Abstract
The c-kit receptor tyrosine kinase (KIT) is constitutively activated by naturally occurring mutations in either the juxtamembrane domain or the kinase domain. Although the juxtamembrane domain mutations led to ligand-independent KIT dimerization, the kinase domain mutations (Asp814 → Val or Tyr) did not. In an effort to determine if the kinase domain mutant could transfer oncogenic signaling without receptor dimerization, we have constructed the truncated types of c-kitWild and c-kitTyr814 cDNAs (c-kitDel-Wild and c-kitDel-Tyr814 cDNAs, respectively), in which ligand-binding and ligand-induced dimerization domains were deleted. When c-kitDel-Wild and c-kitDel-Tyr814 genes were introduced into a murine interleukin-3 (IL-3)–dependent cell line Ba/F3, KITDel-Tyr814 was constitutively phosphorylated on tyrosine and activated, whereas KITDel-Wild was not. In addition, Ba/F3 cells expressing KITDel-Tyr814(Ba/F3Del-Tyr814) grew in suspension culture without the addition of exogenous growth factor, whereas Ba/F3 cells expressing KITDel-Wild (Ba/F3Del-Wild) required IL-3 for growth. The factor-independent growth of Ba/F3Del-Tyr814 cells was virtually abrogated by coexpression of KITW42 that is a dominant-negative form of KIT, but not by that of KITWild, suggesting that KITDel-Tyr814 may not function as a monomer but may require receptor dimerization for inducing factor-independent growth. Furthermore, KITDel-Tyr814 was found to be coimmunoprecipitated with KITWild or KITW42 by an ACK2 monoclonal antibody directed against the extracellular domain of KIT. Moreover, KITW42 was constitutively associated with a chimeric FMS/KITTyr814 receptor containing the ligand-binding and receptor dimerization domain of c-fmsreceptor (FMS) fused to the transmembrane and cytoplasmic domain of KITTyr814, but not with a chimeric FMS/KITWildreceptor even after stimulation with FMS-ligand. These results suggest that constitutively activating mutation of c-kit at the Asp814 codon may cause a conformation change that leads to receptor self-association not in the extracellular domain and that the receptor self-association of the Asp814 mutant may be important for activation of downstream effectors that are required for factor-independent growth and tumorigenicity.
THE PROTO-ONCOGENE c-kit encodes a receptor tyrosine kinase (RTK) that is a member of the same RTK subfamily (type III RTK) as the receptors for platelet-derived growth factor and macrophage colony-stimulating factor (M-CSF)/colony-stimulating factor-1.1,2 This RTK subfamily is characterized by the presence of five Ig-like repeats in the extracellular domain and an insert that splits the cytoplasmic kinase domain into the adenosine triphosphate (ATP)-binding and phosphotransferase regions.1-6 The ligand for c-kitreceptor tyrosine kinase (KIT) has been identified and variously designated as stem cell factor (SCF), mast cell growth factor,kit ligand, or steel factor7-10 (hereafter, we refer to the ligand as SCF). The binding of SCF promotes dimerization of KIT and activates intrinsic tyrosine kinase of KIT, resulting in transphosphorylation at critical tyrosine residues. The tyrosine-phosphorylated KIT can then bind a unique array of intracellular signaling molecules, including phosphatidylinositol 3′-kinase, phospholipase C-γ1, and protein tyrosine phosphatase SHP1, thereby initiating a signaling cascade that leads to various cellular responses.11-15 This KIT-mediated signal transduction is known to play a crucial role in proliferation, differentiation, migration, and survival of hematopoietic stem cells, mast cells, melanocytes, primordial germ cells, and interstitial cells of Cajal (ICCs).6 16
Although the enzymatic activity of KIT is tightly regulated by the binding of SCF, we have found that KIT is constitutively activated by point mutations of c-kit gene in human, rat, and murine mast cell lines. A human mast cell leukemia cell line (HMC-1) carried two types of constitutively activating mutations of c-kit gene: the Val560 to Gly mutation in the juxtamembrane domain and the Asp816 to Val mutation in the phosphotransferase domain.17 Constitutively activating mutation in the corresponding Asp of the phosphotransferase domain was also detected in a rat mast cell leukemia cell line (RBL-2H3; Asp817→ Tyr mutation)18 and a murine mastocytoma cell line (P-815; Asp814 → Tyr mutation),19 whereas constitutive activation of KIT in a murine mastocytoma cell line (FMA3) resulted from deletion of seven amino acids (Thr573-His579) in the juxtamembrane domain.20 The constitutive activation mutations of c-kit in the phosphotransferase and juxtamembrane domains, particularly at the Asp814 codon in the phosphotransferase domain, were found to confer the factor-independent growth and tumorigenicity of murine interleukin-3 (IL-3)–dependent cell lines and normal hematopoietic stem cells.21-23
In addition to hematopoietic cell lines, the Asp816mutation in the phosphotransferase domain of human c-kit gene has also been found in peripheral blood mononuclear cells from patients with myelodysplastic disorders accompanying mastocytosis, in mast cells from patients with urticaria pigmentosa and aggressive mastocytosis, and in leukemia cells from patients with acute myelocytic leukemia.24-26 Furthermore, we have recently demonstrated that activating mutations of c-kit are detected in gastrointestinal stromal tumors (GISTs), the most common mesenchymal tumors in the human digestive tract, that may originate from ICCs expressing both KIT and CD34.27 However, all of the c-kit activating mutations detected in GISTs were located only in the juxtamembrane domain. The occurrence of the activating mutations at the same Asp codon in human hematologic disorders as well as mouse, rat, and human tumor mast cell line implied that the Asp codon may be a hot spot for activating mutation in hematopoietic systems and suggested that the Asp mutation may be involved in neoplastic transformation of mast cells and hematopoietic stem cells. However, the precise mechanisms by which the c-kit mutations activate KIT tyrosine kinase and transmit oncogenic signals are not fully understood.
The ligand-stimulated receptor dimerization is known to be a key event in the activation of intrinsic protein kinase activity and signal transduction of RTK,4,5,28,29 and constitutive activation of RTK oncogenes tends to involve changes that mimic ligand-stimulated activation, such as receptor dimerization. We have previously shown that activating mutations within the juxtamembrane domain of c-kit, such as the murine Val559(Val560 in human) → Gly mutation, led to constitutive dimerization of KIT in the absence of SCF, but the activation mutation of murine Asp814 (Asp816 in human) caused constitutive activation of KIT without receptor dimerization.20,21 Therefore, it was suggested that the Asp814 mutation might be a unique activating mutation that induced factor-independent growth and tumorigenesis independently of receptor dimerization. However, it was also possible that the Asp814 mutation might cause receptor self-association not in the extracellular domain, thereby leading to constitutive activation and cell transformation. In this study, we have constructed the truncated mutants of c-kitWild and c-kitAsp814→Tyr cDNA, in which both ligand-binding and ligand-induced dimerization sites in the extracellular domain were deleted,29-32 and examined as to how the Asp814 mutation yields constitutive activation and cell-transforming potential.
MATERIALS AND METHODS
Reagents.
