The p73 gene, a member of the p53 family, is a new candidate tumor suppressor gene. To investigate the possibility of genetic alteration of p73 in leukemia and lymphoma, we examined 55 cell lines and 39 patient samples together with 17 nonhematopoietic cancer cell lines. Gene expression of p73 was detected by reverse transcriptase-polymerase chain reaction (RT-PCR) in cell lines (5 of 7 pre B/B-acute lymphoblastic leukemia [ALL], 13 of 21 T-ALL/lymphoblastic lymphomas [LBL], 9 of 10 B–non-Hodgkin’s lymphomas [B-NHL], 8 of 9 acute myelogenous leukemias [AML], 2 of 2 T-NHL, 3 of 3 multiple myeloma), and in patient samples (16 of 23 pre B-ALL, 5 of 8 T-ALL/LBL, 5 of 8 B-NHL). PCR–single-strand conformation polymorphism (SSCP) of cDNAs showed no mutation in 43p73-expressing cell lines within the regions that corresponded to the 5 mutational hotspots of the p53 gene. Neither homologous deletion nor rearrangement of the p73 gene were found by Southern blot analysis in any of the cell lines that lack expression of p73. In contrast to prior published data, analysis of a polymorphic site showed that the p73 gene was expressed biallelically in cell lines and normal peripheral blood. Notably, the p73-negative cell lines were hypermethylated at a CpG island in the 5′ untranslated region of the p73 mRNA, and treatment of these cell lines with 5-azacytidine (5-AC), a demethylation reagent, induced p73 expression. Taken together, we found that a sizable proportion (32%) of ALL/B-NHL cell lines and primary tumors had negligible or limited expression of the p73gene associated with hypermethylation of the gene. These findings suggest that silencing of the p73 gene by hypermethylation may contribute to development and/or progression of lymphoid neoplasms.
MORE THAN HALF of all human cancers possess alterations of the p53 gene.1-5 Wild-type p53 is considered to be the guardian of the genome because the protein markedly increases with DNA injury, resulting in G0/G1 and G2/M cell-cycle arrest. If DNA injury is too severe, p53 helps to mediate apoptosis. In contrast, mutant p53 cannot limit cells to a G0/G1 arrest resulting in unrepaired DNA being bequethed to daughter cells.5 Recently, a new member of the p53 gene family has been isolated, known as p73.6 The DNA binding, transactivation, and oligomerization domains of p73are similar to those of p53. Overexpression of p73 can induce the cyclin-dependent kinase inhibitor known as p21waf1 and cause apoptosis in the p53-negative osteosarcoma cell line, SAOS-2.7 p73 can also activate p53-responsive promotors, and mutant, but not wild-type, p53 can bind to p73 and inhibit the ability of p73 to both activate p53-responsive promotors and induce apoptosis.8 Taken together, investigators have suggested that p73 may be a new tumor suppressor.9
Key tumor suppressor genes altered in cancers include p53,p15INK4B, p16INK4A, andRb. These genes can be inactivated by mutation, deletion, and hypermethylation. Each of these tumor suppressors has been reported to be altered in lymphoid malignancies.10-13 Furthermore, alterations of the p53, p16INK4A, andRb genes have been reported to be associated with an unfavorable prognosis of acute lymphoblastic leukemias (ALLs)14-16 and B–non-Hodgkin’s lymphomas (B-NHLs).17-19 To investigate whether p73 is altered in lymphoid malignancies, we examined cell lines from a variety of hematopoietic malignancies, and because we detected abnormalities in lymphoid cell lines, patient samples of ALL/B-NHL were also examined. A sizable number of lymphomas and leukemias had negligible or only limited expression of the p73 gene. This report implicates hypermethylation as an important mechanism in regulating p73expression in ALLs/B-NHLs.
MATERIALS AND METHODS
Cell lines and primary samples.
