Abstract
Recently, a novel phosphatase designated PTEN/MMAC1/TEP1 and located on chromosome 10q23.3 has been implicated as a new tumor suppressor gene in human cancer. Allelic loss and mutation of this gene has been reported in epithelial derived tumors, including breast cancer and prostate cancer, and in glioblastoma multiforme. The present study was designed to evaluate the potential involvement of PTEN in the pathogenesis of lymphoid neoplasms. We analyzed 27 hematopoietic cell lines (representing a variety of lymphoid lineages), 65 primary lymphoid tumors (including 24 lymphoblastic leukemia/lymphoma [LBL], 30 large B-cell lymphoma [LBCL], 7 Burkitt’s lymphoma [BL], and 4 anaplastic large cell lymphoma [ALCL]), and 25 nonmalignant lymph node controls. Gene deletion and gross rearrangement were evaluated using Southern blot analysis, and mutations were studied by polymerase chain reaction (PCR)-single-strand conformation polymorphism (SSCP) (PCR-SSCP) and sequencing. Six of 27 cell lines (22.2%) and 3 of 65 primary lymphomas (4.6%) contained alterations of this gene. A large homozygous deletion spanning exons 2 through 5 was detected in one LBL cell line, and two insertions potentially resulting in premature termination, were detected in a second LBL cell line. Nonconservative nucleotide variations were found in two other cell lines (one LBCL and one BL) and in one primary case of LBCL. In addition, two other cell lines (one BL and one myeloma) and two primary lymphomas, both LBCL, contained small deletions within intron 7. These deletions mapped to a poly-T–rich tract just 5′ to the intron 7/exon 8 spice site. Their significance is unclear, as they may represent polymorphisms. Overall, our results suggest that abnormalities of the PTEN gene can contribute to pathogenesis in a small percentage of malignant lymphomas.
This is a US government work. There are no restrictions on its use.
RECENTLY, A NEW CANDIDATE tumor suppressor gene designated PTEN/MMAC1/TEP1 and located on chromosome 10q23.3 was reported by three groups independently.1-3 PTEN encodes a novel 403–amino acid, dual-specificity phosphatase with homology both to the protein tyrosine phosphatase (PTP) family and to two cytoskeletal proteins, tensin and auxilin.1-3 The protein contains the canonic PTP motif, and recombinant PTEN has been shown to be capable of catalyzing the hydrolysis of phosphoseryl, phosphothreonyl, and phosphotyrosyl residues.4
PTEN is reportedly deleted and/or mutated in a significant fraction of breast cancers,5 prostate cancers,6endometrial carcinomas,7-9 glioblastomas,10,11and sporadic melanomas,12 suggesting that it functions as a tumor suppressor gene in these cancers. In addition, it has also been implicated in Cowden disease,13 an inherited cancer syndrome characterized by benign adenomas and malignant neoplasms of the breast, thyroid, and skin, and in Bannayan-Zonana syndrome,14 an autosomal dominant disorder characterized by microcephaly, vascular malformations, and benign neoplasms such as lipomas and intestinal hamartomatous polyps. Studies in other types of cancer such as thyroid15 and pancreatic16carcinoma have shown an absence or a low frequency of mutation or deletion of the PTEN gene. Although there are many examples of protein tyrosine kinases that function as oncogenes in tumorigenesis, this is the first protein tyrosine phosphatase implicated as a tumor suppressor gene.
To date, there is little information regarding the potential involvement of PTEN in non-Hodgkin’s lymphoma (NHL). At the chromosomal level, abnormalities of chromosome 10q have been reported in a modest percentage of NHLs. Juneja et al17 noted recurrent breaks at 10q22 and 10q26 and Offit et al18 found breaks in the region of 10q22-24 in 5% of NHL. Most recently, abnormalities of 10q23-25 were reported in 10.7% of NHL.19
The cytogenetic findings plus the lack of studies investigating PTEN involvement in NHL encouraged us to examine a series of lymphoid neoplasms for potential involvement of this gene in lymphomagenesis. In this report, we present data on 27 hematopoietic cell lines and 65 primary high-grade NHLs for mutations, deletions, or rearrangements of the PTEN gene.
