR3 as coreceptor.36 Recent reports showed that in vitro eotaxin is also a potent chemoattractant for human basophils.20 37 All of these results suggest that eotaxin may have some unknown functions besides chemotaxis for eosinophils.
The generation of mutant mice with a loss of eotaxin function could provide a definitive answer to the role of this chemokine in the recruitment of eosinophils in inflammation and, possibly, other biological processes. A recent report on the targeted disruption of the mouse eotaxin gene indicates a partially reduced eosinophil recruitment in the experimental lung allergic inflammation model and in a model of parasite-derived antigen-induced stromal keratitis.38 We report here a different approach to generate eotaxin knockout mice by replacing the eotaxin gene with the Escherichia coliβ-galactosidase gene, allowing the identification of the cells that express eotaxin.
MATERIALS AND METHODS
Construction of targeting vector.
Mouse genomic DNA containing the eotaxin gene was cloned from a mouse library prepared from D3 (129/sv) embryonic stem (ES) cell DNA. The pPNT vector containing the neo (neomycin) and tk(thymidine kinase) genes was used to construct the targeting vector. A 3.5-kb mouse genomic DNA containing exon 3 and the 3′ untranslated region of the eotaxin gene was cloned into theXba I-Kpn I sites of pPNT vector to generate pPNT-Ex3. A 3.2-kb BamHI-Sac I mouse genomic DNA containing the 5′ untranslated region and the first four amino acids of eotaxin precursor protein was fused in-frame to the E coliβ-galactosidase gene, and this fragment was then subcloned into theNot I-Xho I sites of pPNT-Ex3 to generate the targeting vector pPNT-Ex.
Generation of knockout mice.
The targeting vector (25 μg) was linearized by Not I digestion and transfected into 1 × 107 129/Sv-derived ES cells (CJ7) by electroporation using a Bio-Rad gene pulser (Bio-Rad, Hercules, CA). The ES cells were grown under positive-negative selection in G418 for neomycin-resistant and fialuridine (FIAU) for thymidine kinase-negative clones. About 400 double-resistant clones were screened by Southern blot analysis using a 320-bp external probe (immediately outside the 5′ arm of the targeting vector) generated by polymerase chain reaction (PCR), and three true recombinant clones were obtained. All three clones were injected into blastocysts of ICR strain of mice and the resulting male chimeras were mated with female mice of ICR strain for germline transmission of the targeted allele. Heterozygous offsprings were interbred to generate homozygous mice that had two alleles of the eotaxin gene disrupted. Siblings from heterozygote matings were used to control for strain background effects. Genotyping was performed by Southern blot analysis of mouse tail/toe DNA using the 320-bp probe. Routinely, genotyping was performed by PCR analysis using a pair of primers specific for the neo gene to identify the targeted allele and a pair of primers to amplify a 400-bp genomic DNA that is deleted in knockout mice. The sequence for the neo-specific primer pair used is as follows: sense, 5′-CGGCCACAGTCGATGAATCCAGAAA-3′, and antisense, 5′-CCATTCGACCACCAAGCGAAACATC-3′. The sequence for the eotaxin-specific primer pair used is as follows: sense, 5′-AGTCCATCCCCAAGTTAGAG-3′, and antisense, 5′-ACATTGTAGGAATGGCATGGCT-3′. PCR was performed with 25 μmol/L of each of the sense and antisense primers under the following amplification conditions: 1 minute at 94°C, 1 minute at 55°C, and 1.5 minutes at 72°C for 30 cycles.
Southern and Northern blots and reverse transcription-PCR (RT-PCR).
Mouse genomic DNA was digested with Spe I (New England BioLabs, Beverly, MA), electrophoresed, and transferred to a GeneScreen Plus nylon membrane (DuPont, Wilmington, DE). The membrane was hybridized with the 32P-labeled 320-bp probe overnight at 65°C in a hybridization solution (5× SSPE/0.1% pyrophosphate [PPiESS], 1× Denhardt’s solution, 1% sodium dodecyl sulfate [SDS]), washed once for 15 minutes at room temperature, and washed twice at 65°C for 20 minutes in 0.2× PPiESS with 0.5% SDS.
RNA was extracted from different tissues with Trizol reagent (GIBCO BRL, Gaithersburg, MD) as per the supplier’s instructions. Ten micrograms of RNA was electrophoresed on a 1% agarose gel containing 0.7% formaldehyde and then transferred in 20× SSC to a GeneScreen Plus nylon membrane and hybridized with a32P-labeled mouse eotaxin full-length cDNA or a probe specific for GAPDH. The hybridization was performed as in the Southern blot, except that the hybridization solution contained 100 μg/mL of salmon sperm DNA.
For RT-PCR, first-strand cDNAs were generated by reverse transcription with poly(dT) primers from 5 μg of total RNA using the SuperScript reverse transcriptase kit (GIBCO BRL). Five percent of the product was then used as the template for PCR amplification using primers specific for mouse eotaxin, fic, or β-actin.
Experimental lung inflammation model.
The ovalbumin (OVA) challenge of mice was performed as described before.34 Briefly, 10 μg of OVA (Sigma Chemical Co, St Louis, MO) in aluminum hydroxide (5 mg/mL in phosphate-buffered saline [PBS]) was injected into mice intraperitoneally on days 0, 7, and 14. Sham-immunized mice were injected with aluminum hydroxide only. On any day between days 21 and 24, mice were restrained in a plexiglass box and challenged once with aerosolized OVA (50 mg/mL in PBS) generated by an ultrasonic nebulizer (DeVilbiss Co, Somerset, PA) for 20 minutes. Control mice (sham-immunized) were also challenged with aerosolized OVA. At 6, 12, 18, 24, and 48 hours after challenge, mice were anesthetized with 0.6 mL of avertin (200 mg/mL of tribromoethanol; Aldrich, Milwaukee, WI) by intraperitoneal injection and perfused with 2% paraformaldehyde. Tissues were harvested, postfixed in the same perfusion solution for 1 hour, and then stored in 30% sucrose/PBS solution overnight. The tissues were sectioned at 30 to 100 μm and stained with hematoxylin and eosin (H&E). For paraffin sections, tissues were postfixed in 10% neutral-buffered formalin, embedded in paraffin, sectioned at 4 to 6 μm, and stained with H&E.
BAL was performed at 18 hours after ovalbumin aerosol challenge. Mice were anesthetized and the trachea was surgically exposed. The lungs were lavaged via the trachea with 0.5-mL aliquots of PBS containing 0.6 mmol/L EDTA (8 × 0.5 mL at 37°C). The total cells were determined with a hemacytometer. Fifty thousand cells were collected by cytospin and stained with Diff-Quick stain set (Dade Diagnostic, Newark, DE). The percentages of eosinophils, neutrophils, macrophages, and lymphocytes were determined from a count of six different high-power (100× objective) views for each mouse. These percentages were multiplied by total cell number to obtain the number of different cell populations.