Cell lines were cultured in RPMI 1640 or Dulbecco’s modified Eagle medium (DMEM) with 10% fetal bovine serum (Life Technologies, Gaithersburg, MD). The cell lines listed in Tables 1 and 2 (21 T-ALL/lymphoblastic lymphoma [LBL], 5 pre B-ALL, 2 B-ALL, 2 T-NHL, 10 B-NHL, 3 multiple myeloma, 9 acute myelogenous leukemia [AML], 3 CML-blastic crisis [CML-BC], and 17 nonhematopoietic cancers) were either established by our labs20,21 or obtained from American Type Culture Collection (ATCC; Rockville, MD). Bone marrow and peripheral blood samples were obtained after informed consent from individuals with lymphoproliferative disorders including T-ALL/LBL (8 samples) and pre B-ALL (23 samples). Mononuclear cells were purified by density gradient centrifugation from bone marrow and peripheral blood cells, and immediately frozen in liquid nitrogen. Lymph node tissues were obtained at the time of surgery from the surgical pathology division of UCLA Center for the Health Sciences. Tissue blocks containing malignant lymphoma (confirmed by histologic evaluation of parallel sections) were snap frozen in liquid nitrogen immediately after excision, and were used for RNA extraction. In most cases, the lymphoma had nearly completely effaced the normal architecture of the lymph node. Lymphomas were classified according to the Revised European-American classification of lymphoid neoplasms (REAL),22 and included 2 cases of follicle center lymphoma, follicular, grade I, 1 case of follicle center lymphoma, follicular, grade II, 2 cases of small lymphocytic lymphoma, 1 case of lymphoplasmacytoid lymphoma, and 1 case of mantle cell lymphoma.
Plasmid vectors.
A 2-kbp fragment of the p73 gene was amplified by reverse transcriptase-polymerase chain reaction (RT-PCR) using Pfu polymerase (Stratagene, La Jolla, CA) from the cDNA of the osteosarcoma cell line, U2OS. The primers used for amplification were 5′-GGGGTACCGGCGTGGGGAAGATGGCCCAGT-3′ and 5′-GCTCTAGATCAGTGGATCTCGGCCTCCGT-3′. The amplified fragment was gel purified and cloned into the KpnI-XbaI site of the pBS-M13 vector (Stratagene).
RT-PCR.
Total RNA was extracted using the acid-guanidinium-phenol-chloroform (AGPC) method,23 and cDNA was synthesized from 5 μg of total RNA using Moloney murine leukemia virus (M-MLV) Reverse Transcriptase (Life Technologies) in 50 μL of reaction solution according to the manufacturer’s instructions. RT-PCR was performed using 10 pmol of each primer, 250 μmol/L of dNTP mix, 10% dimethyl sulfoxide, 2 U of Taq Polymerase (Life Technologies), 1X PCR buffer (Life Technologies), 150 μmol/L of MgCl2, and 1 μL of cDNA. Primers used for p73 amplification were 5′-GACGGAATTCACCACCATCCT-3′ (primer C) and 5′-CCAGGCTCTCTTTCAGCTTCA-3′ (primer D). The PCR conditions were 1 cycle of 3 minutes at 95°C, 35 cycles of 30 seconds at 95°C, 40 seconds at 60°C, 40 seconds at 72°C, and 1 cycle of 3 minutes at 72°C. The quality of cDNA was confirmed by parallel PCR amplification of the GAPDH gene. Primers for GAPDHwere 5′-CCATGGAGAAGGCTGGGG-3′ and 5′-CAAAGTTGTCATGGATGACC-3′, and the PCR conditions were 1 cycle of 3 minutes at 95°C, 28 cycles of 30 seconds at 95°C, 40 seconds at 58°C, 40 seconds at 72°C, and 1 cycle of 3 minutes at 72°C. PCR products were run on ethidium-bromide (Et-Br)-stained 2% agarose gels and then transferred to Hybond N+ filters (Amersham, Buckingamshire, UK). Membranes were probed with a [32P]αdCTP-labeled 2-kbp XbaI/KpnI fragment excised and purified from pBS-M13-p73 for p73 and a 0.2-kbp fragment corresponding to the PCR product for GAPDHusing random priming method, and visualized by autoradiography. For standardization, the photodensity of each p73 band was divided by that of the corresponding GAPDH band using an Alpha Imager system (Alpha Innotech, San Leandro, CA), and the expression ratio ofp73/GAPDH for each sample was compared to the ratio of these two transcripts in HL60. We used HL60 as a standard because thep73 expression of HL60 was close to that of peripheral blood mononuclear cells from 4 normal volunteers.
