Abstract
A variety of previously published studies have shown the presence of autoantibodies directed against oncogenic proteins in the sera of patients with tumors. Generally the underlying genetic aberration responsible for the induction of an immune response directed against an abnormal protein is unknown. In our studies we analyzed the role of gene amplification in the production of autoantibodies in squamous cell lung carcinoma. We screened a cDNA expression library with autologous patient serum and characterized the isolated cDNA clones encoding tumor expressed antigens termed LCEA (lung carcinoma expressed antigens). As determined by sequence analysis, the 35 identified cDNA clones represent 19 different genes of both known and unknown function. The spectrum of different clones were mapped by polymerase chain reaction (PCR) and fluorescence in-situ hybridization, showing that a majority are located on chromosome 3, which is frequently affected by chromosomal abnormalities in lung cancer. Gene amplification of 14 genes was analyzed by comparative PCR. Nine genes (65% of all analyzed genes) were found to be amplified; furthermore, most of them are also overrepresented in the pool of cDNA clones, suggesting an overexpression in the corresponding tumor. These results strongly suggest that gene amplification is one possible mechanism for the expression of immunoreactive antigens in squamous cell lung carcinoma.
HUMORAL IMMUNE RESPONSES to oncogenic proteins have been reported for different types of cancer and are continuing to be defined. The oncogenic proteins are expressed by cancer cells themselves and act as tumor-expressed antigens. Oncogenes become activated by a variety of mechanisms including point mutation, translocation, and overexpression. Each mechanism of activation can result in the expression of proteins with the potential to serve as tumor antigens.
Autoantibodies directed against mutated oncogenic proteins have been reported for many different tumors. The ras oncogenes, for example, are cancer-related genes that become activated by specific point mutations.1 Studies have shown the existence of antibodies directed against the ras protein containing a single amino acid substitution in patients with colon cancer.2 In sera of patients with various types of cancer, eg, breast cancer, colorectal cancer or lung cancer, antibodies against abnormal p53 protein containing multiple mutational hot spots have been detected.3-5 Besides point mutations, translocations can result in the generation of fusion genes expressing chimeric proteins, which could be the target of an immune system attack. The SSX2 gene, for example, which is involved in a translocation in synovial sarcomas, encodes a tumor antigen.6 Furthermore, overexpressed oncogenic proteins represent potential tumor antigens. C-erbB-2 is one of the genes overexpressed in human tumors, especially in breast carcinoma. Antibodies against the overexpressed oncogenic protein could be detected in several patient sera.7 Autoantibodies against the L-myc oncogene, which is known to be overexpressed in tumors with neuroendocrine characteristics, could be identified in patient sera with small cell lung cancer.8 The overexpression of L-myc in human tumors has frequently been correlated with gene amplification, but an amplified gene itself has never been identified together with the presence of antibodies in the same patient. In contrast, we were able to show previously that the translation initiation factor eIF-4γ, which is encoded by an amplified gene, acts as a tumor expressed antigen in squamous cell lung carcinoma.9
To further elucidate the role of gene amplification as a genetic aberration evoking the production of autoantibodies, we selected squamous cell lung cancer as a model system. Squamous cell lung carcinomas constitute 40% to 50% of lung cancers. This is the most frequent lethal tumor in Western societies, with a median survival time of approximately 6 months and a 5-year survival rate of less than 10%.10 Because of the high frequency of gene amplifications, lung cancer is an ideal neoplasm to analyze downstream effects of gene amplification, especially with regard to the expression of immunoreactive antigens in cancer patients.11 12
For investigation of these effects, we used an immunoscreening approach. A cDNA expression library generated from a squamous cell lung carcinoma was screened with autologous patient serum, and cDNA clones representing tumor-expressed antigens were isolated. Both new and known gene sequences were identified and analyzed for gene amplification. Gene amplification is among the most common genetic changes in human cancer cells.13 There are only a few examples of programmed gene amplification in early development of lower eukaryotic animals, but gene amplification does not occur in normal mammalian cells.14 15 Over the last decade amplification analysis has been largely restricted to known oncogenes. Using the immunoscreening approach it is also possible to detect new immunoreactive tumor antigens whose genes are amplified in cancer cells.
MATERIALS AND METHODS
Tumor tissue.