Inflammation induced by thioglycollate and Sephadex.
Mice were injected intraperitoneally with 3 mL of 3% Brewer’s thioglycollate (DIFCO, Detroit, MI) in PBS, and 24 hours postinjection, peritoneal cells were lavaged with Dulbecco’s modified Eagle’s medium (DMEM)/5% fetal bovine serum (FBS), and total cell numbers were determined with a hemacytometer. Cells were collected by centrifugation and resuspended at 1 × 106 cells/mL. Fifty thousand cells were collected by cytospin and stained with Diff-Quick stain set (Dade Diagnostic). Different type of cells were counted at six different high-power views for each mouse. The number of different cell populations was calculated by multiplying the percentage by total cell counts.
Sephadex G50 beads (superfine; Sigma Chemical Co) were suspended in PBS. Each mouse received 0.3 mg of these beads by intravenous injection of the tail vein. At 6 or 24 hours after injection, mice were killed and perfused with 2% paraformaldehyde. Lungs were harvested, postfixed in the same perfusion solution for 1 hour, and then stored in 30% sucrose/PBS solution overnight. Then tissues were processed as described above.
β-Galactosidase and immunohistochemical staining.
To determine β-galactosidase activity, frozen sections were stained in a solution containing 1 mg/mL of X-gal (GIBCO BRL) overnight and then counter-stained with eosin. For immunohistochemical analysis, frozen tissue sections were first stained in X-gal solution as described above and then treated in 0.03% hydrogen peroxide in PBS, blocked with Vector Avidin/Biotin blocking kit SP-2001 (Vector Laboratories, Burlingame, CA), and blocked with 10% normal rabbit serum. The primary anti-CD34 antibody (Pharmigen, San Diego, CA; 1:100 dilution) was added and incubated for 90 minutes at room temperature. After washing in PBS, the sections were treated with a secondary antibody Vector biotinylated antirat (1:100) for 30 minutes and rinsed in PBS. The sections were then treated with the Vector ABC Elite Peroxidase (PK6100) and Vector DAB (SK-4100) kits (Vector Laboratories). Finally, the sections were counter-stained with eosin.
RESULTS
Eotaxin-deficient mice show no phenotypic abnormalities.
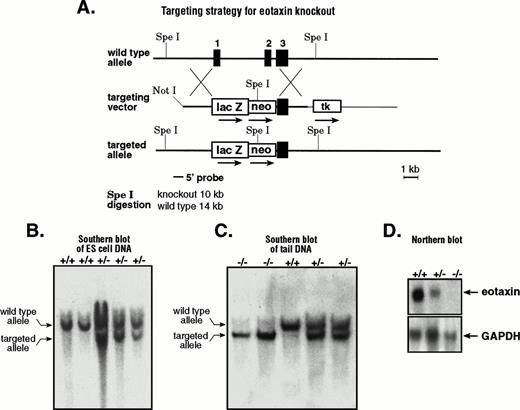
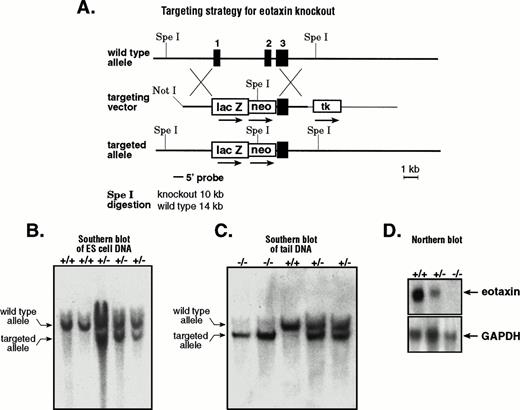
Like most chemokine genes, eotaxin consists of three exons and two introns.12 To knockout eotaxin, we constructed a targeting vector by disrupting the first two exons (Fig 1A). As a marker for the activity of the eotaxin promoter, we fused the E coliβ-galactosidase gene in-frame to the first four amino acids of the leader sequence of the eotaxin precursor. The targeted allele disrupts the eotaxin gene by deleting most of exon 1 and all of exon 2 with the insertion of the E coli β-galactosidase gene and the neomycin-resistance gene. The ES cell clones that underwent homologous recombination were screened by Southern blot analysis using a 5′ external probe. After Spe I digestion, the targeted allele generated a 10-kb fragment, whereas the wild-type allele generated a 14-kb fragment (Fig 1B). The positive clones were further confirmed by a 3′ external probe that distinguishes the Spe I digestion by 6-kb and 14-kb fragments for the targeted and wild-type alleles, respectively (data not shown). Positive ES cell clones were injected into blastocysts, and the male chimeras obtained were used to generate heterozygous animals. These were interbred to generate homozygous mice for the disrupted eotaxin allele. Figure 1C shows the result of Southern blot analysis of the SpeI-digested genomic DNA from wild-type (+/+), heterozygous (+/−), and homozygous (−/−) mice. Because eotaxin mRNA is abundant in lymph nodes,12 we performed Northern blot analysis of total RNA from lymph nodes of wild-type and mutant mice with a probe that is specific for eotaxin. As expected, there is no eotaxin mRNA expression in knockout mice (Fig 1D).
Generation of eotaxin null mice. (A) Strategy for generating eotaxin knockout mice by homologous recombination. The three-exon and two-intron genomic structure of the eotaxingene is shown on the top. The targeting vector is shown in the middle. The 5′ arm contains 3.2 kb of mouse genomic DNA with the sequence encoding the first 4 amino acids of eotaxin fused in-frame to the E coli β-galactosidase gene. The 3′ arm of 3.5 kb of mouse genomic DNA containing exon 3 and the 3′ untranslated sequence of the eotaxin gene is inserted between the neomycin-resistance gene (neo) and the herpes simplex virus thymidine kinase (tk) gene. The targeted allele after homologous recombination and the screening strategy are shown at the bottom. The black boxes represent the exons, with their respective number above. Southern blot analysis of SpeI-digested genomic DNA from ES cells (B) and from wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice (C). The upper band (14 kb) shows the wild-type allele, and the lower band (10 kb) shows the targeted allele. (D) Northern blot of total RNA (10 μg) from lymph nodes of wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice. The blot was probed with32P-labeled mouse eotaxin cDNA (upper panel), stripped, and tested with a probe specific for mouse GAPDH (bottom).