To examine the allelic expression of p73, RT-PCR products were phenol/chloroform extracted, digested with NlaIII (New England Biolab, Beverly ,MA), run on a 2% gel, and visualized by staining with Et-Br.
Southern blot analysis.
Genomic DNAs were extracted by standard methods.24 To detect the deletion of the p73 gene locus, 10 μg of genomic DNA from each sample was digested with 30 U of EcoRI (Life Technologies), BamHI (Life Technologies), or HindIII (Life Technologies) at 37°C overnight. After fractionation on a 0.8% agarose gel, DNA was transferred onto a Hybond N+ membrane. For probes, a 0.8-kbp EcoRI fragment of pBS-M13-p73, which includes exons 2 through 6, and a 390-bp fragment of p73 amplified by PCR with primers C and D (probe CD) , which includes exons 7 through 9, were labeled by random priming with [32P]αdCTP. After hybridization, the membranes were washed stringently and exposed to a Kodak X-OMAT film (Eastman Kodak, Rochester, NY) for 1 to 5 days.
PCR–single-strand conformation polymorphism (SSCP) and nucleotide sequencing.
Forty-three cDNA samples from p73-positive cell lines were screened for mutations by PCR-SSCP analysis. The RT-PCR conditions described above were used to amplify the cDNA in the presence of [32P]αdCTP. This RT-PCR product spans exons 7 through 9, and includes the codons homologous to 5 mutational hotspots (codons 245, 248, 249, 273, and 282) of the p53 gene.1 The PCR products were denatured and run in a 6% Hydrolink MDE gel (J.T. Baker, Phillipsburg, NJ) as described by the manufacturer. The PCR products that showed representative migration patterns on the gel were purified, and cloned into a pGEM vector (Promega, Madison, WI). Sequencing was performed using the Original Taq DyeDeoxy Terminator Cycle Sequencing Kit (Perkin-Elmer, Warrington, UK) and analyzed using an ABI automated sequencer (Perkin-Elmer).
Methylation analyses.
To assess the methylation status of the p73 gene locus, 0.2 μg of genomic DNA from cell lines or normal PBMC was digested with 20 U of either methylcytosine sensitive enzyme, HpaII (Promega), or its methylation resistant isoschizomer, MspI (Promega), for 3 hours at 37°C. The digests were phenol/chloroform extracted, ethanol precipitated, dried, and resuspended to 10 μL with 1X Tris-EDTA (TE) buffer. One tenth of the solution (20 ng of DNA) was amplified by PCR using primers: 5′-GGGGACGCAGCGAAACCG-3′ and 5′-CTGCAGCCGTCGCAGCC-3′, which can amplify the potential CpG island in exon 1. The PCR conditions were 1 cycle of 3 minutes at 95°C, 30 cycles of 30 seconds at 95°C, 40 seconds at 61°C, 40 seconds at 72°C, and 1 cycle of 3 minutes at 72°C. PCR products were run on a 3% gel and visualized by staining with Et-Br.
Cell lines that did not express p73 were cultured with 5-azacytidine (5-AC; Sigma, St Louis, MO; a demethylation reagent), to attempt to induce expression of p73. The U2OS, HL60, CEM, ALL-Sil, and KOPT-K1 cell lines were cultured either with or without 5-AC at 0.5 to 3 μmol/L for 3 to 5 days.
RESULTS
Expression of p73 in hematopoietic malignancies.