Informed consent was obtained from the patients for use of their tumors and sera. Tissue samples of squamous cell lung carcinoma were frozen in liquid nitrogen immediately after biopsy or surgery and stored at −70°C. The pathological stage of the tumor L10 was T2, N0, M0, stage I.
DNA and RNA isolation.
Genomic DNA was isolated by standard methods.16 In brief, after proteinase K digestion at 55°C, proteins were extracted with chloroform and DNA was precipitated with isopropanol. The DNA concentration was determined by optical density measurement at 260 nm.
RNA isolation was according to the manufacturer’s instructions (Stratagene, Heidelberg, Germany). Frozen tissue was homogenized, proteins were phenol-chloroform extracted, and RNA was precipitated twice with isopropanol and finally resuspended in diethylpyrocarbonate (DEPC)-treated H2O. Integrity and concentration of RNA was evaluated using formaldehyde gels.
cDNA expression library.
Total RNA was applied to oligo(dT) cellulose push columns and poly(A) mRNA was eluted according to the Poly(A) Quick Kit (Stratagene). cDNA synthesis was performed with the ZAP Express cDNA synthesis kit (Stratagene). In brief, 4.5 μg of poly(A) mRNA was reverse transcribed by Moloney murine leukemia virus (MMLV) reverse transcriptase using an oligo(dT) primer with a 5′XhoI restriction site. The cDNA was ligated toEcoRI adapters, XhoI-digested, size-fractionated with Sephacryl S-500 columns (Stratagene, Heidelberg, Germany) and cloned into ZAP Express vector arms. The vector was packaged using the Gigapack III Gold Packaging Extract (Stratagene). After one round of amplification of the ZAP Express library, the bacteriophage titer was determined to be 1.3 × 1010 plaque-forming units (pfu)/mL.
Preabsorption of patient serum.
Serum was prepared from 10-mL blood samples using serum gel monovettes and stored at −70°C. Serum samples were diluted 1:10 in 1× TBS (Tris buffered saline), 0.5% (wt/vol) low-fat milk, and 0.01% thimerosal before use. Mechanical preabsorption columns were prepared by incubation of sonicated Escherichia coli XL1blue MRF′cells in 1× TBS with Affinity Absorbent (Glutaraldehyde-activated; Boehringer Mannheim, Mannheim, Germany) overnight in BioRad Polyprep chromatography columns (BioRad, München, Germany). Lytic preabsorption columns were prepared in the same way using instead bacteria lysed by nonrecombinant ZAP express phages. The patient serum was preabsorbed using each column type five times. The preabsorbed serum was diluted to a final concentration of 1:100 in 1× TBS, 0.5% (wt/vol) low-fat milk, and 0.01% thimerosal.
Serological screening.
E coli XL1-Blue MRF′ cells were transfected with recombinant bacteriophage containing lung carcinoma cDNA and plated onto NZCYM-Tetracyclin-agar plates (Greiner, Frickenhausen, Germany). After incubation at 42°C for approximately 4.5 hours, isopropyl-β-D-thiogalactopyranoside (IPTG) (10 mmol/L) pretreated filters were layered on the bacteriophage plaques to induce protein expression. After a second incubation at 37°C for 4 to 5 hours, plates were cooled at 4°C overnight. Filters were removed and washed twice for 30 minutes with TBS/0.05% Tween 20. After blocking with 5% (wt/vol) low-fat milk, filters were incubated with autologous patient serum diluted 1:100 for about 4 hours. For the detection of antigen-antibody complexes, filters were overlayered with a solution containing a secondary antibody directed against the constant region of human IgG heavy chain and conjugated with alkaline phosphatase. After the filters were washed three times in TBS, serum-positive clones were visualized by the addition of 5-bromo-4-chloro-3-indoyl-phosphate-p-toluidine salt (BCIP) and nitroblue tetrazolium-chloride (NBT), which are substrates of alkaline phosphatase, resulting in a blue precipitate. For further analysis, serum-positive clones were isolated and inserted into the pBK-CMV phagemid vector by in vivo excision. Some of the clones, for example clone 9-1, were also isolated using a 1:500 dilution of the autologous patient serum.
Sequence analysis.