Recombinant murine (rm) SCF and rmIL-3 were generous gifts of Kirin Brewery Co Ltd (Tokyo, Japan). Recombinant human (rh) M-CSF was a generous gift of Yoshitomi Pharmaceutical Industries Ltd (Osaka, Japan). Rat antimouse c-kit (ACK2) monoclonal antibody (MoAb)33 and full-length of murine c-kit cDNA were kindly provided by Dr S.-I. Nishikawa (Kyoto University, Kyoto, Japan). The plasmid, pSMc-fms, containing the human c-fms cDNA that encodes FMS/M-CSF receptor, was kindly provided by Dr C.J. Sherr (Howard Hughes Medical Institute Research Laboratories, Memphis, TN).34,35 Rabbit antiserum against the kinase domain of murine KIT (anti-KITKinase serum)36 was kindly provided by Dr A. Bernstein (Samuel Lunenfeld Research Institute, Toronto, Ontario, Canada) and rabbit antiserum against a C-terminal peptide corresponding to the last 10 amino acids of murine KIT (anti-KITC-terminal serum)8 by Dr D.E. Williams (Immunex Corp, Seattle, WA). Rabbit polyclonal antibody (Ab-1) against a synthetic peptide of C-terminal of human KIT was purchased from Oncogene Science, Inc (New York, NY); this antibody reacted with both murine and human KITs. Antiphosphotyrosine MoAb, a murine MoAb generated against phosphotyramine, was generously supplied by Dr B.J. Druker (Oregon Health Sciences University, Portland, OR). Rat MoAb (3-4A4) against the extracellular domain of human FMS/M-CSF receptor was purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA).
Cell lines.
The 293T cell line was derived from human embryonic kidney cells transformed by DNA from human adenovirus type 5,37 and 293T cells were maintained in Dulbecco’s modification of Eagle’s medium (DMEM; ICN Biomedicals, Costa Mesa, CA) supplemented with 10% fetal bovine serum (FBS; Irvine Scientific, Santa Ana, CA). The Ba/F3 murine IL-3–dependent cell line was cultured in α-minimal essential medium (α-MEM; ICN Biomedicals) supplemented with 10% FBS and rmIL-3 at a concentration of 10 ng/mL.
Construction of transgene and transfection.
Bluescript I KS(−) plasmids (Stratagene, La Jolla, CA) containing the whole coding regions of wild-type c-kit cDNA (c-kitWild) and activating mutant (Asp814 → Tyr; Tyr814) c-kitcDNA (c-kitTyr814) were constructed in our laboratory as reported previously.19 TheBamHI-DraI fragments (nucleotide 281 to 1561; the nucleotide numbers refer to the region of c-kit cDNA sequence reported by Qiu et al1) of the c-kitWild and c-kitTyr814 cDNAs were deleted to exclude five Ig-like repeats in the extracellular domain of KIT; the c-kitWild and c-kitTyr814 cDNAs lacking the extracellular domain were hereafter designated as c-kitDel-Wild and c-kitDel-Tyr814, respectively. To generate c-kitW42 cDNA, a single transition mutation (GC → AT) at nucleotide position 2396 was introduced into c-kitWild cDNA by exchanging the NdeI-Nhe I fragment (nucleotide 1850 to 2459) of c-kitWild cDNA for the corresponding fragment of c-kit cDNA obtained fromW42/W42 mice.38 The full coding sequences of c-kitWild, c-kitTyr814, c-kitDel-Wild, c-kitDel-Tyr814, or c-kitW42cDNAs were released, isolated, and inserted into a blunted XbaI site of expression vector pEF-BOS.39
For the construction of plasmids (pEF-BOS containing c-fms/c-kitWild and pEF-BOS containing c-fms/c-kitTyr814) that express a chimeric receptor composing of the ligand-binding and receptor dimerization domain of FMS fused to the transmembrane and cytoplasmic domain of KITWild or KITTyr814 (FMS/KITWildor FMS/KITTyr814), the BamHI-Dra I fragment (nucleotide 281 to 1561 of c-kit cDNA) of pEF-BOS containing c-kitWild or c-kitTyr814 cDNA was exchanged with the fragment (nucleotide 373 to 1836; the nucleotide numbers refer to the region of c-fms cDNA sequence reported by Coussens et al34) of human c-fms cDNA obtained from pSMc-fms.35
The expression vector pEF-BOS containing c-kitWild, c-kitTyr814, c-kitDel-Wild, c-kitDel-Tyr814, c-kitW42, c-fms/c-kitWild, or c-fms/c-kitTyr814 cDNA (each 10 μg) was transfected into 293T cells by the calcium phosphate methods as described previously.40 In some experiments, two types of cDNAs (c-kitDel-Wild and c-kitWild; c-kitDel-Wild and c-kitW42; c-kitDel-Tyr814 and c-kitWild; c-kitDel-Tyr814 and c-kitW42; c-fms/c-kitWild and c-kitW42; c-fms/c-kitTyr814 and c-kitW42) were cotransfected into the cells to examine their association in the cytoplasmic region. Forty hours after transfection, the cells were collected and used for further analyses.
For gene transfer into Ba/F3 cells, the linearized expression vector pEF-BOS containing various types of c-kit cDNA (100 μg) in combination with either pSTneoB (1 μg) or pPGKhyg (1 μg) was added to cell suspension (1 × 107) in 0.7 mL phosphate-buffered saline (PBS), and then the electroporation (975 mF, 350 V) was performed by Gene Pulser II (Bio-Rad Laboratories, Hercules, CA). Two days after the electroporation, 1,000 μg/mL of G418 sulfate (geneticin; GIBCO BRL, Grand Island, NY) or 1,500 μg/mL of HygromycinB (Boehringer Mannheim GmbH Biochemica, Mannheim, Germany) was added to the complete culture medium to select neomycin- or HygromycinB-resistant cells. Cells expressing wild-type or mutant KIT were selected by limiting dilution assay.
Metabolic labeling by [35S]-methionine.
Metabolic labeling was performed as previously described.41Briefly, cells were incubated in methionine-free DMEM (Life Technologies, Grand Island, NY) containing [35S]-methionine (DuPont/NEN Research Products, Boston, MA; 100 mCi/mL), 5 mmol/L glutamine, 1 mmol/L sodium pyruvate, and 10% dialyzed FBS for 5 hours. Radiolabeled KIT was precipitated with protein-G Sepharose beads (Pharmacia, Uppsala, Sweden) and either rat ACK2 MoAb, rabbit anti-KITKinase serum, or rabbit anti-KITC-terminal serum. The immunoprecipitates were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 5% to 20% gradient polyacrylamide. The gel was dried, and radioactive proteins were detected by autoradiography.
Flow cytometry.
To detect cell surface expression of KIT, cells were incubated with ACK2 MoAb at 4°C for 30 minutes, stained with fluorescein isothiocyanate (FITC)-conjugated rabbit antirat Ig antibody (DAKO A/S, Glostup, Denmark), and analyzed on a FACScan (Becton Dickinson, Los Angeles, CA). To detect the expression of KITDel-Wild and KITDel-Tyr814, cells were fixed with ice-cold methanol-aceton (1:1) for 10 minutes, incubated with rabbit anti-KITKinase serum, stained with FITC-conjugated donkey antirabbit Ig antibody (Jackson Immuno Research Laboratories, West Grove, PA), and analyzed on a FACScan.
Immunoblotting.