MATERIALS AND METHODS
Cell line studies.
Twenty-seven human hematopoietic cell lines representing a variety of lymphoma subtypes were selected for this study. These included 6 lymphoblastic leukemia/lymphoma (LBL) cell lines (Molt3, Molt4, HSB, Jurkat, HUT102, and CEM), 4 anaplastic large cell lymphoma (ALCL) cell lines (KARPAS299, JB6, SR786, and KIJK), 6 B-cell lymphoma cell lines (SUDHL4, SUDHL5, SUDHL6, SUDHL7, SUDHL10, and NUDHL1), 6 Burkitt’s lymphoma (BL) cell lines (JD38, CA46, Raji, PA682PB, WMN, and Defaw), 3 myeloma cell lines (KMS5, KMS11, and Jim3), 1 Hodgkin’s lymphoma cell line (L428), and 1 leukemia cell line (YT) with natural killer–like phenotype.
Cases studied.
A total of 65 samples from patients with aggressive lymphoma referred to the National Institutes of Health were selected for this study based on the availability of sufficient tissue for molecular analysis. In addition, 25 samples from patients with nonmalignant lymphoid hyperplasias were also studied to assist in identifying polymorphisms. The lymphoma samples included 24 LBL, 30 large B-cell lymphoma (LBCL), 7 BL, and 4 ALCL. The rationale behind the selection of these particular lymphoma subtypes was (1) to analyze neoplasms of related subtype to the cell lines with PTEN alterations and (2) to focus on aggressive lymphoma variants, since PTEN/MMAC1 was initially described in advanced cancers. The majority of samples used in this study are from patients enrolled in clinical studies who provided informed consent according to the guidelines of the Institutional Review Board of the National Institutes of Health.
DNA extraction and Southern blot analysis.
High-molecular-weight DNA was extracted from frozen tissue samples or cell suspensions using a standard phenol/chloroform extraction procedure as previously described.20 Placental DNA (Oncor, Gaithersburg, MD) was used as control DNA. After restriction enzyme digestion with EcoRI or HindIII, the DNA was size-fractionated by agarose gel electrophoresis and transferred onto nylon membranes (GENE Screen Plus; New England Nuclear Research Products, Boston, MA). The filters were sequentially hybridized with random primed32P-labeled probes for 20 to 24 hours at 42°C and washed under stringent conditions according to the recommendations of the supplier. Autoradiographs were developed after 1 to 7 days.
Probes.
PTEN gene deletions (or rearrangements) were assessed with a genomic 2.8-kb EcoRI fragment of PTEN, JL25. A probe to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used to assess equal loading of DNA in the Southern blot analysis. The JL25 probe was a gift from Dr Ramon Parsons (Columbia University Cancer Center, New York, NY). The GAPDH probe was purchased from the American Tissue Culture Collection (Rockville, MD).
Single-strand conformation polymorphism analysis.