Sequencing was performed using the Perkin Elmer ABIPrism Cycle sequencing kit (Perkin Elmer, Überlingen, Germany). Clone inserts were sequenced with an automatic sequencer (373A DNA Sequencer; Applied Biosystems, Weiterstadt, Germany). For sequence alignment, clone sequences were compared with known sequences by BLASTN search (BCM search launcher, Human Genome Center, Baylor College of Medicine, Houston, TX).
Chromosomal localization.
Chromosomal localization of isolated cDNA clones was performed by hybrid panel mapping or fluorescence in situ hybridization (FISH). For FISH, 12 ng of biotinylated clone DNA was hybridized against normal metaphase chromosome spreads. Before hybridization, slides with chromosomes were treated with RNase A for 1 hour and pepsin for 10 minutes. After dehydration of chromosomes in ethanol, both chromosomes and biotinylated probe were denatured in 50% formamide solution at 80°C. Slides were incubated at 37°C overnight. After washes in 50% formamide solution at 45°C and in 0.1× sodium chloride-sodium citrate (SSC) at 60°C, biotinylated probes were detected using avidin conjugated to fluorescein isothiocyanate. Three rounds of amplification were performed by using goat anti-avidin antibodies. Slides were counterstained with 4,6-diamino-2-phenyl-indol (DAPI). Fluorescence signals were visualized in an Olympus microscope (Olympus, Hamburg, Germany) and analyzed and documented with the program ISIS3 of Metasystems (Altussheim, Germany).
Alternatively, panels of multichromosomal somatic cell hybrids (hybrid mapping panel 1) and monochromosomal somatic cell hybrids (hybrid mapping panel 2) from the Coriell Institute for Medical Research (Camden, NJ) were used for chromosomal localization by polymerase chain reaction (PCR). The human chromosome content of these cell hybrids has been characterized by Drwinga et al17 and Dubois and Naylor.18 The PCR conditions for each sequence specific primer pair are summarized in Table 2.
Comparative PCR.
The concentration of peripheral blood DNA, as control DNA, and tumor DNA was determined by optical density measurement at 260 nm. After preparation of a master solution containing 5 ng/μL of each DNA sample, three different DNA dilutions (2 ng/μL, 1 ng/μL, 0.5 ng/μL) were generated to monitor the linear range of amplification during PCR.19 Before amplification analysis, the three dilutions of each DNA sample were calibrated using sequence-specific primers for the MUC gene. A successful calibration results in PCR products of equal intensity in tumor and blood DNA. After successful calibration of template DNA, the amplification status of the LCEA clones was determined using specific primer pairs. Gene amplification results in increased signal intensities of all three dilutions of a tumor DNA sample. PCR was performed with 5 minutes of initial denaturation at 94°C, for 25 cycles, with 1 minute of denaturation at 94°C, 1 minute of annealing at temperatures given in Table 2, 1 minute of extension at 72°C followed by 10 minutes of final extension at 72°C in an M.J. Research Minicycler PTC-150 and PTC-100 (M.J. Research, Watertown, MA). All PCR reactions were performed in a total volume of 50 μL with 0.5 μmol/L of each primer, 200 μmol/L deoxynucleotide triphosphates, and 2.5 U of Taq polymerase (Pharmacia, Freiburg, Germany) in PCR buffer. The PCR conditions are summarized in Table 2.
RESULTS
Identification of cDNA clones representing tumor-expressed antigens.
A cDNA expression library was generated from a squamous cell lung carcinoma, which has previously been shown to harbor amplification units on several different chromosomes by reverse chromosome painting and comparative genomic hybridization.16 Therefore, enriched poly(A)mRNA was reverse transcribed into cDNA, inserted into the ZAP Express expression vector (Stratagene) in the sense orientation with respect to the lacZ promoter and transfected into E colicells. After induction of polypeptide synthesis, this expression library was screened with autologous patient serum to identify cDNA clones representing tumor expressed antigens. Antigen-antibody complexes were detected by a secondary antibody binding to the constant region of the human IgG heavy chain. By screening a total of 800,000 clones, we identified 35 positive cDNA clones. The positive clones that encode immunoreactive antigens, termed lung carcinoma expressed antigens (LCEA), were isolated and subjected to a second round of screening for verification and enrichment. Figure1 shows the identification of positive clones in the primary screening and the enrichment during the second round of screening.