The procedures of cell lysis, SDS-PAGE, and immunoblotting were performed according to the methods described previously.42Briefly, after depletion of serum and factors, cells were treated with or without either rmSCF (100 ng/mL) or rhM-CSF (10,000 U/mL) for 15 minutes at 37°C. The cells were then washed with cold PBS and lysed in lysis buffer (20 mmol/L Tris-HCl, 137 mmol/L NaCl, 10% glycerol, 1% Nonidet P-40, pH 8.0, and protease and phosphatase inhibitors). After removal of insoluble materials by centrifugation, cell lysates were incubated with rabbit anti-KITKinase serum and protein-G Sepharose beads. The immunoprecipitates were subjected to SDS-PAGE with 5% to 20% gradient polyacrylamide, and proteins were electrophoretically transferred from the gel onto a polyvinylidene difluoride membrane (Immobilon; Millipore Corp, Bedford, MA). Immunoblotting was performed with either antiphosphotyrosine MoAb or rabbit Ab-1.
Immune complex kinase assay.
The immune complex kinase assay was performed as previously described.17-20 43 Briefly, cell lysates were incubated with rabbit anti-KITKinase serum, ACK2 MoAb, or 3-4A4 MoAb followed by the addition of protein-G Sepharose beads to collect the antigen-antibody complexes. After washing, the immune complexes were incubated in kinase buffer (10 mmol/L MnCl2, 20 mmol/L Tris-HCl, pH 7.4) containing 1 μL of γ-[32P]-ATP (Dupont/NEN Research Products; 10 mCi/mL) for 20 minutes at 25°C and separated by SDS-PAGE with 5% to 20% gradient polyacrylamide. The gel was dried, and radioactive proteins were detected by autoradiography.
Cell proliferation assays.
Proliferation of cells was quantified by [3H]-thymidine incorporation as previously described.44 The exponentially growing cells were washed twice with α-MEM, and triplicate aliquots of cells (5 × 104) suspended in 200 μL of Cosmedium-001 (Cosmo Bio Co, Tokyo, Japan) were cultured in 96-well microtiter plates for 72 hours at 37°C with various concentrations of rmIL-3 or rmSCF. At 72 hours after initiation of the culture, 0.5 μCi [3H]-thymidine (specific activity, 5 Ci/mmol; Amersham, Arlington Heights, IL) was added to each well. Five hours after the addition of [3H]-thymidine, the cells were harvested with a semiautomatic cell harvester (Pharmacia LKB Biotechnology, Uppsala, Sweden) and the incorporation of [3H]-thymidine was measured with a liquid scintillation counter. In some experiments, cell proliferation was assessed by cell enumeration. Triplicate aliquots of cells (2 × 104) suspended in 100 μL of Cosmedium-001 were cultured in 96-well microtiter plates, and viable cells were counted at daily intervals by trypan blue dye exclusion.45
Statistical analysis.
The Student’s t-test was used to evaluate the significance of the [3H]-thymidine incorporation and the number of viable cells.
RESULTS
Preparation of mutant c-kit genes lacking the extracellular domain and introduction into 293T and murine IL-3–dependent Ba/F3 cells.
The Ig-like repeats in the extracellular domain of KIT are known to be indispensable for ligand-binding and receptor dimerization. To examine the role of extracellular domain in constitutive activation of mutant KIT bearing the Asp814 mutation, we constructed the truncated c-kitWild and c-kitTyr814 cDNAs, in which the extracellular Ig-like repeats of c-kit were deleted (Fig 1A). The expression vector pEF-BOS containing c-kitDel-Wild and c-kitDel-Tyr814 cDNAs was first transfected into 293T cells to determine if KITDel-Wild and KITDel-Tyr814 were efficiently expressed in cells and were detected with anti-KIT antibodies. The transfected 293T cells were radiolabeled with [35S]-methionine, and cell lysates were subjected to immunoprecipitation experiments with either ACK2 MoAb against the KIT extracellular domain (ECD), rabbit antiserum against the KIT kinase domain (anti-KITKinase serum; Kinase), or rabbit antiserum against the KIT C-terminal domain (anti-KITC-terminal serum; C-terminal) (Fig 1B). As expected, both KITDel-Wild and KITDel-Tyr814were immunoprecipitated with anti-KITKinase serum and anti-KITC-terminal serum, but not with ACK2 MoAb.
Construction of deletion-type c-kit cDNA. (A) Schematic representation of deletion-type KIT. Location of signal peptide (SP), transmembrane (TM), and tyrosine residue of autophosphorylation (Y) are indicated. KITTyr814 and KITDel-Tyr814 carries a point mutation (Asp to Tyr) in codon 814. (B) Expression of KITDel-Wild and KITDel-Tyr814. 293T cells were transfected with c-kitDel-Wild and c-kitDel-Tyr814 cDNA, and then cells were labeled for 5 hours with [35S]-methionine and lysed. KIT was immunoprecipitated with ACK2 MoAb (ECD), rabbit anti-KITKinase serum (Kinase), and rabbit anti-KITC-terminal serum (C-terminal) and analyzed by SDS-PAGE and autoradiography. The similar results were obtained from three independent experiments.
Construction of deletion-type c-kit cDNA. (A) Schematic representation of deletion-type KIT. Location of signal peptide (SP), transmembrane (TM), and tyrosine residue of autophosphorylation (Y) are indicated. KITTyr814 and KITDel-Tyr814 carries a point mutation (Asp to Tyr) in codon 814. (B) Expression of KITDel-Wild and KITDel-Tyr814. 293T cells were transfected with c-kitDel-Wild and c-kitDel-Tyr814 cDNA, and then cells were labeled for 5 hours with [35S]-methionine and lysed. KIT was immunoprecipitated with ACK2 MoAb (ECD), rabbit anti-KITKinase serum (Kinase), and rabbit anti-KITC-terminal serum (C-terminal) and analyzed by SDS-PAGE and autoradiography. The similar results were obtained from three independent experiments.
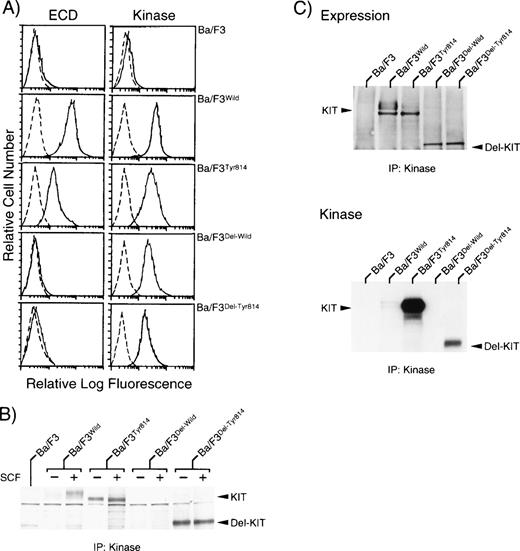
The expression vector pEF-BOS containing either c-kitWild, c-kitTyr814, c-kitDel-Wild, or c-kitDel-Tyr814 cDNA and pST2neo were then transfected into murine IL-3–dependent Ba/F3 cells by electroporation. After selection in a medium containing G418 and rmIL-3 for 3 weeks, Ba/F3 clones expressing KITWild, KITTyr814, KITDel-Wild, or KITDel-Tyr814 were isolated by limiting dilution assays and were designated as Ba/F3Wild, Ba/F3Tyr814, Ba/F3Del-Wild, or Ba/F3Del-Tyr814, respectively. Flow cytometric analysis using an ACK2 MoAb (ECD) showed the cell surface expression of KIT on Ba/F3Wild and Ba/F3Tyr814 cells, but not on Ba/F3 (parent), Ba/F3Del-Wild, or Ba/F3Del-Tyr814 cells (Fig 2A, left panels). However, when each type of the cells was stained with anti-KITKinase serum (Kinase) after fixation with methanol and aceton, KIT expression was easily detected in Ba/F3Wild, Ba/F3Tyr814, Ba/F3Del-Wild, and Ba/F3Del-Tyr814 cells at an almost similar level (Fig 2A, right panels).