Generation of eotaxin null mice. (A) Strategy for generating eotaxin knockout mice by homologous recombination. The three-exon and two-intron genomic structure of the eotaxingene is shown on the top. The targeting vector is shown in the middle. The 5′ arm contains 3.2 kb of mouse genomic DNA with the sequence encoding the first 4 amino acids of eotaxin fused in-frame to the E coli β-galactosidase gene. The 3′ arm of 3.5 kb of mouse genomic DNA containing exon 3 and the 3′ untranslated sequence of the eotaxin gene is inserted between the neomycin-resistance gene (neo) and the herpes simplex virus thymidine kinase (tk) gene. The targeted allele after homologous recombination and the screening strategy are shown at the bottom. The black boxes represent the exons, with their respective number above. Southern blot analysis of SpeI-digested genomic DNA from ES cells (B) and from wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice (C). The upper band (14 kb) shows the wild-type allele, and the lower band (10 kb) shows the targeted allele. (D) Northern blot of total RNA (10 μg) from lymph nodes of wild-type (+/+), heterozygous (+/−), and knockout (−/−) mice. The blot was probed with32P-labeled mouse eotaxin cDNA (upper panel), stripped, and tested with a probe specific for mouse GAPDH (bottom).
The eotaxin−/− mice appear healthy and normal. All tissues examined of 6-week-old animals were unremarkable. A total number of 206 mice born from heterozygous matings showed normal 1:2:1 Mendelian ratio with 46 wild-type (22.3%), 112 heterozygous (54.4%), and 48 knockout (23.3%) mice. Fluorescence-activated cell sorting (FACS) analysis of cell surface markers for T cells, B cells, granulocytes, and macrophages from lymphoid organs, including thymus, lymph nodes, spleen, and bone marrow, showed no differences between wild-type and knockout mice (data not shown). The markers examined included CD4, CD8, TCRαβ, and CD25 in thymus; CD4, CD8, TCRαβ, CD25, Thy-1.2, and B220 in lymph nodes; CD4, CD8, TCRαβ, CD25, Thy-1.2, B220, Ter-119, and Mac-1 in spleen; and Ter-119, Mac-1, B220, and IgM in bone marrow. Therefore, it appears that mice deficient for eotaxin have no hematopoietic abnormalities.
The expression of β-galactosidase in eotaxin heterozygous and null mice.
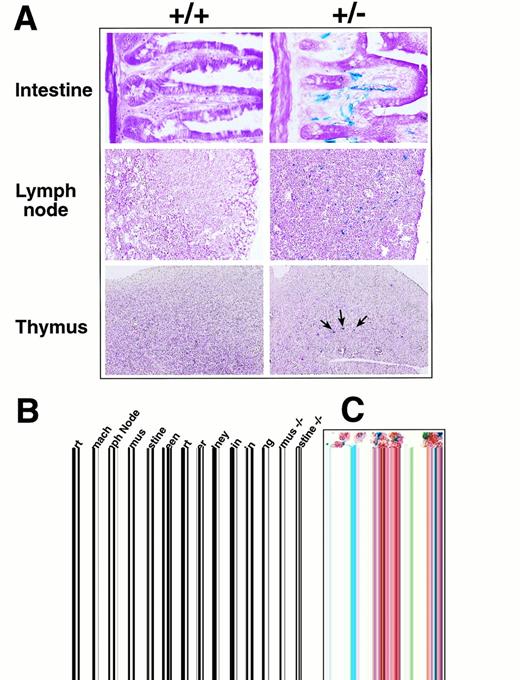
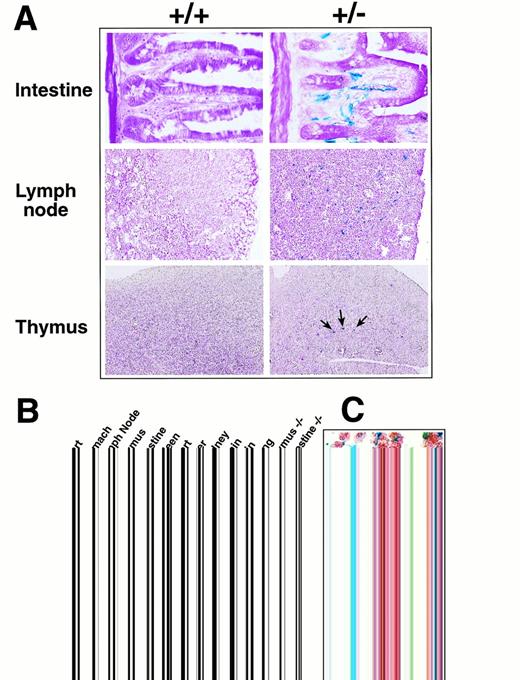
Because the targeted allele contained the E coliβ-galactosidase gene, the activity of the eotaxin promoter could be examined in heterozygous and knockout mice by X-gal staining to detect the β-galactosidase activity. Sections of different tissues from wild-type and heterozygous mice were stained with X-gal as described in Materials and Methods. Tissues such as lymph nodes, thymus, intestine, and stomach were found to contain cells with high β-galactosidase activity as detected by the X-gal blue staining (Fig 2A). In the small intestine, the staining localized to the vasculature in the lamina propria surrounding the intestinal crypts. The β-galactosidase staining in lymph nodes localized to vessels and was most prominent in parafollicular areas in the cortex with less prominent staining of medullary sinuses. In thymus, the β-galactosidase staining was localized only in the medullary region and observed in scattered large (20 to 25 μm) cells with a centrally located nucleus and angular cytoplasmic profiles. The histomorphology of the cells in the thymic medulla was most consistent with dendritic cells. Most of the blue staining in lymph nodes and small intestine localized to cells with the morphology of endothelial cells. Furthermore, the β-galactosidase staining in lymph nodes and small intestine colocalized with immunohistochemical staining for CD34, a marker for endothelial cells (Fig 2C). Several markers specific for T, B, and monocyte/macrophage cells showed no colocalization with β-galactosidase staining. These results suggest that, in lymph nodes and small intestine, eotaxin is expressed by endothelial cells. Other tissues had either very weak or no detectable X-gal staining (data not shown). Wild-type tissues showed no background in staining, as shown in Fig 2A, with the exception of the stomach, which had a slight background staining (not shown). Eotaxin-deficient mice with two copies of the β-galactosidase gene have the same pattern of X-gal staining in different tissues (data not shown).
Expression of β-galactosidase and eotaxin in different tissues in eotaxin+/− mice. (A) Expression of β-galactosidase detected by X-gal staining. Frozen tissue sections from wild-type (+/+) and heterozygous (+/−) mice were stained with X-gal overnight and counter-stained with eosin. Shown are the tissues with strong β-galactosidase staining. (B) Expression of eotaxin and fic in different tissues detected by RT-PCR. Total RNA was isolated from eotaxin heterozygous and null mice and amplified by RT-PCR with primers specific for mouse eotaxin, fic, or β-actin. (C) Costaining of X-gal with CD34 markers in lymph nodes and small intestine. Frozen sections were stained with X-gal overnight, followed by immunohistochemical staining with anti-CD34 antibody and then counter-stained with eosin as described in Materials and Methods. The blue staining illustrates the β-galactosidase activity and the brown staining is for CD34 marker.