Screening of a cDNA expression library generated from a squamous cell lung carcinoma with autologous serum. Recombinant proteins were screened with preabsorbed patient serum (see Materials and Methods) and antigen-antibody complexes were detected by a color reaction. As shown for clones 11-4 and 26-4, the serum-positive clones were identified in a primary screening (left-hand side). The enrichment of serum-positive clones by replating and subjection to a second round of screening is shown on the right-hand side.
Screening of a cDNA expression library generated from a squamous cell lung carcinoma with autologous serum. Recombinant proteins were screened with preabsorbed patient serum (see Materials and Methods) and antigen-antibody complexes were detected by a color reaction. As shown for clones 11-4 and 26-4, the serum-positive clones were identified in a primary screening (left-hand side). The enrichment of serum-positive clones by replating and subjection to a second round of screening is shown on the right-hand side.
Positive clones were isolated and partially or completely sequenced from both ends using vector- and sequence-specific primers. Sequences were analyzed by comparison with each other and known sequences of different databases.
Grouping of identified cDNA clones.
Sequence analysis showed that the 35 isolated cDNA clones represent 19 different genes including both known and unidentified sequences. As documented in Table 1, the 35 LCEA encoding cDNA clones were grouped depending on their sequence homology to known genes or to each other. Group 1 is represented by six clones encoding the gene for human translation initiation factor eIF-4γ.20 These clones represent almost 20% of all isolated cDNA clones, indicating the relevance of this translation initiation factor as a tumor-expressed antigen in squamous cell lung carcinoma. Our preliminary expression data indicate an mRNA overexpression of the eIF-4γ gene. The second group is represented by 5 clones with homology to the gene for human DnaJ-protein homologue heat-shock protein.21 Group number 3 consists of 3 clones identical with the MGEA5 gene, group number 4 of 2 clones homologous to the RBPJK gene, group number 5 of 2 clones homologous to the NNP-1 mRNA, and group number 6 of 2 clones identical with the human transcriptional repressor (GCF2) mRNA.22-25 In group 7 we summarized both individual cDNA clones representing different genes of known function, for example the gene for protein kinase p160ROCK, the gene for human transcription factor NFATc.b and the gene for a human ring zinc-finger protein.26 27 The clones 9-1/46-1 and 11-2/54-2 represent homologous cDNA sequences encoding new genes, whereas the cDNA sequences of the successive clones show homology to known genes with unknown function.
Chromosomal mapping of identified cDNA clones.
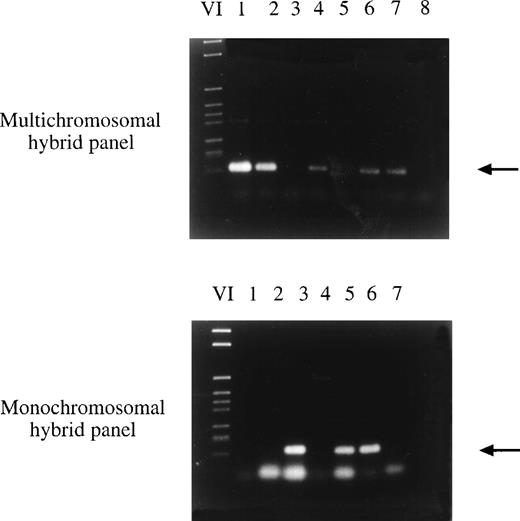
The newly isolated LCEA encoding clones were assigned to human chromosomes by somatic cell hybrid panel mapping. Using sequence-specific primer pairs and DNA of hybrid cell lines with one or several human chromosomes in front of a rodent background, genomic mapping was possible by PCR (Fig 2). The PCR conditions are listed in Table 2. Additionally, the exact chromosomal localization was determined by FISH. The results of the chromosomal localization are summarized in Table 3. In total, 11 cDNA clones representing 4 different genes were mapped on chromosome 3. For 3 clones representing 3 different genes, the exact localization has been determined by FISH showing in all cases a localization to the long arm of chromosome 3, namely at chromosomal bands 3q25, 3q26.3-qter, and 3q27. The fact that 11 of 32 clones representing 4 of 16 genes are located on chromosome 3 and, as shown by FISH, in the terminal region of the chromosomal q-arm, suggests a correlation between this region on chromosome 3 and the production of tumor expressed antigens. Furthermore, two different cDNA clones were mapped to chromosome 14 and two were mapped to chromosome 10. The chromosomal localization of 8 additional clones was mapped to chromosomes 2, 6, 9, 15, 17, 18, 20, and 21. For the remaining cDNA clones the localization has not yet been determined because of the difficulty in finding appropriate primer pairs for hybrid panel mapping.