Constitutive tyrosine phosphorylation and activation of KITDel-Tyr814, but not of KITDel-Wild. (A) Flow cytometric analysis on the expression of KIT. To detect the expression of KIT on the cell surface, cells were incubated with either ACK2 MoAb (ECD) (—) or negative control antibody (---). To detect the whole expression of KIT, cells were fixed with methanol-aceton (1:1) and then incubated with either rabbit anti-KITKinaseserum (Kinase) (—) or negative control antibody (---). (B) Constitutive tyrosine phosphorylation of KITDel-Tyr814. The state of tyrosine phosphorylation of KIT before and after stimulation with rmSCF was examined by immunoblotting using antiphosphotyrosine MoAb. The mobilities of KIT and deletion type of KIT are indicated at right. Three independent experiments were performed with comparable results. (C) Constitutive activation of KITDel-Tyr814. KIT was immunoprecipitated from cell lysates without rmSCF stimulation using anti-KITKinase serum. The immunoprecipitated KIT was examined by immunoblotting using rabbit Ab-1 (upper panel) and was subjected to immune complex kinase assay (lower panel). Three independent experiments were performed with comparable results.
Constitutive tyrosine phosphorylation and activation of KITDel-Tyr814, but not of KITDel-Wild. (A) Flow cytometric analysis on the expression of KIT. To detect the expression of KIT on the cell surface, cells were incubated with either ACK2 MoAb (ECD) (—) or negative control antibody (---). To detect the whole expression of KIT, cells were fixed with methanol-aceton (1:1) and then incubated with either rabbit anti-KITKinaseserum (Kinase) (—) or negative control antibody (---). (B) Constitutive tyrosine phosphorylation of KITDel-Tyr814. The state of tyrosine phosphorylation of KIT before and after stimulation with rmSCF was examined by immunoblotting using antiphosphotyrosine MoAb. The mobilities of KIT and deletion type of KIT are indicated at right. Three independent experiments were performed with comparable results. (C) Constitutive activation of KITDel-Tyr814. KIT was immunoprecipitated from cell lysates without rmSCF stimulation using anti-KITKinase serum. The immunoprecipitated KIT was examined by immunoblotting using rabbit Ab-1 (upper panel) and was subjected to immune complex kinase assay (lower panel). Three independent experiments were performed with comparable results.
Activation state of KITWild and KITTyr814after removal of their extracellular domains.
To examine the state of KIT-tyrosyl phosphorylation in the transfected Ba/F3 cells, the cells were deprived of serum and growth factors for 12 hours and stimulated with or without rmSCF (100 ng/mL) for 15 minutes. KIT was then immunoprecipitated with anti-KITKinase serum and subjected to immunoblotting with either antiphosphotyrosine MoAb or anti-KIT polyclonal antibody (Ab-1). In accord with our previous findings, immunoblotting with an antiphosphotyrosine MoAb showed that KITTyr814 was constitutively phosphorylated on tyrosine residues, whereas tyrosine phosphorylation of KITWild was induced in a ligand-dependent manner (Fig 2B). In the case of deletion mutants, KITDel-Wild was not phosphorylated on tyrosine before and after stimulation with rmSCF; KITDel-Tyr814 was found to be significantly phosphorylated on tyrosine regardless of rmSCF stimulation (Fig 2B). We further examined the kinase activity of KIT by immune complex kinase assay. KIT was immunoprecipitated with anti-KITKinase serum from cell lysates of Ba/F3, Ba/F3Wild, Ba/F3Tyr814, Ba/F3Del-Wild, and Ba/F3Del-Tyr814 cells that were not stimulated with rmSCF, and the kinase activity was examined. Almost identical levels of KIT proteins were immunoprecipitated from the cell lysates of Ba/F3Wild, Ba/F3Tyr814, Ba/ F3Del-Wild, and Ba/F3Del-Tyr814 cells (Fig 2C, upper panel). Consistent with the data on immunoblotting analysis, both KITTyr814 and KITDel-Tyr814exhibited a striking kinase activity, whereas little kinase activity was detected in KITWild and KITDel-Wild (Fig2C, lower panel).
The effects of KITDel-Wild and KITDel-Tyr814on the growth of Ba/F3 cells.
To determine if KITTyr814 could induce factor-independent growth of Ba/F3 cells after removal of its extracellular domain, Ba/F3, Ba/F3Wild, Ba/F3Tyr814, Ba/F3Del-Wild, and Ba/F3Del-Tyr814 cells were cultured in the presence of 0 to 100 ng/mL rmIL-3 or 0 to 1,000 ng/mL rmSCF for 72 hours, followed by measurement of cell proliferation using a [3H]-thymidine incorporation assay (Fig 3). The parental Ba/F3 cells showed rmIL-3–dependent proliferation, but did not proliferate in response to rmSCF. As in the case of parental Ba/F3 cells, both Ba/F3Wild and Ba/F3Del-Wild cells proliferated in response to rmIL-3 in a dose-dependent manner, and rmSCF-dependent proliferation was observed in Ba/F3Wild cells, but not in Ba/F3Del-Wild cells that lacked SCF-binding region of KIT. By contrast, Ba/F3Tyr814 and, albeit to a lesser degree, Ba/F3Del-Tyr814 cells proliferated in the absence of exogenous rmIL-3 or rmSCF. These findings suggested that the extracellular region required for ligand-binding and receptor dimerization may not be involved in the constitutive activation and growth-promoting signaling of KITTyr814.
Factor-independent proliferation of Ba/F3Del-Tyr814 cells. Proliferation of Ba/F3, Ba/F3Wild, Ba/F3Tyr814, Ba/F3Del-Wild, and Ba/ F3Del-Tyr814 cells at various concentrations of rmIL-3 (upper panel) or rmSCF (lower panel) was measured with [3H]-thymidine incorporation assay. Each point represents the mean of data from three experiments. Bars are the standard error. In some points, the standard error was too small to be shown by bars.
Factor-independent proliferation of Ba/F3Del-Tyr814 cells. Proliferation of Ba/F3, Ba/F3Wild, Ba/F3Tyr814, Ba/F3Del-Wild, and Ba/ F3Del-Tyr814 cells at various concentrations of rmIL-3 (upper panel) or rmSCF (lower panel) was measured with [3H]-thymidine incorporation assay. Each point represents the mean of data from three experiments. Bars are the standard error. In some points, the standard error was too small to be shown by bars.
Effect of dominant-negative KITW42 on the factor-independent growth of Ba/F3Del-Tyr814 cells.