Expression of β-galactosidase and eotaxin in different tissues in eotaxin+/− mice. (A) Expression of β-galactosidase detected by X-gal staining. Frozen tissue sections from wild-type (+/+) and heterozygous (+/−) mice were stained with X-gal overnight and counter-stained with eosin. Shown are the tissues with strong β-galactosidase staining. (B) Expression of eotaxin and fic in different tissues detected by RT-PCR. Total RNA was isolated from eotaxin heterozygous and null mice and amplified by RT-PCR with primers specific for mouse eotaxin, fic, or β-actin. (C) Costaining of X-gal with CD34 markers in lymph nodes and small intestine. Frozen sections were stained with X-gal overnight, followed by immunohistochemical staining with anti-CD34 antibody and then counter-stained with eosin as described in Materials and Methods. The blue staining illustrates the β-galactosidase activity and the brown staining is for CD34 marker.
It has been shown that eotaxin mRNA is abundant in mouse tissues such as skin and skeletal muscle as well as in thymus, lymph node, and intestine.12 However, different reports showed different mRNA expression results in some mouse tissues such as skin and intestine.12,13 In humans, eotaxin mRNA is abundant in the heart.14 We did not detect β-galactosidase expression by X-gal staining in the skin, skeletal muscle, or heart, where abundant eotaxin mRNA was previously reported. To examine whether this may be due to differences in mouse genetic background, we performed RT-PCR to check the eotaxinexpression in our animals. Figure 2B shows that the expression ofeotaxin detected by RT-PCR in the thymus, lymph node, and intestine of heterozygous mice is consistent with the β-galactosidase expression detected by X-gal staining. We also detected abundanteotaxin expression in the heart (results from 2 different mice are shown), but in the heart tissue of eotaxin heterozygous and null mice, there was no detectable β-galactosidase expression (data not shown). Because the other chemokine gene fic(fibroblast-induced cytokine), the mouse homolog of MCP-3, is adjacent to eotaxin (unpublished data), we examined whether the replacement of the eotaxin gene with the β-galactosidase and neomycin genes could affect the expression offic. As shown in Fig 2B, the expression of fic in thymus and intestine is not affected in the eotaxin null mice.
Eosinophil recruitment into lung tissue after ovalbumin challenge.
Eotaxin was originally purified from BAL fluid of guinea pig sensitized and challenged with aerosol ovalbumin.11 In a similar mouse model, eotaxin mRNA in lung was induced rapidly and eosinophils infiltrated the lung after ovalbumin aerosol challenge.13,34 Prechallenge treatment of mice with anti-CD3 MoAb inhibited the induced eotaxin mRNA expression and significantly reduced eosinophil infiltration in the lung.34 These results implicate that eotaxin may be responsible for the recruitment of eosinophils into the lung in this antigen-induced lung inflammation model. A recent report using eotaxin-deficient mice generated by an approach different from ours indicated that, at 18 hours after ovalbumin challenge, the number of eosinophils in the BAL was reduced by 70% in theeotaxin−/− mice compared with wild-type mice.38 However, at a later time (48 hours) after ovalbumin challenge, there was no difference in eosinophil numbers between wild-type and eotaxin null mice. This result suggests that eotaxin is important in the early recruitment of eosinophils in this antigen-induced lung eosinophilia model.
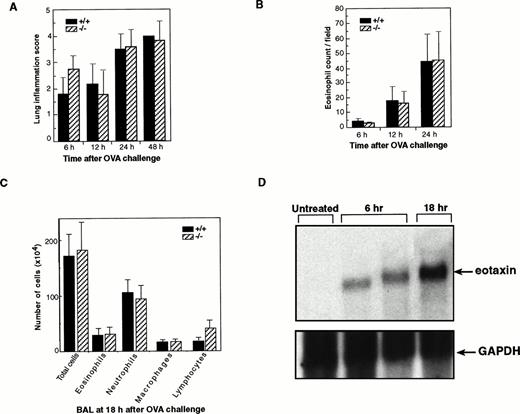
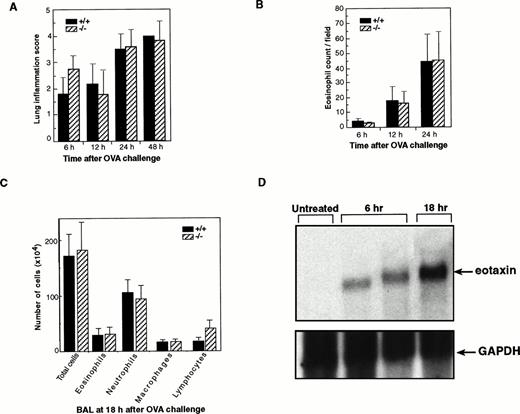
To further determine the role of eotaxin in the early recruitment of eosinophils in this model, we examined our eotaxin null mice for eosinophil infiltration in lung tissues at different time points after the challenge with ovalbumin aerosol. In this inflammation model,eotaxin mRNA in lung was induced rapidly after ovalbumin aerosol challenge (peaking at 6 hours) and a significant number of eosinophils were detected in lung tissues at 6 hours postchallenge.34 Therefore, we examined lung tissues starting at 6 hours after ovalbumin challenge. Lung tissues from wild-type and eotaxin knockout mice were analyzed at 6, 12, 24, and 48 hours after ovalbumin aerosol challenge. Lung inflammation and eosinophil numbers were examined without knowledge of the genotype of each mouse. Inflammation was characterized by aggregates of granular leukocytes (neutrophils and eosinophils) in the peribronchial adventitia with lesser number of macrophages and lymphocytes. Inflammation severity scores were highest in the 24- and 48-hour groups as compared with the 6- and 12-hour groups (Fig 3A). There was no significant difference in the severity of inflammation at different time points between wild-type and knockout mice. As controls, sham-immunized mice show no or minor inflammation at all time points (data not shown). The number of eosinophils in the lung tissues of wild-type and eotaxin null mice was determined microscopically. The numbers of infiltrated eosinophils at different time points, even at the early time point of 6 hours after ovalbumin challenge, between wild-type and knockout mice