Chromosomal localization of clone 59-2 by hybrid panel mapping. Sequence-specific primers were used in a PCR reaction with DNA of hybrid cell lines. In the first PCR reaction, DNA of multichromosomal somatic cell hybrids with several human chromosomes in a rodent chromosome background was amplified in the PCR reaction (top part of the figure). Finally the exact localization was determined using DNA of monochromosomal somatic cell hybrids with one or several human chromosomes in a rodent chromosome background (bottom part of the figure). Clone 59-2 was mapped to chromosome 6. (Top) VI, DNA marker; lane 1, NA09927; lane 2, NA09929; lane 3, NA 09931; lane 4, NA09935A; lane 5, NA00347A (hamster DNA); lane 6, NAIMR91 (human genomic DNA); lane 7, human blood DNA; lane 8, no DNA. (Bottom) VI, DNA marker; lane 1, NA10253 (chromosome 3); lane 2, NA10115 (chromosome 4); lane 3, NA10629 (chromosome 6); lane 4, NA00347A (hamster DNA); lane 5, NAIMR91 (human genomic DNA); lane 6, human blood DNA; lane 7, no DNA.
Chromosomal localization of clone 59-2 by hybrid panel mapping. Sequence-specific primers were used in a PCR reaction with DNA of hybrid cell lines. In the first PCR reaction, DNA of multichromosomal somatic cell hybrids with several human chromosomes in a rodent chromosome background was amplified in the PCR reaction (top part of the figure). Finally the exact localization was determined using DNA of monochromosomal somatic cell hybrids with one or several human chromosomes in a rodent chromosome background (bottom part of the figure). Clone 59-2 was mapped to chromosome 6. (Top) VI, DNA marker; lane 1, NA09927; lane 2, NA09929; lane 3, NA 09931; lane 4, NA09935A; lane 5, NA00347A (hamster DNA); lane 6, NAIMR91 (human genomic DNA); lane 7, human blood DNA; lane 8, no DNA. (Bottom) VI, DNA marker; lane 1, NA10253 (chromosome 3); lane 2, NA10115 (chromosome 4); lane 3, NA10629 (chromosome 6); lane 4, NA00347A (hamster DNA); lane 5, NAIMR91 (human genomic DNA); lane 6, human blood DNA; lane 7, no DNA.
In summary, the localization of the isolated cDNA clones correlates well with the localization of amplified regions in the tumor sample used to generate the cDNA library. Previously, we have been able to identify, in this tumor sample, amplifications on chromosome 3q26, 6p21.1, 1q, 5p, and 14 by reverse chromosome painting and one-color comparative genomic hybridization.28
Analysis of gene amplification of identified cDNA clones.
To analyze the amplification status of the identified and isolated cDNA clones, we chose comparative PCR, which is a very sensitive technique to detect gene amplifications from small amounts of tumor DNA.19 In the first step of comparative PCR one control DNA and several tumor DNA samples are calibrated using sequence-specific primers from an unamplified chromosomal region. In the second step, primers specific for a potentially amplified gene are used in the PCR. Amplifications are indicated by increased signal intensities in the tumor DNA compared with the control DNA, as shown in Fig3. A gene is considered to be amplified if the signal intensities in all three tumor samples are increased in comparison to the signal intensity of the corresponding dilutions of the blood DNA. Although this method is highly suited to detect gene amplification, it does not allow a quantification of the amplification level. Of the 14 analyzed LCEA encoding clones, 9 were found to be amplified in the tumor DNA which had been used for generating the cDNA library. This translates into an amplification frequency of 65%. Most of the amplified genes are represented by more than one cDNA clone, for example the 6 cDNA clones homologous to the gene for human translation initiation factor eIF-4γ (group 1), the 5 cDNA clones of group 2 encoding a human heat shock protein, the two clones of group 4 showing homology to the gene for Jk-recombination signal binding protein, and the clones 9-1/46-1 and 11-2/54-2 (group 7) encoding tumor-expressed genes of unknown function. With one exception, all clones that did not show gene amplification of tumor DNA are representated by a single cDNA clone isolated from the expression library. Representative results of the amplification analysis are shown in Fig 3. The results concerning the determination of the amplification status of the various clones are summarized in Table 3. The gene for eIF-4γ has been found to be amplified in independent tumor samples (data not shown).