The Asp790 → Asn mutation in theW42 mutant form of c-kit gene abolishes kinase activity.38 The c-kitW42 product (KITW42) was previously shown to confer dominant-negative effect, because the kinase-defective KITW42 would sequester KITWild into nonfunctional heterodimers, leading to a marked reduction in the efficiency of signal transduction.41 To examine whether KITTyr814could mediate ligand-independent signaling as a monomer, the pEF-BOS expression vector containing c-kitW42 or c-kitWild cDNA and pPGKhyg were introduced into Ba/F3Del-Wild or Ba/ F3Del-Tyr814 cells by electroporation. After selection in a medium containing rmIL-3 and HygromycinB for 3 weeks, Ba/F3 clones coexpressing KITDel-Wild and KITWild(KITDel-Wild/KITWild), KITDel-Wild and KITW42(KITDel-Wild/KITW42), KITDel-Tyr814and KITWild(KITDel-Tyr814/KITWild), or KITDel-Tyr814 and KITW42 (KITDel-Tyr814/KITW42) were isolated by limiting dilution assays and were named Ba/F3Del-Wild/Wild, Ba/F3Del-Wild/W42, Ba/F3Del-Tyr814/Wild, or Ba/F3Del-Tyr814/W42, respectively. Flow cytometric analysis using ACK2 MoAb (ECD) showed that each type of the clones expressed KITWild or KITW42 on their surface (Fig 4A).
Dominant negative effect of KITW42 in Ba/F3Del-Tyr814 cells. (A) Flow cytometric analysis of the surface binding of ACK2 MoAb (ECD). Cells were incubated with either ACK2 MoAb (—) or negative control antibody (---). (B) Viability assays. After plating cells (2 × 104), viable cells were counted at daily intervals using trypan blue dye. Three independent experiments were performed with comparable results, and the result of a representative experiment is shown. Each point represents the mean of triplicate samples. Bars are the standard error. In some points, the standard error was too small to be shown by bars. (C) Incorporation of [3H]-thymidine. Proliferation of cells for rmIL-3 (10 ng/mL) and rmSCF (100 ng/mL) was measured with [3H]-thymidine incorporation assay. Three independent experiments were performed with comparable results, and the result of a representative experiment is shown. Each value represents the mean of triplicate samples, and the standard error was less than 5%. Asterisks indicate the presence of the statistical significance (P < .01) when compared with the value of either Ba/F3Del-Wildor Ba/F3Del-Tyr814 cells at the absence of rmIL-3 and rmSCF.
Dominant negative effect of KITW42 in Ba/F3Del-Tyr814 cells. (A) Flow cytometric analysis of the surface binding of ACK2 MoAb (ECD). Cells were incubated with either ACK2 MoAb (—) or negative control antibody (---). (B) Viability assays. After plating cells (2 × 104), viable cells were counted at daily intervals using trypan blue dye. Three independent experiments were performed with comparable results, and the result of a representative experiment is shown. Each point represents the mean of triplicate samples. Bars are the standard error. In some points, the standard error was too small to be shown by bars. (C) Incorporation of [3H]-thymidine. Proliferation of cells for rmIL-3 (10 ng/mL) and rmSCF (100 ng/mL) was measured with [3H]-thymidine incorporation assay. Three independent experiments were performed with comparable results, and the result of a representative experiment is shown. Each value represents the mean of triplicate samples, and the standard error was less than 5%. Asterisks indicate the presence of the statistical significance (P < .01) when compared with the value of either Ba/F3Del-Wildor Ba/F3Del-Tyr814 cells at the absence of rmIL-3 and rmSCF.
To determine the effect of KITW42 on cell proliferation, we initially examined changes in the viable cell numbers after culture of Ba/F3Del-Wild/Wild, Ba/F3Del-Wild/W42, Ba/F3Del-Tyr814/Wild, and Ba/F3Del-Tyr814/W42cells in a serum-free medium without any added growth factors (Fig 4B). Like Ba/F3Del-Wild cells, Ba/ F3Del-Wild/Wildand Ba/F3Del-Wild/W42 cells did not proliferate in the absence of growth factors. The expression of KITWild did not affect the factor-independent growth of Ba/F3Del-Tyr814cells, and the viable cell numbers of Ba/F3Del-Tyr814 and Ba/F3Del-Tyr814/Wild cells increased similarly with the lapse of culture periods. By contrast, the factor-independent growth of Ba/F3Del-Tyr814 cells was almost completely abrogated by the introduction of KITW42.
In addition to cell enumeration, cell proliferation was also examined by a [3H]-thymidine incorporation assay after culture with or without 10 ng/mL rmIL-3 or 100 ng/mL rmSCF for 72 hours (Fig 4C). Ba/F3Del-Wild, Ba/F3Del-Wild/Wild, and Ba/ F3Del-Wild/W42cells did not proliferate in the absence of any added growth factors, but did in response to rmIL-3. Among these cells, only Ba/F3Del-Wild/Wild cells showed proliferative response to rmSCF. In the case of Ba/F3Del-Tyr814 and Ba/ F3Del-Tyr814/Wild cells, both types of cells could proliferate even in the absence of growth factors; and the proliferation of these cells was not affected by rmSCF, but significantly augmented by the addition of rmIL-3, possibly due to synergistic effect of KITDel-Tyr814 and IL-3–mediated signaling. In contrast, [3H]-thymidine incorporation of Ba/F3Del-Tyr814/W42 cells was only minimal when cultured with or without rmSCF, but they could proliferate in response to rmIL-3 at an almost similar level to Ba/F3Del-Wild, Ba/F3Del-Wild/Wild, or Ba/F3Del-Wild/W42 cells.
Self-association of KITTyr814.
The dominant-negative effect of KITW42 on the proliferation of Ba/F3Del-Tyr814 cells raised the possibility that KITTyr814 may yield homodimeric and heterodimeric association of KIT not in the extracellular region. To test this possibility, 293T cells were transfected with c-kitWild, c-kitW42, c-kitDel-Wild, or c-kitDel-Tyr814 genes or with the combinations of c-kitWild and c-kitDel-Wildgenes, c-kitW42 and c-kitDel-Wild genes, c-kitWildand c-kitDel-Tyr814 genes, or c-kitW42 and c-kitDel-Tyr814genes, and then the expression and association of KIT was examined. Immunoblotting with anti-KIT polyclonal antibody (Ab-1), followed by immunoprecipitation with anti-KITKinase serum (Kinase), showed that each type of transfectant expressed full-length KIT (KIT: KITWild and KITW42) and/or deleted-form KIT (Del-KIT: KITDel-Wild and KITDel-Tyr814) at a nearly similar level (Fig 5A, upper panel). When KIT was immunoprecipitated with ACK2 MoAb (ECD) and subjected to immune complex kinase assay before treatment with rmSCF (Fig 5A, lower panel), phosphorylation bands of KITWild and KITW42 were barely detectable. In addition, because KITDel-Wild and KITDel-Tyr814 were not recognized by ACK2 MoAb, phosphorylation bands of KITDel-Wild and KITDel-Tyr814 were undetectable. When the cells expressing KITDel-Wildtogether with KITWild or KITW42 were used, phosphorylation bands of Del-KIT and KIT were also barely detected. By contrast, when the cells expressing KITDel-Tyr814 together with KITWild or KITW42 were used, phosphorylation bands of both Del-KIT (KITDel-Tyr814) and KIT (KITWild or KITW42) were detected, suggesting that KITDel-Tyr814 could be coimmunoprecipitated with KITWild or KITW42.