were similar (Fig 3B). As a direct comparison to the result of Rothenberg et al,38 lung lavage was performed and the number of different cells in the BAL fluid was assessed at 18 hours after the ovalbumin aerosol challenge. As shown in Fig 3C, no significant differences in the number of total cells, neutrophils, macrophages, and lymphocytes as well as eosinophils were detectable between wild-type and eotaxin null mice. The eosinophil counts were 2.9 ± 1.1 × 105 and 3.1 ± 1.2 × 105 for wild-type and eotaxin null mice, respectively. We noticed a slight but not significant increase (about 2-fold) in the number of lymphocytes in eotaxin null mice (Fig 3C). A representative result of the staining of the cells from the BAL is shown in Fig 4. The majority of the infiltrated cells in the lung tissues at 6 and 12 hours and in the BAL fluid at 18 hours after challenge were neutrophils (Figs 3C and 4). In a similar model in guinea pigs, using an assay to measure eosinophil peroxidase (EPO), Humbles et al39 observed that eosinophils appeared in BAL mainly in the late phase (24 hours after OVA challenge), whereas, in lung tissues, eosinophil accumulation elevated at 6 hours and remained high up to 24 hours. We also estimated the number of eosinophils in the BAL fluid using an assay that specifically detects mouse eosinophil peroxidase40 and found no difference in the level of eosinophil peroxidase between wild-type and eotaxin null mice (data not shown). Furthermore, we examined eosinophil degranulation and found no difference between wild-type and eotaxin null mice (data not shown). To determine that eotaxin is expressed in wild-type mice after ovalbumin challenge, we performed Northern blot analysis of total lung RNA from wild-type mice at 6 and 18 hours after ovalbumin challenge. As shown in Fig 3D, the eotaxin mRNA was induced in agreement with previous reports using the same model.34 38
Normal infiltration of eosinophils in lung tissues ofeotaxin−/− mice challenged with ovalbumin. (A) Inflammation scores of lung tissues of wild-type (+/+) and eotaxin null (−/−) mice at different time points after ovalbumin challenge. Inflammation severity was scored to account for different infiltrated cell types with 0 as the least and 4 as marked inflammation. (B) Quantification of eosinophils in lung tissues of wild-type (+/+) and eotaxin null (−/−) mice at different time points after aerosol challenge. Data represent the mean ± SEM of eosinophils from 6 to 8 high-power fields for each mouse; n = 7 for both +/+ and −/− at 6 hours; n = 4 for both +/+ and −/− at 12 hours; n = 6 for +/+ and n = 8 for −/− at 24 hours. (C) Quantification of different cells recovered from the BAL fluid of wild-type (+/+) and eotaxin null (−/−) mice at 18 hours after OVA aerosol challenge. Data represent the mean ± SEM of cells; n = 5 for wild-type and n = 6 for eotaxin null mice. (D) Northern blot analysis of total lung RNA from untreated and OVA-challenged wild-type mice. The blot was probed with 32P-labeled mouse eotaxin cDNA (upper panel), stripped, and tested with a probe specific for mouse GAPDH (bottom).
Normal infiltration of eosinophils in lung tissues ofeotaxin−/− mice challenged with ovalbumin. (A) Inflammation scores of lung tissues of wild-type (+/+) and eotaxin null (−/−) mice at different time points after ovalbumin challenge. Inflammation severity was scored to account for different infiltrated cell types with 0 as the least and 4 as marked inflammation. (B) Quantification of eosinophils in lung tissues of wild-type (+/+) and eotaxin null (−/−) mice at different time points after aerosol challenge. Data represent the mean ± SEM of eosinophils from 6 to 8 high-power fields for each mouse; n = 7 for both +/+ and −/− at 6 hours; n = 4 for both +/+ and −/− at 12 hours; n = 6 for +/+ and n = 8 for −/− at 24 hours. (C) Quantification of different cells recovered from the BAL fluid of wild-type (+/+) and eotaxin null (−/−) mice at 18 hours after OVA aerosol challenge. Data represent the mean ± SEM of cells; n = 5 for wild-type and n = 6 for eotaxin null mice. (D) Northern blot analysis of total lung RNA from untreated and OVA-challenged wild-type mice. The blot was probed with 32P-labeled mouse eotaxin cDNA (upper panel), stripped, and tested with a probe specific for mouse GAPDH (bottom).
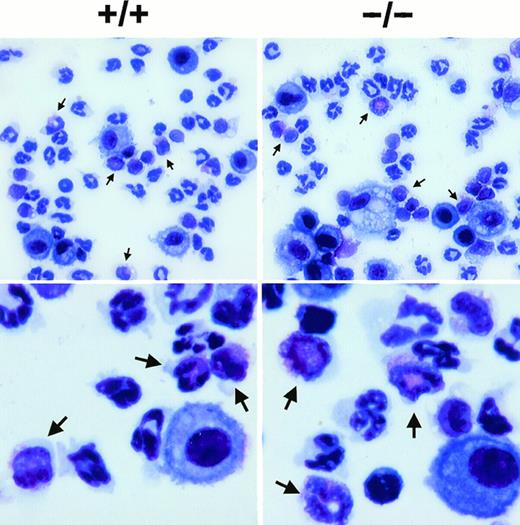
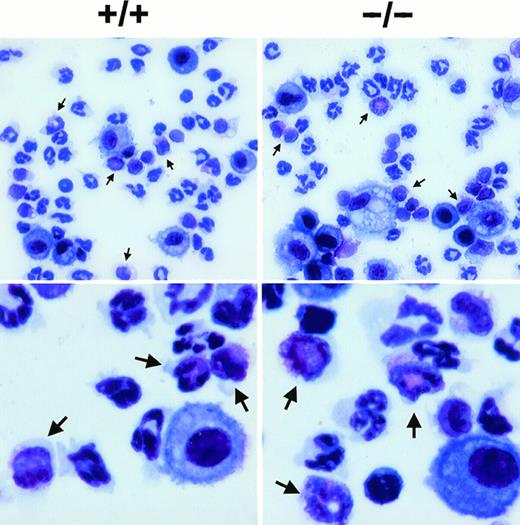
Staining of eosinophils from the BAL fluid of wild-type (+/+) and eotaxin null (−/−) mice. Lung lavage was performed at 18 hours after OVA aerosol challenge and the cells were stained by Diff-Quick staining after cytospin. Representative pictures are shown for each genotype in lower (upper panel) and higher (lower panel) magnification. Arrowheads indicate eosinophils.
Staining of eosinophils from the BAL fluid of wild-type (+/+) and eotaxin null (−/−) mice. Lung lavage was performed at 18 hours after OVA aerosol challenge and the cells were stained by Diff-Quick staining after cytospin. Representative pictures are shown for each genotype in lower (upper panel) and higher (lower panel) magnification. Arrowheads indicate eosinophils.
The expression of β-galactosidase in eotaxin null mice after ovalbumin challenge.