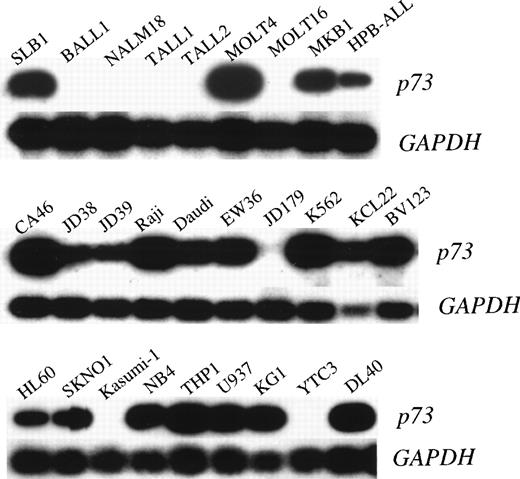
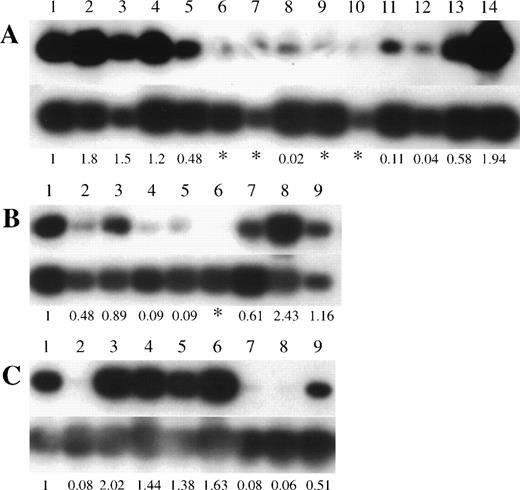
To investigate the possibility of genetic alteration of p73 in leukemias/lymphomas, we performed semiquantitative RT-PCR in cell lines and clinical samples. Each RT-PCR was repeated 2 or 3 times for each sample with consistent results. Representative results are shown in Fig 1 and the data are summarized in Tables 1 and 2. Thirteen of 21 T-ALL/LBL, 5 of 7 pre-B/B-ALL, 9 of 10 B-NHL, and 8 of 9 AML cell lines were positive for p73 expression by RT-PCR, whilep73 expression was detectable in all nonhematopoietic cancer cell lines (17 of 17). We also examined the expression of p73mRNA in fresh ALLs and B-NHLs from patients. Representative results are shown in Fig 2 and the data are summarized in Table 3. We found p73-positive samples in 16 of 23 pre–B-ALL and 5 of 8 T-ALL/LBL, and 5 of 8 B-NHL, while 13 of 39 samples (33%) of ALLs/B-NHLs had negligible or low expression of p73 in total. Low expression was arbitrarily set at less than 1/10 of the level in HL60 cells (<0.1), and an expression level of less than 1/100 of that in the HL60 cells (<0.01) was regarded as negligible. Because most samples had some contamination with normal lymphocytes that express p73 mRNA, this cutoff selected against false positives. Taken together, a sizable proportion (32%, 25 of 77) of ALL/B-NHL cell lines and primary tumors had negligible or limited expression of p73 gene.
p73 expression in pre B/B-ALL, T-ALL/LBL, and B-NHL cell lines. Autoradiography of RT-PCR products (390 bp) of thep73 gene is shown in the top panels and GAPDH in the bottom panels.
p73 expression in pre B/B-ALL, T-ALL/LBL, and B-NHL cell lines. Autoradiography of RT-PCR products (390 bp) of thep73 gene is shown in the top panels and GAPDH in the bottom panels.
p73 gene expression in pre B/B-ALL, T-ALL/LBL, and B-NHL clinical samples. Autoradiography of RT-PCR products (390 bp) of the p73 gene is shown in the top panels and GAPDH in the bottom panels. (A) Lane 1, HL60; lanes 2 through 5, peripheral blood mononuclear cells from 4 volunteers; lanes 6 through 14, pre B-ALL samples. (B) Lane 1, HL60; lanes 2 through 9, T-ALL/LBL samples. (C) Lane 1, HL60; lanes 2 through 9, B-NHL samples (lane 2, follicle center lymphoma, follicular, grade I; lane 3, diffuse large B-cell lymphoma; lane 4, lymphoplasmacytiod lymphoma; lane 5, small lymphocytic lymphoma; lane 6, small lymphocytic lymphoma; lane 7, mantle cell lymphoma; lane 8, follicle center lymphoma, follicular, grade II; lane 9, follicle center lymphoma, follicular, grade I). Numbers below the lanes represent the ratio ofp73 expression compared with expression in HL60 cells. The asterisk (*) indicates p73 expression was less than 1/100 of the level observed in HL60 cells.