Self-association of KITTyr814. (A) 293T cells were transfected with pEF-BOS containing c-kitWild, pEF-BOS containing c-kitW42, pEF-BOS containing c-kitDel-Wild, or pEF-BOS containing c-kitDel-Tyr814 and cotransfected with pEF-BOS containing c-kitDel-Wild and pEF-BOS containing c-kitWild, pEF-BOS containing c-kitDel-Wild and pEF-BOS containing c-kitW42, pEF-BOS containing c-kitDel-Tyr814 and pEF-BOS containing c-kitWild, or pEF-BOS containing c-kitDel-Tyr814 and pEF-BOS containing c-kitW42. The expression of KIT is shown in upper panel. Cell lysates were immunoprecipitated with rabbit anti-KITKinase serum (Kinase), and then the immunoprecipitates were subjected to SDS-PAGE for 90 minutes and immunoblotting with rabbit Ab-1. The phosphorylation bands of immune complex kinase assay are shown in lower panel. KIT was immunoprecipitated from cell lysates with ACK2 MoAb (ECD), and the immunoprecipitates were incubated with γ-[32P]-ATP. Incorporation of 32P into KIT and/or Del-KIT was visualized by autoradiography. (B) 293T cells were transfected with pEF-BOS containing c-fms/c-kitWild or pEF-BOS containing c-fms/c-kitTyr814 and cotransfected with pEF-BOS containing c-fms/c-kitWild and pEF-BOS containing c-kitW42 or with pEF-BOS containing c-fms/c-kitTyr814 and pEF-BOS containing c-kitW42. The expression of FMS/KIT and/or KITW42 is shown in upper panel. Cell lysates were immunoprecipitated with rabbit anti-KITKinase serum (Kinase) before and after treatment with rhM-CSF, and then the immunoprecipitates were subjected to SDS-PAGE for 3 hours and immunoblotting with rabbit Ab-1. The phosphorylation bands of immune complex kinase assay are shown in lower panel. KIT was immunoprecipitated from cell lysates with 3-4A4 MoAb (FMS-ECD), and the immunoprecipitates were incubated with γ-[32P]-ATP. Incorporation of 32P into FMS/KIT and/or KITW42 was visualized by autoradiography. The similar results were obtained from three independent experiments.
Self-association of KITTyr814. (A) 293T cells were transfected with pEF-BOS containing c-kitWild, pEF-BOS containing c-kitW42, pEF-BOS containing c-kitDel-Wild, or pEF-BOS containing c-kitDel-Tyr814 and cotransfected with pEF-BOS containing c-kitDel-Wild and pEF-BOS containing c-kitWild, pEF-BOS containing c-kitDel-Wild and pEF-BOS containing c-kitW42, pEF-BOS containing c-kitDel-Tyr814 and pEF-BOS containing c-kitWild, or pEF-BOS containing c-kitDel-Tyr814 and pEF-BOS containing c-kitW42. The expression of KIT is shown in upper panel. Cell lysates were immunoprecipitated with rabbit anti-KITKinase serum (Kinase), and then the immunoprecipitates were subjected to SDS-PAGE for 90 minutes and immunoblotting with rabbit Ab-1. The phosphorylation bands of immune complex kinase assay are shown in lower panel. KIT was immunoprecipitated from cell lysates with ACK2 MoAb (ECD), and the immunoprecipitates were incubated with γ-[32P]-ATP. Incorporation of 32P into KIT and/or Del-KIT was visualized by autoradiography. (B) 293T cells were transfected with pEF-BOS containing c-fms/c-kitWild or pEF-BOS containing c-fms/c-kitTyr814 and cotransfected with pEF-BOS containing c-fms/c-kitWild and pEF-BOS containing c-kitW42 or with pEF-BOS containing c-fms/c-kitTyr814 and pEF-BOS containing c-kitW42. The expression of FMS/KIT and/or KITW42 is shown in upper panel. Cell lysates were immunoprecipitated with rabbit anti-KITKinase serum (Kinase) before and after treatment with rhM-CSF, and then the immunoprecipitates were subjected to SDS-PAGE for 3 hours and immunoblotting with rabbit Ab-1. The phosphorylation bands of immune complex kinase assay are shown in lower panel. KIT was immunoprecipitated from cell lysates with 3-4A4 MoAb (FMS-ECD), and the immunoprecipitates were incubated with γ-[32P]-ATP. Incorporation of 32P into FMS/KIT and/or KITW42 was visualized by autoradiography. The similar results were obtained from three independent experiments.
To further determine if the receptor association did not require the extracellular domain and was observed only in KITTyr814 but not in ligand-activated KITWild, we constructed plasmids that express a chimeric receptor composing of the ligand-binding and receptor dimerization domain of human FMS fused to the transmembrane and cytoplasmic domain of mouse KITWild or KITTyr814 (FMS/KITWild or FMS/KITTyr814). 293T cells were transfected with c-fms/c-kitWild or c-fms/c-kitTyr814 gene or with the combinations of c-fms/c-kitWild and c-kitW42 genes or c-fms/c-kitTyr814 and c-kitW42 genes. Immunoblotting with anti-KIT polyclonal antibody (Ab-1), followed by immunoprecipitation with anti-KITKinase serum (Kinase), showed that chimeric receptors (FMS/KITWild or FMS/KITTyr814) and/or KITW42 were expressed in each type of transfectants (Fig 5B, upper panel). When FMS/KITWild was immunoprecipitated with anti-FMS (3-4A4) MoAb against the extracellular domain of FMS (FMS-ECD) and subjected to immune complex kinase assay, the phosphorylation band of FMS/KITWild was barely detectable before stimulation with rhM-CSF, but was visualized after rhM-CSF stimulation (Fig 5B, lower panel), indicating a ligand-dependent activation of FMS/KITWild. On the other hand, FMS/KITTyr814 was constitutively activated, because the phosphorylation band of FMS/KITTyr814 was observed before and after stimulation by rhM-CSF (Fig 5B, lower panel). When the cells expressing FMS/KITWild together with KITW42 were used, anti-FMS MoAb could detect a FMS/KITWild band after stimulation with rhM-CSF. By contrast, when the cells expressing FMS/KITTyr814 together with KITW42 were used, both FMS/KITTyr814 and KITW42 bands were detected by anti-FMS MoAb, regardless of stimulation with rhM-CSF (Fig 5B, lower panel). These results suggested that KITW42 could be coimmunoprecipitated with constitutively active FMS/KITTyr814, but not with ligand-activated FMS/KITWild.
DISCUSSION
It is known that enzymatic activity of RTKs is deregulated by a myriad of structural alterations, named loss-of-function and gain-of-function mutations. We have previously shown that KIT can be constitutively activated by gain-of-function mutations in either the juxtamembrane domain or the phosphotransferase (kinase) domain.17-20,27However, the molecular mechanism regulating constitutive activation appears to be different between the juxtamembrane domain and the kinase domain mutants, because the juxtamembrane domain mutants of KIT were organized in the plasma membrane in a dimerized form without the addition of exogenous rmSCF, whereas a dimeric form of the kinase domain mutants (KITAsp814 → Tyr or Val) was not detectable in the absence of rmSCF.20,21 Regarding the ligand-induced dimerization, Blechman et al32 have reported that the fourth of five Ig-like domains on the extracellular domain contains determinants required for ligand-induced dimerization and activation, whereas Lemmon et al46 have recently demonstrated that the first three Ig-like domains can bind to SCF and dimerize in a manner identical to the complete extracellular domain of KIT. These results suggested that the Asp814 mutations of the kinase domain may not stabilize a conformation equivalent to that induced by ligand binding and that the KIT mutant may not require its extracellular Ig-like domains for the constitutive activation and growth-promoting signals. In accord with this hypothesis, we found that KITTyr814 devoid of the extracellular Ig-like domains was constitutively activated and capable of conferring factor-independent growth of murine IL-3–dependent Ba/F3 cells.