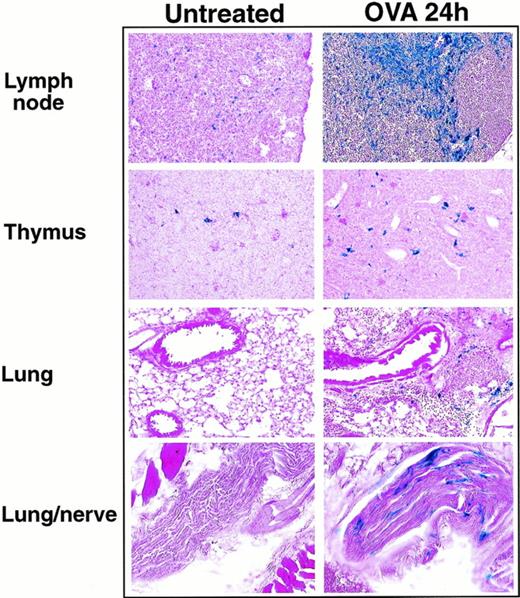
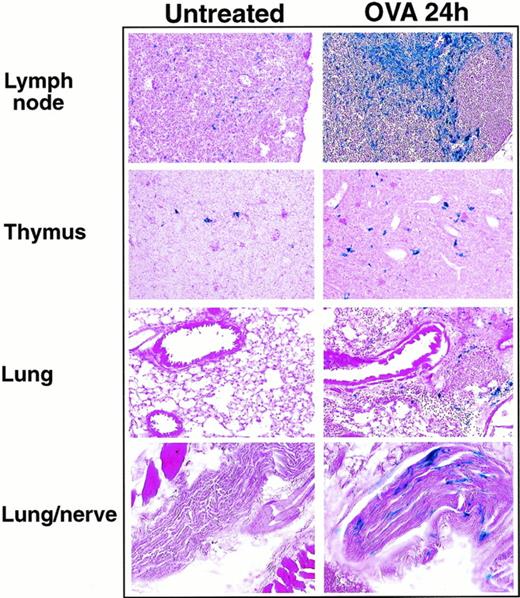
With the replacement of eotaxin by the β-galactosidase gene in our knockout mice, eotaxin expression could be examined indirectly by detecting the β-galactosidase activity after ovalbumin aerosol challenge of eotaxin null mice. Previous reports suggested that resident epithelial cells and alveolar macrophages appear to be the primary source of eotaxin in response to ovalbumin challenge.13 35 The fact that β-galactosidase is not secreted after expression facilitated the determination of the cell types responsible for eotaxin expression during inflammation. We examined the expression of β-galactosidase in different tissues from eotaxin null mice at different time points after ovalbumin challenge. Table 1 shows the level of β-galactosidase activity detected by X-gal staining in different tissue sections. At 6 and 12 hours after the ovalbumin challenge, no β-galactosidase staining was detected in the lung tissues. The β-galactosidase staining began to appear in the lung tissues at 18 hours after challenge. The staining was more evident in the lung tissues of eotaxin null mice at 24 and 48 hours after aerosol challenge (Fig 5). The positive staining localized to the peribronchial and perivascular connective tissue and was present in cells with histomorphologic features of macrophages or stromal cells (fibroblasts/fibrocytes). In addition, peripheral nerves surrounding the lungs of eotaxin null mice at 24 and 48 hours after aerosol challenge had positive staining in cells with histomorphologic features of Schwann cells. In thymus and lymph nodes, there was no significant difference in the β-galactosidase staining in eotaxin null mice at 6, 12, and 18 hours after ovalbumin challenge compared with untreated mice (Table 1). There was a slight increase in β-galactosidase staining in thymus and a strong increase in lymph nodes in mice 24 and 48 hours after ovalbumin challenge (Table 1 and Fig 5). In addition to the cells with histomorphological features of dendritic cells, positive staining was also observed in vessels of the thymic medulla. Besides endothelial cells, macrophages in the subcapsular sinuses of lymph nodes also stained positively for β-galactosidase. As control, the β-galactosidase staining in the intestine (positive) and spleen (negative) in eotaxin null mice did not change after ovalbumin challenge (Table 1).
The expression of βgalactosidase in lung, thymus, and lymph nodes 24 hours after ovalbumin aerosol challenge. Frozen sections of lung, thymus, and lymph nodes from untreated or ovalbumin aerosol-challenged (OVA 24 hours) mice were stained with X-gal overnight followed by counter-stain with eosin. Shown are results from a representative experiment.
The expression of βgalactosidase in lung, thymus, and lymph nodes 24 hours after ovalbumin aerosol challenge. Frozen sections of lung, thymus, and lymph nodes from untreated or ovalbumin aerosol-challenged (OVA 24 hours) mice were stained with X-gal overnight followed by counter-stain with eosin. Shown are results from a representative experiment.
Eosinophil recruitment in other inflammation models.
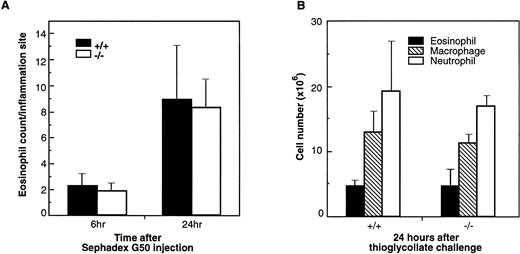
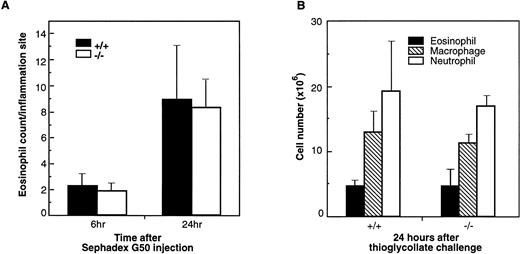
Lung eosinophilia induced by Sephadex beads has been developed in rats and guinea pigs.41,42 Eosinophils corresponded to more than 40% of the cell population in BAL fluid in Sephadex bead-treated animals.42 In guinea pigs treated with Sephadex beads, eotaxin protein was shown to be induced in the BAL fluid and paralleled the eosinophil infiltration in lung tissue and the infiltration of eosinophils was significantly inhibited by a polyclonal antieotaxin antibody.43 Because this type of experiment has not been reported in mice, we examined the role of eotaxin in this inflammation model using eotaxin−/− mice. Lung tissues from wild-type and mutant mice were collected at 6 and 24 hours after Sephadex G50 injection. At both time points, there was mild to moderate interstitial inflammation in lung tissue with intralesional foreign material. The inflammation was more severe at the 24-hour time point as compared with the 6-hour time point. However, the inflammation was composed predominantly of neutrophils, with lesser numbers of macrophages, lymphocytes, and eosinophils (data not shown). In most areas, eosinophils constituted less than 5% to 10% of the inflammatory cell population, and there was no difference in the severity or composition of the inflammation in eotaxin null mice as compared with wild-type controls. Figure 6A shows the result of the eosinophil count in lung tissues of wild-type and eotaxin null mice 6 and 24 hours after Sephadex bead injection. As shown, there are very few eosinophils and no significant differences between wild-type and knockout mice.