p73 gene expression in pre B/B-ALL, T-ALL/LBL, and B-NHL clinical samples. Autoradiography of RT-PCR products (390 bp) of the p73 gene is shown in the top panels and GAPDH in the bottom panels. (A) Lane 1, HL60; lanes 2 through 5, peripheral blood mononuclear cells from 4 volunteers; lanes 6 through 14, pre B-ALL samples. (B) Lane 1, HL60; lanes 2 through 9, T-ALL/LBL samples. (C) Lane 1, HL60; lanes 2 through 9, B-NHL samples (lane 2, follicle center lymphoma, follicular, grade I; lane 3, diffuse large B-cell lymphoma; lane 4, lymphoplasmacytiod lymphoma; lane 5, small lymphocytic lymphoma; lane 6, small lymphocytic lymphoma; lane 7, mantle cell lymphoma; lane 8, follicle center lymphoma, follicular, grade II; lane 9, follicle center lymphoma, follicular, grade I). Numbers below the lanes represent the ratio ofp73 expression compared with expression in HL60 cells. The asterisk (*) indicates p73 expression was less than 1/100 of the level observed in HL60 cells.
Mutation, deletion, and polymorphism of the p73 gene.
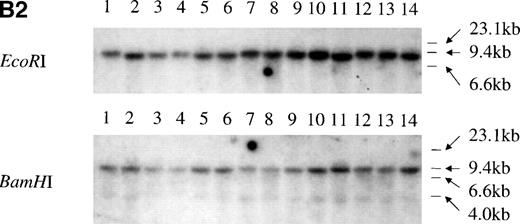
We performed PCR-SSCP analysis with cDNAs to identify possible point mutations. The primers encompassed the regions corresponding to the 5 mutational hotspots of the p53 gene, which are frequently altered in various cancers, including hematopoietic malignancies.1 Representative data are shown in Fig 3A. No mutations in 43p73-positive cell lines from both hematopoietic and nonhematopoietic lineages were detected, strongly suggesting that mutation of the p73 gene might be rare. Two previously identified polymorphic sites, codon 336 (GCC to GCT) and codon 349 (CAT to CAC), were identified by sequencing of the RT-PCR products.25 26 For the p73-nonexpressing cell lines, we performed Southern blot analysis of the p73 gene locus using 2 different probes, but no alteration of the p73gene structure was observed (Fig 3B). Taken together, the data suggest that mutation or structural alteration of the p73 gene locus are rare in hematological malignancies.
Structural analysis of p73 in leukemia and lymphoma. (A) PCR-SSCP analysis of p73-cDNA from cell lines. Sequencing of RT-PCR products of representative samples showed that no mutations were present and the differences in migration of the bands were derived from the polymorphisms at codons 336 and 349. Arrow (a) indicates the band appearing when codons 336 and 349 nucleotides are GCC and CAT, and arrow (b) indicates the band appearing when codons 336 and 349 nucleotides are GCT and CAC, respectively. (B) Southern blot analysis of the p73 gene from p73 nonexpressing lymphoid cell lines. (B1) A 0.8-kb EcoRI fragment of pBS-M13-p73, which covers exons 2 through 6, was used as a probe. Lanes 1 and 7, peripheral blood mononuclear cells; lanes 2 and 8, Hela; lanes 3 and 9, TALL-1; lanes 4 and 10, RPMI8402; lanes 5 and 11, ALL-Sil; lanes 6 and 12, BALL-1. (B2) Probe CD, which covers exons 7-9, was used as a probe. Lane 1, Raji; lane 2, HL60; lane 3, HUT78; lane 4, U937; lane 5, Jurkat; lane 6, SKW3; lane 7, TALL-1; lane 8, CEM; lane 9, ALL-Sil; lane 10, RPMI8402; lane 11, KOPT-K1; lane 12, DS179; lane 13, NALM18; lane 14, BALL-1. Lanes 6, 7, 9, 10, 12, 13, and 14 are p73-negative cell lines as measured by RT-PCR.
