Abstract
The rate of transcription of several genes encoding proteins involved in O2 and energy homeostasis is controlled by hypoxia-inducible factor-1 (HIF-1), a heterodimeric DNA binding complex composed of and β subunits. HIF-1 is considered the primarytrans-acting factor for the erythropoietin (EPO) and vascular endothelial growth factor (VEGF) genes. Since EPO gene expression is inhibited by the proinflammatory cytokines interleukin-1β (IL-1β) and tumor necrosis factor- (TNF-), while no such effect has been reported with respect to the VEGF gene, we investigated the effects of IL-1β and TNF- on the activation of the HIF-1 DNA-binding complex and the amount of HIF-1 protein in human hepatoma cells in culture. Under normoxic conditions, both cytokines caused a moderate activation of HIF-1 DNA binding. In hypoxia, cytokines strongly increased HIF-1 activity compared with the effect of hypoxia alone. Only IL-1β increased HIF-1 protein levels. In transient transfection experiments, HIF-1–driven reporter gene expression was augmented by cytokines only under hypoxic conditions. In contrast to their effect on EPO synthesis, neither IL-1β nor TNF- decreased VEGF production. The mRNA levels of HIF-1 and VEGF were unaffected. Thus, cytokine-induced inhibition of EPO production is not mediated by impairment of HIF-1 function. We propose that HIF-1 may be involved in modulating gene expression during inflammation.
ADAPTATION to reduced O2availability involves the synthesis of proteins that systemically or locally protect the organism from hypoxic damage.1,2 Thus, at low pO2, the production of the glycoprotein erythropoietin (EPO) increases to stimulate the proliferation and differentiation of erythrocytic progenitors in bone marrow, resulting in a greater O2 capacity of the blood.3,4 The kidneys and liver are the main sites of production of the hormone. Apart from renal failure, chronic inflammatory and malignant diseases are often associated with normocytic and normochromic anemia, which is partly caused by a lack of EPO.5,6 In vitro studies using human hepatoma cells7-9 or isolated perfused rat kidneys8 10 have shown that the proinflammatory cytokines interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α) decrease EPO mRNA levels and inhibit hypoxia-induced EPO production, respectively.
Besides erythropoiesis, angiogenesis is also stimulated by hypoxia. The most specific inducer of angiogenesis is vascular endothelial growth factor (VEGF), which is produced by normal and malignant cells at low pO2.11 Encoded in a single gene, VEGF mRNA may exist as 4 or 5 different splice variants, with the 165–amino acid isoform (VEGF165) being the predominant secreted form. VEGF and EPO synthesis share several regulatory mechanisms,12 including the participation of a hemoprotein as the O2 sensor and cis-acting elements that bind to hypoxia-inducible factor-1 (HIF-1). However, while erythropoiesis is suppressed in inflammatory states, angiogenesis is locally stimulated as in wound-healing and solid-tumor formation. IL-1β and other proinflammatory cytokines increase VEGF mRNA levels in several cell lines.11
HIF-1 is a heterodimeric transcription factor composed of the basic helix-loop-helix-PAS-domain (period, arylhydrocarbon receptor, single-minded) containing proteins HIF-1α and aryl hydrocarbon receptor nuclear translocator ([ARNT] = HIF-1β).13,14 The main pO2-sensitive component of the complex appears to be HIF-1α.15 Its rapid degradation under normoxic conditions is probably mediated by the ubiquitin-proteasome system.16Although ARNT may also increase in response to hypoxic stress,14 its abundance is more constitutive than that of HIF-1α. The fact that HIF-1α and ARNT transcripts and HIF-1 DNA binding and trans-activating activity have been demonstrated in all tissues investigated thus far underlines the importance of HIF-1 in pO2-dependent gene expression.1,2 17
In the present investigation, the effects of IL-1β and TNF-α on HIF-1 DNA binding activity, HIF-1α protein content, reporter gene expression, and HIF-1α mRNA and VEGF mRNA levels in HepG2 cells were studied. This human hepatoma cell line was chosen because it expresses both the EPO and VEGF genes in a pO2-dependent manner.12,18 19
MATERIALS AND METHODS
Cell culture.