It has been reported that the deletion of the extracellular domain is predisposed to generate ligand-independent activation of RTKs, because this event removes some negative regulatory constraints imposed by the extracellular domain.47,48 However, the deletion of extracellular domain is insufficient to activate the full transforming potential of RTKs, and their oncogenic activation occurs in combination with additional mutations. For example, the v-kit oncogene differs from the c-kit proto-oncogene by deletion of the extracellular domain and by additional mutations in the cytoplasmic regions, including deletion of tyrosine-569 and valine-570, substitution of glycine-761 for aspartate, and replacement of the C-terminal 50 amino acids by five unrelated residues; and it was suggested that the deletion of tyrosine-569 and valine-570 is crucial for the oncogenic potential of the v-kit oncogene product.49 50 In this study, we demonstrated that deletion of extracellular Ig-like domains did not yield tyrosine phosphorylation and activation of KITWild and that KITWilddevoid of the extracellular region did not induce proliferation of Ba/F3 cells. This finding, including the data on KITDel-Tyr814, suggested that deletion of the extracellular domain may not be sufficient for constitutive activation of KIT and that the extracellular domain may not be directly involved in constitutive activation and oncogenic potential of KIT with Asp814 mutations.
Although the Asp814 mutation did not render constitutive receptor dimerization in the extracellular domain, the data presented here suggested that mutant KIT with the Asp814 mutation may not function as a monomeric form. When KITDel-Tyr814 was expressed on Ba/F3 cells, they showed factor-independent growth, albeit to a slightly lesser degree than those expressing KITTyr814. However, the factor-independent growth of Ba/F3 cells by KITDel-Tyr814 was almost completely abrogated by coexpression of KITW42. The dominant-negative effect of KITW42 on the proliferation of Ba/F3Del-Tyr814cells suggested the requirement of receptor association for proper biological function of KITDel-Tyr814. Furthermore, KITDel-Tyr814 was coimmunoprecipitated with either KITWild or KITW42 by ACK2 MoAb directed against the extracellular domain of KIT. Moreover, KITW42 was found to be coimmunoprecipitated with a constitutively activated chimeric FMS/KITTyr814 receptor composed of the ligand-binding and receptor dimerization domain of FMS fused to the transmembrane and cytoplasmic domain of KITTyr814, but not with a ligand-activated chimeric FMS/KITWild receptor. It is therefore possible that self-association of KITTyr814may result from the 814 mutation itself and that the 814 mutation may activate KIT signaling by creating a novel receptor self-association domain that is located not in the extracellular domain, but possibly in the cytoplasmic region. This possibility may be supported by the recent findings by Murali et al51; they analyzed the potential interactions of the cytoplasmic kinase domains of the epidermal growth factor receptor and p185c-neu tyrosine kinases by homology molecular modeling and proposed that their kinase domains can associate as homodimers and heterodimers.
In our previous studies in which the effects of wild-type and mutant KITs on normal hematopoietic stem cells were investigated in vitro and in vivo, the kinase domain mutant of KIT was suggested to transmit signals other than those mediated by an ordinary ligand-receptor interaction and showed a higher tumorigenic activity than the juxtamembrane domain mutant.23 Furthermore, it was reported that c-kitVal814 mutation altered the sites of receptor autophosphorylation and peptide substrate selectivity and also resulted in the degradation of src homology 2 (SH2)-containing protein tyrosine phosphatase SHP1 as well as KITVal814itself.40,52 Because the SHP1 negatively regulates signaling from a number of cytokine receptors, including erythropoietin receptor, IL-3 receptor, and KIT,53-55 the oncogenic activity of KITVal814 and KITTyr814 can be explained in part by the downregulation of SHP1. However, many questions still remain as to how the KIT kinase domain mutants transfer oncogenic signaling into hematopoietic cells. By generating a variety of the Tyr → Phe substitution mutants of this oncoprotein and by using the yeast two-hybrid system, current efforts are directed to identify the downstream effectors that are essential for the transformed phenotype. It is hoped that a better understanding of the signaling cascades delivered from the constitutively activating mutants of KIT will lead to greater insights into normal and abnormal growth controls of hematopoietic cells.
ACKNOWLEDGMENT
The authors thank Dr D. Baltimore (Rockefeller University, New York, NY) for providing the 293T cell line, Dr S. Nagata (Osaka University, Osaka, Japan) for pEF-BOS expression vector, Dr S.-I. Nishikawa (Kyoto University, Kyoto, Japan) for ACK2 MoAb and full length of murine c-kit cDNA, Dr C.J. Sherr (Howard Hughes Medical Institute Research Laboratories, Memphis, TN) for pSMc-fms, Dr D.E. Williams of Immunex Corp (Seattle, WA) for rabbit antiserum against a C-terminal peptide corresponding to the last 10 amino acids of the murine KIT, Dr A. Bernstein (Samuel Lunenfeld Research Institute, Toronto, Ontario, Canada) for rabbit antiserum against a kinase domain of the murine KIT, Dr B. Druker (Oregon Health Sciences University, Portland, OR) for anti-phosphotyrosine MoAb, Kirin Brewery Co Ltd for rmIL-3 and rmSCF, and Yoshitomi Pharmaceutical Industries Ltd for rhM-CSF.
Supported in part by grants from the Japanese Ministry of Education, Science and Culture, the Japanese Ministry of Health and Welfare, the Mochida Memorial Foundation for Medical and Pharmaceutical Research, and the Ryoichi Naito Foundation for Medical Research.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Yuzuru Kanakura, MD, PhD, The Department of Hematology and Oncology, Osaka University Medical School, 2-2, Yamada-oka, Suita, Osaka 565-0871, Japan; e-mail:kanakura@bldon.med.osaka-u.ac.jp.