Structural analysis of p73 in leukemia and lymphoma. (A) PCR-SSCP analysis of p73-cDNA from cell lines. Sequencing of RT-PCR products of representative samples showed that no mutations were present and the differences in migration of the bands were derived from the polymorphisms at codons 336 and 349. Arrow (a) indicates the band appearing when codons 336 and 349 nucleotides are GCC and CAT, and arrow (b) indicates the band appearing when codons 336 and 349 nucleotides are GCT and CAC, respectively. (B) Southern blot analysis of the p73 gene from p73 nonexpressing lymphoid cell lines. (B1) A 0.8-kb EcoRI fragment of pBS-M13-p73, which covers exons 2 through 6, was used as a probe. Lanes 1 and 7, peripheral blood mononuclear cells; lanes 2 and 8, Hela; lanes 3 and 9, TALL-1; lanes 4 and 10, RPMI8402; lanes 5 and 11, ALL-Sil; lanes 6 and 12, BALL-1. (B2) Probe CD, which covers exons 7-9, was used as a probe. Lane 1, Raji; lane 2, HL60; lane 3, HUT78; lane 4, U937; lane 5, Jurkat; lane 6, SKW3; lane 7, TALL-1; lane 8, CEM; lane 9, ALL-Sil; lane 10, RPMI8402; lane 11, KOPT-K1; lane 12, DS179; lane 13, NALM18; lane 14, BALL-1. Lanes 6, 7, 9, 10, 12, 13, and 14 are p73-negative cell lines as measured by RT-PCR.
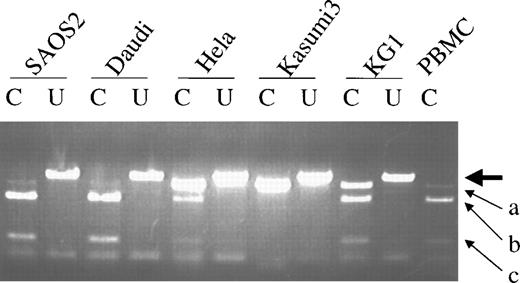
Because a previous study suggested expression of p73 was monoallelic,6 we examined our samples for allelic expression of p73 gene by digesting the RT-PCR products withNlaIII that identifies the polymorphic site at codon 349 by cutting at nucleotides CAT but not CAC. In contrast to the prior report,6 the p73 gene was expressed biallelically in cell lines and normal PBMC (Fig 4), although unbalanced expression of the 2 alleles may exist as seen in the SAOS-2, Daudi, Hela cell lines and peripheral blood mononuclear cells.
Allelic expression of p73. Differential expression was detected by NlaIII digestion of RT-PCR products. The large arrow indicates the uncut band (390 bp). The small arrow (a) indicates the 358-bp product (NlaIII cuts at codon 223) when codon 349 is CAC (not cut with NlaIII). The small arrows (b) and (c) indicate the 277-bp and 89-bp bands, respectively, afterNlaIII cuts at both codon 349 (CAT) and codon 223. C, cut withNlaIII; U, uncut.
Allelic expression of p73. Differential expression was detected by NlaIII digestion of RT-PCR products. The large arrow indicates the uncut band (390 bp). The small arrow (a) indicates the 358-bp product (NlaIII cuts at codon 223) when codon 349 is CAC (not cut with NlaIII). The small arrows (b) and (c) indicate the 277-bp and 89-bp bands, respectively, afterNlaIII cuts at both codon 349 (CAT) and codon 223. C, cut withNlaIII; U, uncut.
Methylation status of p73 gene.