The human hepatoma cell line HepG2 was obtained from the American Type Culture Collection (Rockville, MD). The murine hepatoma cell line Hepa-1 (kindly provided by Dr O. Hankinson, Los Angeles, CA) was used to compare HIF-1 DNA-binding activity with different oligonucleotides. The cells were grown in RPMI 1640 medium (HepG2) or α-minimal essential medium without nucleosides (Hepa-1) containing 10% fetal bovine serum (Sigma, Deisenhofen, Germany) in a humidified atmosphere of 5% CO2 in air at 37°C, on either conventional polystyrene dishes (16- or 94-mm diameter) or petriPERM tissue culture dishes (52-mm diameter; Heraeus, Hanau, Germany). The bottom of the petriPERM dish is made of a gas-permeable fluoroethylene-propylene copolymer Teflon membrane of 25-μm thickness that allows adjustment of the pericellular pO2 to the pO2 level in the gas atmosphere.19 In conventional dishes with a gas-impermeable bottom, the pericellular pO2 decreases with increasing culture density.20 Low-density cultures in polystyrene dishes were placed in an incubator with 3% O2 (21 mm Hg), 5% CO2, and 92% N2 for hypoxic incubation. Cells grown on petriPERM dishes were placed in a gas-tight acrylic chamber at continuous flow of a premixed gas of 2% O2 (14 mm Hg), 5% CO2, and 93% N2 as described previously.19 20 Cells received fresh medium 16 hours before the experiments. The proinflammatory cytokines used for the study were recombinant human IL-1β (Ciba-Geigy, Basel, Switzerland) and recombinant human TNF-α (6.6 × 106 U/mg by L929 bioassay; BASF/Knoll, Ludwigshafen, Germany).
Nuclear extract preparation.
Nuclear extracts were prepared according to the method of Semenza and Wang21 with minor modifications. Hepa-1 cells were grown to 40% to 60% confluence and HepG2 cells to 20% to 30% confluence in conventional cell culture dishes. In total, 4 to 5 × 107cells were washed with ice-cold phosphate-buffered saline (PBS), harvested in 6 mL PBS, and centrifuged at 400 × g for 5 minutes at 4°C. The cell pellets were washed with 4 mL ice-cold buffer A (10 mmol/L Tris, pH 7.8, 1.5 mmol/L MgCl2, and 10 mmol/L KCl), resuspended in 1 mL buffer A, and kept on ice for 10 minutes. Subsequently, cells were lysed by Dounce homogenization, and lysis was controlled with trypan blue. Nuclei were pelleted at 3,500 × g for 5 minutes at 4°C, resuspended in 150 μL ice-cold buffer C (420 mmol/L KCl, 20 mmol/L Tris, pH 7.8, 1.5 mmol/L MgCl2, and 20% glycerol), and incubated for 30 minutes on ice with occasional flicking of the tubes. Just before use, buffers A and C were supplemented with 2 μg/mL aprotinin, 10 μg/mL leupeptin, 20 μg/mL pepstatin, 1 mmol/L Na3VO4, 0.5 mmol/L benzamidine, 2 mmol/L levamisole, 10 mmol/L β-glycerophosphate, 0.5 mmol/L dithiothreitol (DTT), and 0.4 mmol/L phenylmethylsulfonyl fluoride. Nuclei were centrifuged at 12,000 ×g for 30 minutes at 4°C. The supernatant was dialyzed against 1 L buffer D (100 mmol/L KCl, 20 mmol/L Tris, pH 7.8, 2 mmol/L EDTA, and 20% glycerol) overnight at 4°C. After dialysis, nuclear extracts were collected by brief centrifugation at 4°C, divided into aliquots, and frozen in liquid nitrogen and stored at −80°C. Protein concentrations were determined by the Bradford method using bovine serum albumin as standard.
Gel-shift assay.