![Fig. 1. Construction of deletion-type c-kit cDNA. (A) Schematic representation of deletion-type KIT. Location of signal peptide (SP), transmembrane (TM), and tyrosine residue of autophosphorylation (Y) are indicated. KITTyr814 and KITDel-Tyr814 carries a point mutation (Asp to Tyr) in codon 814. (B) Expression of KITDel-Wild and KITDel-Tyr814. 293T cells were transfected with c-kitDel-Wild and c-kitDel-Tyr814 cDNA, and then cells were labeled for 5 hours with [35S]-methionine and lysed. KIT was immunoprecipitated with ACK2 MoAb (ECD), rabbit anti-KITKinase serum (Kinase), and rabbit anti-KITC-terminal serum (C-terminal) and analyzed by SDS-PAGE and autoradiography. The similar results were obtained from three independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/4/10.1182_blood.v93.4.1319/5/m_blod40411001w.jpeg?Expires=1769361446&Signature=sI8Hc-LI0oB3EONDihl5ueGOHbJH22v-XP6NmVfNGMYGgltsy2U6PL9XscZ8ArzDZZTyQfLRokTZaMXkO1rgYy29HJeg2OB5R2e8ggCShNjS-GzfYbJMULZRbZt31ibFnk3Meq2LBStHSBfh~P5y1kpcQ8hYYUYg8tYfXPz~WGRaKVMxlcbNg1j8MO46S2N833uVrgjPVGSujPez7cyggYb0FP-ZjH3qBzW-z4icLkQWZJIe6H~KNHzio-DsAYMR1CRDqtIh6OctjO1D-gIvo7CdEBEfra2tJCnrMCFTyZrCWWf-F4EN-2oxJPUCd5qlcA5No3n5V-D0ICwN5aFzqQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Factor-independent proliferation of Ba/F3Del-Tyr814 cells. Proliferation of Ba/F3, Ba/F3Wild, Ba/F3Tyr814, Ba/F3Del-Wild, and Ba/ F3Del-Tyr814 cells at various concentrations of rmIL-3 (upper panel) or rmSCF (lower panel) was measured with [3H]-thymidine incorporation assay. Each point represents the mean of data from three experiments. Bars are the standard error. In some points, the standard error was too small to be shown by bars.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/4/10.1182_blood.v93.4.1319/5/m_blod40411003x.jpeg?Expires=1769361446&Signature=cVevo6azUmj8JbvdIMak0NHBsmvzac7~DZXLjPN1IeFjDbNVn4M~WYqABcJKFXKTwW31dZ45-Gu1YXJc7jRgrTnPFWcp8p1QjexRzCLcKQf3UsGCL4TTBLp6-nZvtc0y9~3V5Neqpi9Rr53xYtuC6w4E1UAHdfqzKBBsP8N7~ELF5o3xW3ZS9jsyXQ38OD6apjlDyONp4UQpda3CT0~8lF06zU7gxOSD5L~6fB5fM5CtnkEkuJuQ3WjP4GDKAeax~6yErs7m~1eGI7sUb~gjF6RjTIQGLpCYlI1f0GiOXQ97xMousgGNZSaOkvK0g~gH1iQsVenBAaRg2Pa7Stfifw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. Dominant negative effect of KITW42 in Ba/F3Del-Tyr814 cells. (A) Flow cytometric analysis of the surface binding of ACK2 MoAb (ECD). Cells were incubated with either ACK2 MoAb (—) or negative control antibody (---). (B) Viability assays. After plating cells (2 × 104), viable cells were counted at daily intervals using trypan blue dye. Three independent experiments were performed with comparable results, and the result of a representative experiment is shown. Each point represents the mean of triplicate samples. Bars are the standard error. In some points, the standard error was too small to be shown by bars. (C) Incorporation of [3H]-thymidine. Proliferation of cells for rmIL-3 (10 ng/mL) and rmSCF (100 ng/mL) was measured with [3H]-thymidine incorporation assay. Three independent experiments were performed with comparable results, and the result of a representative experiment is shown. Each value represents the mean of triplicate samples, and the standard error was less than 5%. Asterisks indicate the presence of the statistical significance (P < .01) when compared with the value of either Ba/F3Del-Wildor Ba/F3Del-Tyr814 cells at the absence of rmIL-3 and rmSCF.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/4/10.1182_blood.v93.4.1319/5/m_blod40411004x.jpeg?Expires=1769361446&Signature=KdenwGYyYo7WVuomZem1D6DkVgt4N9T36uPcvb0BJzeF-LhUhwgE32D5EKykk0bkptpddOMTtv95d8Dq3ElRN0PjLJFoFxvUWtWEgGIAnag-c2OjB6YLbjmlFHQcfBpXofoTKOjbbCBYcOF8vzcxteq0vLJH1naqbzFMPs1Q2RRtRr8jzCY5I1ugeghOUAV0Oen~yP1qNIgmyRJMYBatQbjhzEXt-KisZr9iCI~h8NAZ0L9HvlVTt3WodA4~~NSeaNIapJHf-qDPvfWhjYOJlry22CMYJczrEtZFOmj58JYtCENa5VJaCyPHaaiFalNEL7I1BkU0x-W7cVRTDa3ICA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. Self-association of KITTyr814. (A) 293T cells were transfected with pEF-BOS containing c-kitWild, pEF-BOS containing c-kitW42, pEF-BOS containing c-kitDel-Wild, or pEF-BOS containing c-kitDel-Tyr814 and cotransfected with pEF-BOS containing c-kitDel-Wild and pEF-BOS containing c-kitWild, pEF-BOS containing c-kitDel-Wild and pEF-BOS containing c-kitW42, pEF-BOS containing c-kitDel-Tyr814 and pEF-BOS containing c-kitWild, or pEF-BOS containing c-kitDel-Tyr814 and pEF-BOS containing c-kitW42. The expression of KIT is shown in upper panel. Cell lysates were immunoprecipitated with rabbit anti-KITKinase serum (Kinase), and then the immunoprecipitates were subjected to SDS-PAGE for 90 minutes and immunoblotting with rabbit Ab-1. The phosphorylation bands of immune complex kinase assay are shown in lower panel. KIT was immunoprecipitated from cell lysates with ACK2 MoAb (ECD), and the immunoprecipitates were incubated with γ-[32P]-ATP. Incorporation of 32P into KIT and/or Del-KIT was visualized by autoradiography. (B) 293T cells were transfected with pEF-BOS containing c-fms/c-kitWild or pEF-BOS containing c-fms/c-kitTyr814 and cotransfected with pEF-BOS containing c-fms/c-kitWild and pEF-BOS containing c-kitW42 or with pEF-BOS containing c-fms/c-kitTyr814 and pEF-BOS containing c-kitW42. The expression of FMS/KIT and/or KITW42 is shown in upper panel. Cell lysates were immunoprecipitated with rabbit anti-KITKinase serum (Kinase) before and after treatment with rhM-CSF, and then the immunoprecipitates were subjected to SDS-PAGE for 3 hours and immunoblotting with rabbit Ab-1. The phosphorylation bands of immune complex kinase assay are shown in lower panel. KIT was immunoprecipitated from cell lysates with 3-4A4 MoAb (FMS-ECD), and the immunoprecipitates were incubated with γ-[32P]-ATP. Incorporation of 32P into FMS/KIT and/or KITW42 was visualized by autoradiography. The similar results were obtained from three independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/93/4/10.1182_blood.v93.4.1319/5/m_blod40411005w.jpeg?Expires=1769361446&Signature=1Hg8wdClJzbEA5-CYME87yyyS9M9E5uCbKa106B8OT1iTuimBaKNmCIv60CGnFlr~O9wq6y7am7C5cU3JJ2RuQO~viKBrFB~BNqTqVSZIwuUrEOcjMc31uOxRxC2KymSa1piLaG5E9E9kBn68bhuw21grEiDeH3rZf8MlJnn10HOdnym6yoc-f2u1leSldFcnamp4Aq~FwJG2yBtlYeFFd3nc5PGSZenjFd0h5Bwi7bglPzhhEjVHogIi9m6OJ-mlGZUbUQcX8TXD8nt71zp3Qh2SaGlk0XUc4kBSUAa0JWi-CsnXF0O3GzeIqZhMemt63qt4fvAa3BejBQZbkf-yQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