We hypothesized that an absence or low level of p73 expression might be caused by hypermethylation of the CpG island located in exon 1. Hypermethylation and silencing of several tumor suppressor genes, including p16INK4A, has previously been noted.27 28 The methylation status of the CG-rich region in exon 1 of p73 (Fig 5A) was examined. The genomic DNA was digested in parallel with a methylcytosine-sensitive and -resistant isoschizomeric enzyme,HpaII and MspI. The digests were amplified by PCR using the primer sets that could amplify exon 1. The p73-negative cell lines (BALL-1, ALL-Sil) were methylated in this region (Fig 5B). Also, the cell lines with relatively low expression of p73(Jurkat, Raji, U937, KOPT-K1) were methylated. Thep73-expressing cell lines (U2OS, HL60, BC-1) and normal cells (PBMC1, 2) were not methylated in this region (Fig 5B), consistent with our hypothesis that methylation of this region was important for the expression of p73.
(A) Nucleotide sequence of the CpG island in exon 1. Putative methylation sites of exon 1 are illustrated. CG sequences are bold, and the HpaII/MspI sites are underlined. Numbers above the nucleotide sequence are based on the cDNA sequence in the European Molecular Biology Laboratory database under the accession number Y11416EMBL. (B) Methylation status of CpG island in exon 1. Digests of 20 ng of genomic DNA with either HpaII orMspI were amplified by PCR using primers that amplify the entire exon 1 DNA (77 bp). U, uncut; H, HpaII; M,MspI.
(A) Nucleotide sequence of the CpG island in exon 1. Putative methylation sites of exon 1 are illustrated. CG sequences are bold, and the HpaII/MspI sites are underlined. Numbers above the nucleotide sequence are based on the cDNA sequence in the European Molecular Biology Laboratory database under the accession number Y11416EMBL. (B) Methylation status of CpG island in exon 1. Digests of 20 ng of genomic DNA with either HpaII orMspI were amplified by PCR using primers that amplify the entire exon 1 DNA (77 bp). U, uncut; H, HpaII; M,MspI.
To strengthen our hypothesis, we determined if 5-AC (a demethylating agent) could enhance p73 expression in selected cell lines. Thep73 non- or low-expressing cell lines were cultured with 5-AC (0.5 to 5 μmol/L) for 3 days. After 3 days of treatment of thep73-negative cell line, ALL-Sil expressed p73 mRNA at a level comparable to untreated CEM. Furthermore, the p73-low expressors (KOPT-K1, CEM) increased their p73 expression when cultured in the presence of 5-AC (Fig 6). In contrast, levels were unchanged in the U2OS and HL60 cells which constitutively expressed p73 (Fig 6).
Induction of p73 expression after culture with 5-AC (RT-PCR). U2OS and HL60 were cultured with 5-AC for 3 and 5 days at 1 and 5 μmol/L, and 0.5 and 3 μmol/L, respectively. CEM, ALL-Sil, and KOPT-K1 were treated with 5-AC for 3 days at 3 μmol/L.
Induction of p73 expression after culture with 5-AC (RT-PCR). U2OS and HL60 were cultured with 5-AC for 3 and 5 days at 1 and 5 μmol/L, and 0.5 and 3 μmol/L, respectively. CEM, ALL-Sil, and KOPT-K1 were treated with 5-AC for 3 days at 3 μmol/L.
DISCUSSION
Mutations, translocations, and deletions can alter or cause loss of expression of gene products. The p73 gene is a homolog of p53 and is located at 1p33.2-3, a region that is frequently deleted in neuroblastoma, melanoma, and other cancers.29-32Forced expression of p73 in p53-negative cells resulted in their apoptosis.7 Furthermore, some investigators have claimed that structural abnormalities of p73 occur in some solid tumors.26,33 Allelic loss of the p73 locus was observed in 42% (11 of 26 informative cases) of lung cancers,26 and 5.3% (2 of 38 cases) of prostatic cancers.33 Because of these observations, p73 is regarded as a potential tumor suppressor.