Oligonucleotide primers were synthesized by the solid-phase triester method. Nine primer sets designed to amplify exons 1 to 9 and corresponding to those previously described by Liaw et al13were used. A second alternative primer set to exon 3 was used in some cases.12 The sequence of the primers listed from the most 5′ set to the most 3′ set is as follows: exon 1: X1R246, 5′-ACTACGGACATTTTCGCATC-3′; CONBF1, 5′-GAGGATTATTCGTCTTCTCCC-3′; exon 2: X2F102, 5′-GTTTGATTGCTGCATATTTCAG-3′; X2R88, 5′-TCTAAATGAAAACACAACATGAA-3′; exon 3: X3F135, 5′-ATTTCAAATGTTAGCTCATTTTG-3′; X3R115, 5′-TTTAGAAGATATTTCAAGCATAC-3′; exon 3 alternate: 3alt-F, 5′-TGTTAATGGTGGCTTTTTG-3′; 3alt-R, 5′-GCAAGCATACAAATAAGAAAAC-3′; exon 4: X4F51, 5′-CATTATAAAGATTCAGGCAATG-3′; X4R108, 5′-GACAGTAAGATACAGTCTATC-3′; exon 5: X5F1, 5′-ACCTGTTAAGTTTGTATGCAAC-3′; X5R1, 5′-TCCAGGAAGAGGAAAGGAAA-3′; exon 6: X6F575, 5′-CATAGCAATTTAGTGAAATAACT-3′; X6R169, 5′-GATATGGTTAAGAAAACTGTTC-3′; exon 7: X7F50, 5′-TGACAGTTTGACAGTTAAAGG-3′; X7R147, 5′-GGATATTTCTCCCAATGAAAG-3′; exon 8: X8F1, 5′-CTCAGATTGCCTTATAATAGTC-3′; X8R1, 5′-TCATGTTACTGCTACGTAAAC-3′; and exon 9: X9F528, 5′-AAGGCCTCTTAAAAGATCATG-3′; X9N1738R, 5′-TTTTCATGGTGTTTTATCCCTC-3′.
The polymerase chain reaction (PCR) was performed in standard buffer conditions as described previously21 with 100 to 200 ng genomic DNA, 200 nmol/L of each primer, 200 mmol/L dNTPs, 1 μCi [α-32P]dCTP (specific activity, 6,000 Ci/mmol), 10 mmol/L Tris hydrochloride (pH 8.3), 1.5 to 2.5 mmol/L MgCl2, 50 mmol/L KCl, 0.01% gelatin, and 0.5 U Ampli Taq DNA Polymerase (Boehringer Mannheim, Indianapolis, IN), plus 6% dimethyl sulfoxide for the primer set of exon 5 and exon 1, in a final volume of 10 μL. After an initial denaturation at 94°C for 3 minutes, 30 cycles of denaturation (94°C for 1 minute), annealing (55°C for the primer sets of exons 3, 5, and 7; 56°C for the additional primer set of exon 3; and 58°C for the primer sets of exons 1, 2, 4, 6, 8, and 9 [for 1 minute]), and extension (72°C for 1 minute) were performed on DNA thermal cycler 480 (Perkin-Elmer Applied Biosystems, Foster City, CA). The final extension was performed for 10 minutes.
Samples of exons 1 to 4, 6, 7, and 9 were loaded onto an MDE gel (FMC Corp, Rockland, ME) containing 10% glycerol, and samples of exons 5 and 8 were analyzed using a 6% acrylamide gel containing 10% glycerol. To obtain optimal separation of the single-stranded conformers, the ratio of methylene-bis-acrylamide to acrylamide was 1:19. Gel electrophoresis was performed at 8W for 12 to 16 hours. Autoradiography was performed using an intensifying screen and an exposure time of 4 to 36 hours at −70°C.
Sequencing strategy.
After detection of a variant allele, DNA was extracted from the excised SSCP gel fragment by overnight incubation in 60 μL distilled water at 4°C or 65°C for 10 minutes and subsequently reamplified by PCR. The PCR products were sequenced directly using a cycle sequencing method (AmpliCycle Sequencing Kit; Perkin-Elmer Applied Biosystems) and [α-32P]dCTP. The product of the exon 8 fragment was sequenced using dye-termination cycle sequencing and an automated sequencer (Perkin-Elmer Applied Biosystems 373) following ligation to pCR2.1 (TA cloning kit; Invitrogen, Carlsbad, CA). The primers for sequence analysis were the same primers already listed. One additional primer for sequence analysis of exon 1 and five additional primers to the exon 8 fragment were also synthesized to assist in sequencing: exon 1-1F, 5′-TCTAAGAGAGTGACAGA-3′; exon 8-1, 5′-CATTCTTCATACCAGGACCAG-3′; exon 8-2, 5′-ATAATGACAAGGAATATCTAG-3′; exon 8-3, 5′-CTTGTCATTATCTGCACGCTCT-3′; exon 8-4, 5′-CACATCACATACATACAAGTC-3′; and exon 8-5, 5′-CTGGTCCTGGTATGAAGAATGT-3′.