Amplification analysis of clones 26-4, 96-5, and 129-1 in squamous cell lung carcinomas L115, L10, and L116 by comparative PCR. Three different concentrations of tumor and blood DNA were amplified each by PCR using primers specific for clones 26-4, 96-5, and 129-1 and the MUC gene. The PCR products were separated by gel electrophoresis and visualized by SYBR green I. While the signal intensities of tumor and peripheral blood DNA (PB) were comparable using primers specific for the MUC gene, the signals of all three clones 26-4, 96-5, and 129-1 were significantly increased in tumor DNA L10 versus normal DNA indicating amplification of the corresponding genes in tumor L10. This tumor was used to generate the cDNA expression library. Lower amplification levels or the absence of amplification are observed for tumors L115 and L116, respectively. Lane 1, DNA marker; lanes 2 through 4, three dilutions of blood DNA; lanes 5 through 7, three dilutions of tumor DNA L115; lanes 8 through 10, three dilutions of tumor DNA L10; lanes 11 through 13, three dilutions of tumor DNA L116; lane 14, no DNA.
Amplification analysis of clones 26-4, 96-5, and 129-1 in squamous cell lung carcinomas L115, L10, and L116 by comparative PCR. Three different concentrations of tumor and blood DNA were amplified each by PCR using primers specific for clones 26-4, 96-5, and 129-1 and the MUC gene. The PCR products were separated by gel electrophoresis and visualized by SYBR green I. While the signal intensities of tumor and peripheral blood DNA (PB) were comparable using primers specific for the MUC gene, the signals of all three clones 26-4, 96-5, and 129-1 were significantly increased in tumor DNA L10 versus normal DNA indicating amplification of the corresponding genes in tumor L10. This tumor was used to generate the cDNA expression library. Lower amplification levels or the absence of amplification are observed for tumors L115 and L116, respectively. Lane 1, DNA marker; lanes 2 through 4, three dilutions of blood DNA; lanes 5 through 7, three dilutions of tumor DNA L115; lanes 8 through 10, three dilutions of tumor DNA L10; lanes 11 through 13, three dilutions of tumor DNA L116; lane 14, no DNA.
In summary, we were able to isolate, together with our previously published results, 35 cDNA clones representing 19 LCEA encoding genes. These data show that squamous cell lung carcinoma expresses a wide variety of immunogenic antigens. Many cDNA clones encoding tumor-expressed antigens could be mapped to chromosome 3, a chromosome that is frequently involved in chromosomal aberrations, especially in lung cancer. Furthermore, a great majority of cDNA clones are amplified in the tumor sample used to generate the expression library. Additionally, most of them are overrepresented in the pool of isolated cDNA clones, suggesting an overexpression at the RNA level. Unfortunately, an expression analysis was impossible because of a lack of tumor tissue. These results strongly suggest that gene amplification may be a general mechanism for overexpression of tumor-expressed antigens, which induce a humoral immune response resulting in the production of autoantibodies.
DISCUSSION
As an early event during lung cancer development, alterations in oncogenes and/or tumor suppressor genes may lead to the production of mutant forms or abnormal levels of proteins within tumor cells or on tumor cell membranes. Recognition by the immune system is possible through presentation on the cell surface. Alternatively, there may be a release of proteins from damaged cancer cells due to spontaneous tumor necrosis resulting in antigen presentation via complex formation with other proteins such as heat shock proteins. To detect the humoral response to oncogenic proteins in lung cancer, we generated a cDNA library from a squamous cell lung carcinoma, expressed the cDNA clones as recombinant fusion proteins, and screened them with autologous patient serum. An important advantage of this screening approach is the possibility of detecting antigens that are encoded by both known and unknown genes. Moreover, this technique enabled us to identify multiple antigens, which correlates with the suggestion that immunogenicity of human tumors is not caused by a single antigen.