Infiltration of eosinophils and other leukocytes after Sephadex beads (A) and thioglycollate injections (B). (A) Lung tissues of wild-type (+/+) and eotaxin null (−/−) mice were examined for eosinophils at 6 and 24 hours after mice received Sephadex G50 (superfine) beads by intravenous injection. Data represent the mean ± SEM from three fields for each mouse and from 5 mice of each group. (B) Cells were collected by peritoneal lavage 24 hours after intraperitoneal injection of thioglycollate into mice and stained by Diff-Quick staining. Data represent the mean ± SEM from at least six high-power fields from 6 mice of each phenotype.
Infiltration of eosinophils and other leukocytes after Sephadex beads (A) and thioglycollate injections (B). (A) Lung tissues of wild-type (+/+) and eotaxin null (−/−) mice were examined for eosinophils at 6 and 24 hours after mice received Sephadex G50 (superfine) beads by intravenous injection. Data represent the mean ± SEM from three fields for each mouse and from 5 mice of each group. (B) Cells were collected by peritoneal lavage 24 hours after intraperitoneal injection of thioglycollate into mice and stained by Diff-Quick staining. Data represent the mean ± SEM from at least six high-power fields from 6 mice of each phenotype.
In a nonspecific inflammation model induced in peritoneal cavity by thioglycollate, eosinophils comprised 15% to 20% of the recruited cells. To examine the role of eotaxin in this model, wild-type and eotaxin-deficient mice were injected with thioglycollate. At 24 hours after injection, cells recovered from the peritoneal lavage were examined for different cell populations. At this time point, neutrophils are the major population of infiltrated cells followed by macrophages and eosinophils and a minor population of lymphocytes and other types of cells. As shown in Fig 6B, compared with wild-type, there is no difference in the number of infiltrated neutrophils, macrophages, and eosinophils in eotaxin knockout mice.
DISCUSSION
Some chemokines have been found to be active on eosinophils, among which eotaxin and the recently identified eotaxin-2 are the most selective as well as most effective for eosinophil chemotaxis.17,20 The eosinophil growth factor IL-5 has also been found to selectively promote eosinophil chemotaxis, as well as eosinophil growth, differentiation, and activation.44-46IL-5 has been shown to play an essential role in the mediation of eosinophil infiltration in a mouse asthma model, because, in IL-5–deficient mice, the lung eosinophilia and airway hyperactivity resulting from ovalbumin aerosol challenge were abolished.46 Like IL-5, eotaxin expression was induced in models of allergic inflammation and other conditions characterized by eosinophil accumulation.12,13,47 The role of eotaxin in the recruitment of eosinophils in these inflammation models was complicated by the fact that the expression of other chemokines, such as RANTES, MCP-4, MCP-3/FIC (fibroblast-induced cytokine), MIP-1α, and MCP-5, which are also active on eosinophils, paralleled with the infiltration of eosinophils.26,34,35,48,49 To determine the role of eotaxin in the recruitment of eosinophils during inflammation, and possible other functions of eotaxin, we generated eotaxin-deficient mice. These mice develop normally with no histologic or hematopoietic defect compared with their wild-type littermates. These results are consistent with another line of eotaxin knockout mice generated by Rothenberg et al38 with a different targeting strategy and different genetic background. Mice deficient for CC chemokine MIP-1α and CC chemokine receptor CCR1 also had no abnormalities in development, histology, and hematopoiesis.50,51 In contrast, mice deficient for the CXC chemokine SDF-1 showed a perinatal lethal phenotype,52 and mice deficient for the murine IL-8 receptor homolog CXCR2 showed histologic defects in spleen and lymph nodes and disordered hematopoiesis.53
Although with no apparent phenotype, mice deficient for MIP-1α or CCR1 had impaired inflammatory responses to microbial or antigen challenge.50,51 The eotaxin knockout mice generated by Rothenberg et al38 showed 50% to 70% reduction in the recruitment of eosinophils at early time points after challenge with parasite-derived antigen (1 day) or ovalbumin aerosol (18 hours). However, at a later time point after parasite-derived antigen (8 days) or ovalbumin aerosol (48 hours) challenge, there is no difference in eosinophil infiltration between eotaxin null mice and wild-type mice. These results led them to suggest that eotaxin is important in the early recruitment of eosinophils in these inflammatory processes. However, in the ovalbumin-induced lung inflammation model, our eotaxin knockout mice did not show any significant differences in the number of infiltrated eosinophils in lung tissues compared with wild-type mice, even at the time points of 6 and 12 hours after ovalbumin aerosol challenge (Fig 3). As a direct comparison, there is no difference in the number of eosinophils in the BAL fluid at 18 hours after ovalbumin aerosol challenge in our eotaxin null mice compared with wild-type (Fig3C). Eotaxin mRNA in lung was shown to be rapidly induced after ovalbumin challenge with levels detectable by 3 hours and peaking by 6 hours.34 The difference seen between the knockout mice might be due to the different genetic background of the mouse strains. There have been reports that the phenotypes of some knockout mice are dependent on genetic background.54,55 Indeed, our mice showed some differences from those of Rothenberg et al12,38 in the expression of eotaxin in tissues like skin, where eotaxin mRNA was not detectable in our mice but abundant in those of Rothenberg et al12,38 (Fig 2B). To make sure that eotaxin is also induced after ovalbumin challenge in our strain, we checked eotaxin mRNA expression in wild-type mice and found it was indeed induced in the lung at 6 and 18 hours after ovalbumin challenge (Fig 3D). On the other hand, Rothenberg et al38 also observed a significant reduction (∼70%) of total eosinophil count in the peripheral blood in their knockout mice compared with wild-type. We did not observe significant differences in the blood eosinophil count between our knockout and wild-type mice (wild-type and knockout mice had 44 ± 16 [mean ± SEM, n = 10] and 36 ± 7 [n = 12] eosinophils/μL of blood, respectively; P = .61). Thus, the reduced eosinophil infiltration in eotaxin-deficient mice at early time points after ovalbumin challenge observed by Rothenberg et al38 could be accounted for by the reduced level of blood eosinophils in their knockout mice. In response to ovalbumin challenge, the eosinophil pool in the bone marrow is mobilized by the signal provided by IL-5.44-46,56Therefore, at later time points after ovalbumin challenge, there is no difference in eosinophil infiltration between eotaxin null and wild-type mice. The absence of eotaxin may be compensated by other chemokines such as RANTES, MIP-1α, and MCP-3/FIC. A chemokine named eotaxin-2 with very similar function to eotaxin has recently been identified in humans.20 It will be interesting to determine whether there is a mouse homolog of eotaxin-2 and, if there is one, whether it is involved in this type of allergic inflammation. Contrary to the general notion, there have been recent reports that eosinophilic airway inflammation develops normally in mice deficient of IgE, B cells, and mast cells.57-60 These results, together with ours, are consistent with the existence of parallel pathways of eosinophil recruitment to the airways after antigen challenge.