Following the sequencing reactions, 8 μL of each termination reaction was added to 10 μL stop solution and heated at 95°C for 5 minutes, and 3 μL was loaded onto a sequencing gel consisting of 6% polyacrylamide and 7 mol/L urea in TBE buffer. Gel electrophoresis was performed at 55W for 2 to 3 hours and subjected to autoradiography for 16 hours at −70°C.
RESULTS
Analysis of cell lines.
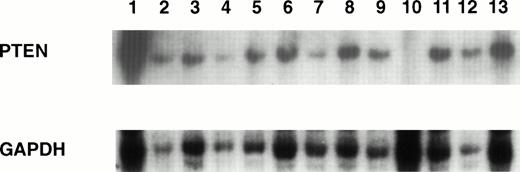
We initially examined 27 hematopoietic cell lines representing a variety of cell lineages for gross deletion or rearrangement of the PTEN gene by Southern blot analysis using probe JL25. Homozygous deletion of the PTEN gene was detected in one LBL cell line (CEM; Fig1). None of the other cell lines showed either a rearrangement or deletion of PTEN. Because the JL25 probe includes only a portion of the PTEN gene, we further mapped the extent of homozygous deletion in CEM using PCR primers to the PTEN exons (the same as used for PCR-SSCP). PCR products were generated from exon primer pairs 1 and 6 to 9 but not from exon primer pairs 2 to 5, indicating that the region of homozygous deletion was limited to exons 2 to 5 (data not shown).
Southern blot analysis of PTEN gene. High-molecular-weight DNA was digested with HindIII, and the blot was hybridized with (top panel) the JL25 probe or (bottom panel) GAPDH to estimate DNA loading. CEM (lane 10) showed homozygous deletion of the PTEN gene. Lane 1, placental DNA; lane 2, SUDHL4; lane 3, SUDHL5; lane 4, SUDHL6; lane 5, SUDHL10; lane 6, NUDHL1; lane 7, KMS5; lane 8, KMS11; lane 9, Jim3; lane 10, CEM; lane 11, HUT102; lane 12, L428; lane 13, YT.
Southern blot analysis of PTEN gene. High-molecular-weight DNA was digested with HindIII, and the blot was hybridized with (top panel) the JL25 probe or (bottom panel) GAPDH to estimate DNA loading. CEM (lane 10) showed homozygous deletion of the PTEN gene. Lane 1, placental DNA; lane 2, SUDHL4; lane 3, SUDHL5; lane 4, SUDHL6; lane 5, SUDHL10; lane 6, NUDHL1; lane 7, KMS5; lane 8, KMS11; lane 9, Jim3; lane 10, CEM; lane 11, HUT102; lane 12, L428; lane 13, YT.