The majority of isolated cDNA clones encoding tumor-expressed antigens represent genes that are known to be involved in tumor formation, supporting the idea that oncogenic proteins induce an immune system response. The translation initiation factor eIF-4γ, for example, is a critical link between mRNAs and the 40s ribosomal subunit during the initial steps of translation.29,30 Overproduction of this factor has been shown to form transformed foci on a monolayer of cells, to cause anchorage-independent cell growth, and to form tumors in nude mice.31 Increased levels of eIF-4γ protein, which is the limiting factor of translation initiation in normal cells, may be responsible for the production of autoantibodies by B cells.9 Our preliminary data indicate a correlation between gene amplification and mRNA overexpression of the eIF-4γ gene. Another interesting antigen we identified is the Rho-associated, coiled-coil containing protein kinase p160ROCK that functionally associates with the guanosine triphosphate (GTP)-bound Rho-protein.32 In addition to organizing specific actin cytoskeletons, recent studies have shown that Rho is involved in cell-cycle progression and in cell transformation.33 34 Because of its functional association with Rho-protein, it can be speculated that p160ROCK is also involved in tumor formation.
In addition to the above-mentioned antigens, we also isolated a DnaJ-homologue heat-shock protein that is an inducer of a humoral immune response. This DnaJ-like protein is involved in the regulation of HSP70 heat-shock protein which plays a major role in antigen presentation.35 It has been shown that in antibody-elicting breast tumors, a 70-kD heat-shock proteins forms complexes with p53, whereas none of the antibody-negative tumors contained any of these complexes.36 This implies that heat-shock proteins are involved in the antigenic presentation of p53. This observation has been supported by further studies which identified antibodies directed against p53 protein complexed with HSP70 protein in sera from oral cancer patients.37 Because of the results of these investigations, we suppose that the DnaJ-homologue heat-shock protein may also be involved in antigen presentation in lung cancer. Additionally, we isolated several cDNA clones encoding unknown proteins. The role of these antigens needs to be determined.
In addition to isolating cDNA clones encoding tumor-expressed antigens in squamous cell lung carcinoma, we also determined their chromosomal localization by hybrid panel mapping and FISH. Most of the isolated cDNA clones could be mapped to the long arm of chromosome 3 (3q25-qter). It is of great significance that this chromosomal region is most frequently involved in chromosomal imbalances in squamous cell lung carcinoma. More than one third of these tumors show DNA sequence amplifications of the distal part of chromosome 3q, which may serve as a source for genes encoding tumor-expressed antigens through overexpression or abnormal protein formation. Recently, we reported an amplicon localized at 3q26 containing the genes BCHE and SCL2A2 which were amplified in 40% of squamous cell lung carcinomas.12,28,38 39
In general, DNA amplification is frequent in squamous cell lung carcinoma and may result in immunoreactive antigens via increased protein levels without additional DNA mutations. In our study, we showed that the majority of immunogenic antigens expressed in the analyzed squamous cell lung carcinoma are encoded by amplified genes. It is likely that at least some of the genes amplified in tumor L10 are also frequently amplified in other squamous cell lung carcinomas. Our results strongly support the hypothesis that DNA amplification is a major mechanism provoking the expression of immunoreactive autoantigens in this type of lung cancer. This is the first report presenting a detailed investigation of a correlation between gene amplification, overrepresentation of cloned tumor antigen genes, and the production of autoantibodies in human cancer. Additional genetic aberrations may be responsible for the expression of antigens which are not encoded by amplified genes.
Newly isolated antigens are likely to be useful as novel prognostic parameters of squamous cell lung carcinoma, especially in the light of a poor prognosis after late diagnosis. Generally, the presence of autoantibodies directed against oncogenic proteins in cancer patient sera predict a poor prognosis.40 However, several studies showed that antibodies are already detectable in sera of patients without obvious tumor formation. The presence of antibodies against p53 have been detected in the serum of patients several months before the diagnosis of a squamous cell lung carcinoma, proposing that the detection of specific autoantibodies may be an ideal tool for early diagnosis, especially for early detection of squamous cell lung carcinoma.5
Supported by grants from the Deutsche Krebshilfe (Grant No. 10-11-1-Me2) and from the Deutsche Forschungsgemeinschaft (SFB 399, A1). A.R. was a recipient of a grant from the Landesgraduierten Förderungsprogramm des Saarlandes.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Eckart Meese, PhD, Department of Human Genetics, Medical School, University of Saarland, Building 60, 66421 Homburg/Saar, Germany; e-mail: hgemee@med-rz.uni-sb.de.