Our experiments with thioglycollate- and Sephadex G50-induced inflammation in which the infiltrated cells are predominantly neutrophils indicated that eotaxin null mice had no defect in the recruitment of neutrophils, macrophages, lymphocytes, and eosinophils (Fig 6). Rothenberg et al38 also examined the thioglycollate model at a later time point (48 hours), when macrophages were the predominant infiltrated cells, and found no defect in their knockout mice. Thus, it appears that eotaxin does not play a role in the recruitment of eosinophils in the inflammatory response induced by thioglycollate. In rats and guinea pigs, Sephadex bead injection induced inflammatory response with predominantly eosinophil infiltration.41,42 In guinea pigs, the expression of eotaxin correlates with the infiltration of eosinophils in lung after Sephadex bead injection and the infiltration of eosinophils was inhibited by antieotaxin antibodies.43 However, in both wild-type and eotaxin−/− mice, the inflammatory response induced by Sephadex bead injection is characterized predominantly by neutrophils with less macrophages, lymphocytes, and eosinophils (Fig 6A). It seems that mice respond to Sephadex beads differently from rats and guinea pigs, probably due to the different number of circulating eosinophils in these animals.
The replacement of the eotaxin gene with E coliβ-galactosidase gene provided a marker for the expression of eotaxin in different tissues under normal conditions or after challenge with antigen. The E coli lacZ reporter gene has been used to target into mouse genome to replace other genes and, in several cases, faithfully reproduced the expression pattern of the targeted genes.61-64 However, the shortcoming of this technique is that the expression of the β-galactosidase may not always faithfully represent the expression of the targeted gene protein product. The expression of many genes is regulated by posttranscription (ie, RNA half-life and/or translation control), which could be altered by gene replacement. In addition, the lacZ mRNA may be differentially regulated and translated in different tissues. Our X-gal staining results are consistent with the expression pattern of eotaxin mRNA in different tissues, with the exception of the heart (Fig 2). We detected high levels of eotaxin expression in the heart by RT-PCR. Another group also reported a high level of eotaxin expression in the human heart but not in the blood cells detected by Northern blot.14 However, in the heart tissue sections of eotaxin null or heterozygous mice, we did not observe any detectable β-galactosidase expression (data not shown). The mRNA level in the heart is comparable to that in the thymus, lymph nodes, and intestine, where the β-galactosidase activity was easily detected in eotaxin null and heterozygous mice (Fig 2B). Thus, the lack of X-gal staining in heart in eotaxin null and heterozygous mice cannot be due to the sensitivity of the assay but rather may be due to the different regulation of the eotaxin mRNA in wild-type mice and the lacZ mRNA in eotaxin+/− and eotaxin−/− mice.
After ovalbumin challenge, β-galactosidase staining was detectable in the lung of eotaxin null and heterozygous mice at 18, 24, and 48 hours but not at 6 or 12 hours after aerosol challenge (Table 1 and Fig 5). This pattern of staining does not parallel the induced eotaxinmRNA expression in the lung, which peaks at 6 hours.34 The protein activity of eotaxin detected in guinea pig BAL fluid also peaked around 3 to 6 hours after ovalbumin challenge.11Because we could not detect β-galactosidase activity in the lung tissue of eotaxin null mice at 6 and 12 hours after ovalbumin challenge, the induced eotaxin expression at early time points may be mainly in the BAL fluid and only at a later time in lung tissue. Our results suggest that eotaxin might also play a role in the later phase of the inflammatory response after ovalbumin challenge. The β-galactosidase staining in lung tissue suggested that, in the late phase of the inflammation, stromal cells (fibroblasts/fibrocytes) and infiltrated macrophages could be the sources for eotaxin expression. Previous reports with in situ hybridization and eotaxin antibody staining indicated that resident lung epithelial cells and alveolar macrophages were the principal cells producing eotaxin.13,35 However, we did not detect any β-galactosidase staining in lung epithelial cells after ovalbumin challenge. The difference may be due to variations in the protocols used to challenge mice. In our experiments, we challenged mice with ovalbumin aerosol only once, whereas mice were repeatedly challenged with ovalbumin aerosol several times in the above-mentioned reports.13 35 Therefore, the level of the induced expression may be quite different.
At the late phase of the ovalbumin-induced inflammation, we also found that there was increased β-galactosidase staining in thymus and lymph nodes (Fig 5). This finding suggested that eotaxin expression was increased in these tissues. However, we did not observe significant infiltration of eosinophils in these tissues (data not shown). In a recent report, a dense infiltration of eosinophils was observed in the tracheobronchial lymph nodes, especially pronounced in the B-cell–deficient mice challenged with ovalbumin aerosol.58It was suggested that these infiltrated eosinophils might be important for T-cell activation in response to antigen challenge. The investigation of the strong induced expression of eotaxin in thymus and lymph nodes may provide clues to other physiological functions of eotaxin. The presence of β-galactosidase staining of Schwann cells in nerves surrounding the lungs after ovalbumin challenge was an intriguing finding. This finding suggested that eotaxin might be expressed by Schwann cells during the inflammation processes in lung. The primary function of Schwann cells is to secrete and maintain the myelin sheath around axons in peripheral nerves. However, Schwann cells are capable of expressing MHC-II molecules and produce various cytokines, including IL-10, IL-12, and nitrite, under certain stimulatory conditions.65-67 Stimulation of the vagus nerve by factors released by mast cells leads to bronchoconstriction and is an important factor in the pathophysiology of asthma. Production of eotaxin by Schwann cells may provide another mechanism whereby the nervous system modifies the immune response in asthma.
ACKNOWLEDGMENT
The authors thank S. Lira, M. Swerdel, and A. Lee in the Transgenic Unit and all the staff in Veterinary Sciences at BMS for generating and maintaining the mice; A. Lewin, D. Barton, M. French, C. Rizzo, C. Ryan and J. Stevens for excellent technical assistance; C. Raventos-Suarez and K. Class for flow cytometry; and N. Thompson and T. Nelson for assistance with DNA sequencing. We thank Drs Y. Zhou, V. Iotsova, and D. Dambach for helpful suggestions.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Rodrigo Bravo, PhD, Department of Oncology, Bristol-Myers Squibb Pharmaceutical Research Institute, PO Box 4000, Princeton, NJ 08543.