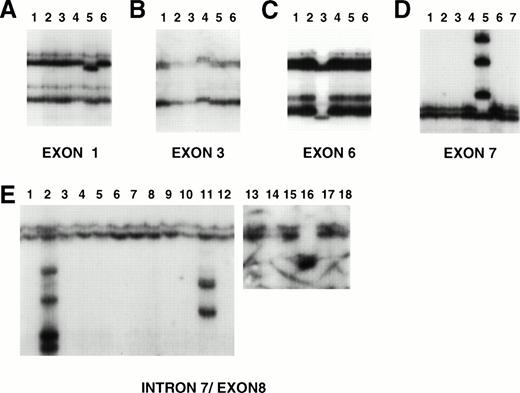
To investigate whether mutations of the PTEN gene were present in the cell lines, we next screened the PTEN coding region (exons 1 through 9) by PCR-SSCP using previously published primer sets as described in Materials and Methods. Five cell lines (Jurkat, SUDHL10, Jim3, Raji, and PA682PB) had variant conformers in PCR-SSCP analysis (Fig2). Sequence analysis showed that small deletions, insertions, and single-nucleotide alterations accounted for these alterations (Table 1). The Jurkat cell line contained two different paired alterations within exon 7 (four variant bands) without “normal” conformers present. Two variant bands corresponded to an allele containing a 9-bp insertion preceded by a 2-bp deletion that disrupted codon 234 (Fig 3). This abnormality resulted in several downstream stop codons. The second pair of variant conformers was the result of a 39-bp insertion after codon 246 that contained a stop codon (TGA) at the new codon 247 (data not shown). The presence of two pairs of variant bands suggests involvement of both alleles or intratumoral heterogeneity. The SUDHL10 cell line showed variant conformers in exons 3 and 5, without normal conformers present. Sequence analysis showed that the exon 3 alteration occurred in codon 68, resulting in the substitution of tyrosine for histidine (TAC to CAC; Tyr to His), and the exon 5 alteration occurred in codon 162, resulting in the substitution of aspartic acid for histidine (GAC to CAC; Asp to His). The alteration in PA682PB occurred in exon 6 and was the result of a G to C substitution in codon 166, resulting in the substitution of valine for leucine (Val to Leu; GTA to CTA; Fig3). Again, the normal conformers were not present. CEM, in which the deletion of the PTEN gene was detected by Southern blot analysis, showed no products for exons 2 through 5 (as previously mentioned) and a normal SSCP pattern for exons 1, 6, 7, 8, and 9. Although Jim3 and Raji displayed variant conformers in the PCR-SSCP of exon 8, no alterations were detected in exon sequences. Instead, Jim3 had a 2-bp deletion in a 13-bp poly-T tract located just proximal to the 5′ exon 8 splice site, and Raji contained a 38-bp deletion involving the proximal two Ts of the same poly-T tract and the preceding 36 bps. In both of the latter cell lines, normal conformers were also present. Altogether, we identified sequence alterations in 2 of 6 LBL cell lines, 1 of 6 LBCL cell lines, 1 of 3 myeloma cell lines, and 2 of 6 BL cell lines (Table2).
Examples of PCR-SSCP analysis of PTEN exons (A, exon 1; B, exon 3; C, exon 6; D, exon 7; E, exon 8). DNA was prepared and subjected to PCR-SSCP analysis. In each analysis, lane 1 contains control DNA (placental). Alterations are present in exon 1, lane 5 (primary LBCL), exon 3, lane 4 (SUDHL10), exon 6, lane 3 (PA682PB), exon 7, lane 5 (Jurkat), and exon 8, lanes 2, 11, and 16 (primary case 1706, Jim3, and primary case 1517, respectively).
Examples of PCR-SSCP analysis of PTEN exons (A, exon 1; B, exon 3; C, exon 6; D, exon 7; E, exon 8). DNA was prepared and subjected to PCR-SSCP analysis. In each analysis, lane 1 contains control DNA (placental). Alterations are present in exon 1, lane 5 (primary LBCL), exon 3, lane 4 (SUDHL10), exon 6, lane 3 (PA682PB), exon 7, lane 5 (Jurkat), and exon 8, lanes 2, 11, and 16 (primary case 1706, Jim3, and primary case 1517, respectively).
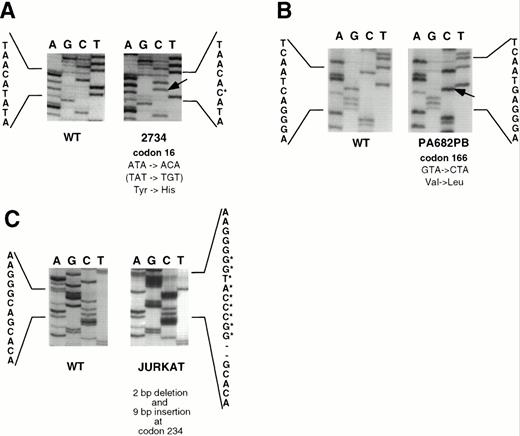
Sequence analysis of SSCP-positive cell lines. (A) A → G transition in codon 16 (TAT → TGT; Tyr → His) in primary LBCL (case 2734) (reverse sequence depicted). (B) G → C transversion in codon 166 (GTA → CTA; Val → Leu) in PA682PB. (C) 2-bp deletion followed by 9-bp insertion (GGCCCATGG) at codon 234 in Jurkat cell line. Control sequences are derived from placental DNA.