To investigate whether alteration of the p73 gene locus occurs in ALLs/B-NHLs/AMLs, we initially examined the expression of thep73 gene by RT-PCR using a large variety of hematopoietic cell lines. Thirty-three percent of ALL/B-NHL cell lines were negative forp73 gene expression. SSCP analysis of RT-PCR products within the region that corresponded to the 5 mutational hotspots of thep53 gene showed no mutation in 43 cell lines (25 hematopoietic) that expressed p73. In contrast, the p53 gene has a high frequency of mutations in leukemia cell lines that leads to an overexpression of the aberrant gene.3,34 Furthermore, we found neither deletions nor translocations of the p73 gene by Southern blot analysis. Deletions in p53 gene locus occur with moderate frequency in several human malignancies.4 35 These results suggest that p53-type mutations rarely occur in thep73 gene in leukemias/lymphomas.
Nevertheless, a number of hematopoietic cell lines and fresh samples from individuals had either negligible or very low expression ofp73. Therefore, we looked for a nonmutational mechanism to explain the abberant expression of p73. Imprinting allows the preferential maternal or paternal allele of a gene to be expressed. For example, retinoblastoma is associated with imprinting of the Rb gene locus.36 A study suggested that the p73 gene was imprinted resulting in expression of only 1 allele.6Quizzically, a second study reported that both alleles were expressed in lung cancers, but only a single allele was expressed in the matched normal lung tissues.25 They suggested that the activation of a silenced allele leading to p73 overexpression may play an important role in lung cancers. In contrast, a third study found that 25 of 26 normal lung tissue samples expressed both alleles.26 These same investigators found that the expression pattern of the 2 alleles varied between various normal tissues from the same individual. These 3 groups examined the same polymorphic site in exon 2, but their conclusions were very disparate. Therefore, we chose to examine another polymorphic site. We found that both the tumor cell lines and normal peripheral blood cells had biallelic p73 expression, although several of the cell lines had an imbalance of expression between the p73 alleles.
The expression of the p16INK4A tumor suppressor gene is silenced by hypermethylation at the 5′ untranslated region in the course of tumor development in various cancers including bladder, brain, breast, colon, lung, prostate, and myelomas.27,37-41 We analyzed the methylation status of the CpG island (Fig 5A) in the 5′ untranslated region of exon 1. Twop73-negative cell lines (BALL-1, ALL-Sil) had hypermethylation as detected by digestion with the methylcytosine sensitive enzyme,HpaII and its isoschizomer MspI, followed by PCR. These data indicate a correlation between hypomethylation and p73gene expression. Interestingly, however, 2 cell lines (KOPT-K1, Raji) expressed p73 but were also hypermethylated in exon 1 ofp73. Possibly other key regions of the gene are hypomethylated in these cell lines. A similar phenomenon has been reported for thep16INK4A gene in several myeloma cell lines and fresh breast cancers.42,43 To explore this further, we took advantage of a well-known demethylating agent. Expression of several tumor suppressor genes (p16, PTEN) has been induced after tumor cells were cultured with 5-AC.27 28 This presumably occurred as a result of hypomethylation of the target genes. We noted a similar phenomenon when we cultured thep73-nonexpressing cell line (ALL-Sil, Fig 6) with 5-AC.
Taken together, these data suggest that hypermethylation of the CpG island of p73 might silence expression of the gene in ALL/B-NHL cell lines and patient samples. Absence or low expression ofp73 may contribute to the development or progression of ALL and B-NHL.
Supported in part by National Institutes of Health Grants, the Parker Hughes Trust, C. and H. Koeffler Fund, and Lymphoma Fundation of America. H.P.K. is a member of the Jonsson Comprehensive Cancer Center and holds the endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center/UCLA School of Medicine. S.K. is supported in part by the Scholarship from Uehara Memorial Foundation.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Seiji Kawano, MD, Hematology/Oncology, Cedars-Sinai Medical Center, UCLA School of Medicine, Davis Bldg, Room 5065, 8700 Beverly Blvd, Los Angeles, CA 90048; e-mail:kawanos@csmc.edu.