Sequence analysis of SSCP-positive cell lines. (A) A → G transition in codon 16 (TAT → TGT; Tyr → His) in primary LBCL (case 2734) (reverse sequence depicted). (B) G → C transversion in codon 166 (GTA → CTA; Val → Leu) in PA682PB. (C) 2-bp deletion followed by 9-bp insertion (GGCCCATGG) at codon 234 in Jurkat cell line. Control sequences are derived from placental DNA.
Analysis of primary cases.
Initial results from the cell lines encouraged us to study a larger series of primary lymphomas and control samples. Because all of the cell lines with PTEN alterations were derived from high-grade neoplasms, we focused our attention primarily on related high-grade primary tumors. PCR-SSCP analysis was performed on 65 primary tumors, including 24 LBL, 30 LBCL, 7 BL, and 4 ALCL. Southern blot analysis was also performed on the 24 LBL cases, since these samples had a sufficiently high number of tumor cells to allow reliable interpretation of potential deletions. None of the LBLs showed deletions or rearrangements of PTEN by Southern blot analysis. Among the entire group of primary cases, 3 of 65, all LBCL, showed SSCP alterations (Table 2 and Fig 2). In one case, sequencing of the variant conformer detected with the exon 1 primer set revealed an A to G transition located 12 bp before the initiation codon plus a second mutation at codon 16 that resulted in the substitution of a tyrosine for a cysteine (Tyr to Cys; TAT to TGT; Fig 3). This case lacked the normal set of conformers. In the other two LBCL cases, variant conformers were detected in the exon 8 SSCP analysis. One retained the normal conformers as well. Similar to the results for Jim3 and Raji cell lines, these cases showed small deletions in and around the intron 7 poly-T tract 5′ to exon 8. Case 1517 showed a 2-bp deletion in the poly-T tract, while case 1706 had the identical 38-bp deletion that was present in the Raji cell line involving the first two bases of the poly-T tract and the preceding 36 bp. None of the 25 nonneoplastic patient samples displayed variant conformers by SSCP.
DISCUSSION
In the present study, we examined 27 hematopoietic cell lines and 65 high-grade lymphoid neoplasms for mutation or deletion of the newly described PTEN/MMAC1/TEP1 tumor suppressor gene. We identified alterations of coding sequences in 4 of 27 hematopoietic cell lines, while another 2 cell lines showed small deletions within intron 7 unaccompanied by alterations of coding or splice-junction sequences. Of 65 primary cases, one showed a single nucleotide alteration in exon 1 and two others showed small deletions in intron 7 outside of classic splice-junction sequences.
Five of the nine alterations identified involved exon sequences, two of which were predicted to interfere with translation of an intact protein. The CEM LBL-derived cell line showed homozygous loss of exons 2 to 5, which encode the critical phosphatase domain of PTEN. The Jurkat LBL cell line showed two variant conformers, one with a 2-bp deletion followed by a 9-bp insertion that resulted in a frameshift, and the other with a 39-bp insertion containing a stop codon. The presence of two different mutated conformers suggests that either both alleles of Jurkat are mutated or there is intratumoral heterogeneity with respect to the PTEN/MMAC1 locus. The SUDHL10 and PA682 cell lines each contained nucleotide variations different from the published germline sequence. SUDHL10 contained two alterations, one resulting in the substitution of tyrosine for histidine at codon 68 (exon 3) and the other resulting in the substitution of an aspartic acid residue for the wild-type histidine at position 162 (exon 5). The nucleotide variant in PA682 was relatively conservative, resulting in the substitution of leucine for the germline valine at position 166 (exon 6). One primary LBCL had a sequence variation at codon 16 (exon 1) resulting in the substitution of tyrosine for the germline cysteine. In addition, this case also had a nucleotide transition (A to G) located 12 bp before the initiation codon. The normal conformers were present but much fainter than the variant conformers in this case. Although we did not perform a loss of heterozygosity analysis for this study, the fact that the normal conformers either were not observed in the cell lines or were of diminished intensity in the primary case raises the possibility that the normal allele was lost in the tumors.
In four samples, we could not identify alterations in the coding sequences, although variant conformers were detected in the exon 8 SSCP analysis. All but one of these variants were associated with the presence of the normal conformer set. These alterations were found to be the result of small deletions (2 to 38 bp) occurring within a poly-T–rich tract in intron 7, which is located just before the start of exon 8. Normal tissue from our patients was not available to determine with certainty whether these deletions were polymorphisms. However, a similar deletion of 39 bp in this region was reported by Okami et al22 in a primary head and neck cancer and shown to be a polymorphism, suggesting that our deletions are likely polymorphisms also. The fact that we did not identify any deletions in 25 control samples indicates that they are rare.
Sequence alterations in the cell lines were not confined to a particular type of aggressive lymphoid neoplasm, indicating that PTEN variants are not specific to a particular subtype of tumor. Two occurred in T-cell LBL cell lines, 2 in cell lines derived from BL, 1 in a cell line derived from LBCL, and 1 other in a myeloma cell line. In the primary tumors, sequence alterations were identified in only 3 LBCLs, and 2 of these 3 were exon 7 deletions. Since the frequency of PTEN variants in the primary tumor group is low, a larger number of cases must be studied to identify prevalence rates with more accuracy.
Our study has identified PTEN sequence variations in a small percentage of lymphoid neoplasms. The frequency of alterations was higher in cell lines than in primary tumors, and this finding may reflect selection for PTEN/MMAC1 mutations during cell line development. Several questions remain regarding the extent and relevance of PTEN alterations in lymphoid neoplasms that are not addressed adequately in the present study. First, in only two LBL cell lines have we shown definitive evidence of mutation (ie, homozygous deletions or nonsense mutations). In all of the other cell lines and cases, the alterations identified were either nucleotide substitutions or intronic deletions. Although the intron deletions may represent rare polymorphisms as discussed earlier, we would argue that the nonconservative nature of several of the missense variants (SUDHL-10 and case 2734), their absence in the normal control population, and the apparent loss of the normal allele in SSCP analysis make these alterations more likely to be somatic mutations rather than polymorphisms. However, since normal tissues from the lymphoma samples and cell lines were not available for study, we cannot formally exclude the possibility that these missense variants represent rare polymorphisms. Second, we did not examine low-grade lymphomas, choosing instead to focus on high-grade neoplasms because they most closely correlated with the types of cell lines we studied, and because initial studies of PTEN/MMAC1 suggested that this gene is preferentially involved in more advanced or aggressive tumors, as the name MMAC itself suggests: mutated in multiple advanced cancers. However, recently, at least one study has suggested that abnormalities of PTEN/MMAC1 may also occur early in some types of neoplasia such as thyroid cancer15 and melanoma.12 Thus, it may also be worthwhile to expand the series of lymphomas studied to include low-grade neoplasms as well. Third, the current study used techniques to specifically identify coding region mutations or, in a subset of the cases studied, larger gene deletions. It is important to note that inactivation of tumor suppressor genes can also occur through mechanisms that do not alter gene coding regions, such as gene methylation, and through other abnormalities involving noncoding sequences such as promoter sequences.
Address reprint requests to Mark Raffeld, MD, Laboratory of Pathology, National Cancer Institute, Bldg 10, Room 2N110, 9000 Rockville Pike, Bethesda, MD 20892; e-mail: mraff@box-m.nih.gov.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.