[γ-32P]adenosine triphosphate (ATP) was obtained from New England Nuclear (Köln, Germany). Oligonucleotides for gel-shift assays were synthesized by EuroGenTec (Seraing, Belgium) or MWG (Ebersberg, Germany). Sequences containing HIF-1 binding sites were derived from the human transferrin gene (TfHBSww) and the EPO enhancer (EpoWT). In the TfHBSww enhancer, two HIF-1 binding sites are present.22 A mutated HIF-1 binding site in another EPO enhancer oligonucleotide (EpoMut) was used to demonstrate specificity. Sequences were as follows: TfHBSww (sense), 5′-TTCCTGCACGTACACACAAAGCGCACGTATTTC-3′; TfHBSww (antisense), 5′-GAAATACGTGCGCTTTGTGTGTACGTGCAGGAA-3′; EpoWT (sense), 5′-GCCCTACGTGCTGCCTCGCATGGC-3′; EpoWT (antisense), 5′-GCCATGCGAGGCAGCACGTAGGGC-3′; EpoMut (sense), 5′-GCCCTAATGTCTGCCTCGCATGGC-3′; and EpoMut (antisense), 5′-GCCATGCGAGGCAGACATTAGGGC-3′. Anti–HIF-1α antibodies OZ12 and OZ15 were generously provided by Dr D. Livingston (Boston, MA). Binding reactions were set up in a volume of 20 μL, and nuclear extracts (5 μg protein) were incubated in a buffer with a final concentration of 50 mmol/L KCl, 10 mmol/L Tris, pH 7.7, 5 mmol/L DTT, 1 mmol/L EDTA, 1 mmol/L MgCl2, 5% glycerol, 0.03% Nonidet P-40, and 400 ng salmon testes DNA. Before addition of the 32P-labeled oligonucleotide, the reactions were preincubated on ice for 30 minutes. The final incubation was overnight at 4°C. Samples were resolved by electrophoresis on 5% polyacrylamide gels (polyacrylamide:bisacrylamide 30:0.8) at room temperature. Gels were dried and analyzed directly by autoradiography. For competition experiments, a 250-fold molar excess of unlabeled annealed oligonucleotides were added before addition of the labeled probes. For supershift experiments, undiluted anti–HIF-1α antibodies (5 μL each) OZ12 and OZ15 were mixed and added to the reactions 2 hours before the gel was run.
Western blot analysis.
For determination of immunoreactive HIF-1α protein in the nuclei of cells, nuclear extracts were prepared as already described and subjected to Western blot analysis. Samples were run on sodium dodecyl sulfate (SDS)/7.5% polyacrylamide gels and transferred to nitrocellulose membranes (Amersham, Braunschweig, Germany) electrophoretically (Trans-Blot SD; BioRad, München, Germany). Equal loading and transfer efficiency were verified by staining with 2% Ponceau S. Membranes were blocked overnight with PBS/5% fat-free skim milk and then incubated with a monoclonal mouse antibody raised against human HIF-1α (Transduction Laboratories, Lexington, KY) diluted 1:500 for 2 hours at room temperature. For detection, a horseradish peroxidase–linked anti–mouse IgG antibody (1:2,000, 1 hour at room temperature; Santa Cruz, Heidelberg, Germany) and enhanced chemiluminescence substrate (Amersham) were used.
Transfection and luciferase assay.
HepG2 cells (1.6 × 105) were seeded onto 8-cm2 cell culture dishes and grown to 25% to 50% confluence. The medium was changed 3 hours before the cells were transfected using the LIPOFECTIN reagent (Life Technologies, Karlsruhe, Germany) and 1 μg of the reporter DNA (pEpo-HIF plasmid) per dish. The pEpo-HIF plasmid was kindly provided by T. Kietzmann (Göttingen, Germany) and corresponds to the pGL3-promoter vector (Promega, Mannheim, Germany) with a 46mer oligonucleotide containing 3 HIF-1 binding sites (HBSs) from the EPO enhancer inserted 5′ to the SV40 promoter. All transfections were performed in duplicate with aliquots of transfection mixture from a single pool. Eighteen hours after transfection, the medium was changed and the cells were exposed to either normoxic or hypoxic conditions with or without cytokines (10 ng/mL TNF-α or 300 pg/mL IL-1β). After 24 hours, cells were washed twice with NaCl (0.9%) and lysed in reporter lysis buffer (Promega). Luciferase activity was determined according to the manufacturer’s instructions (Berthold Detection Systems, Pforzheim, Germany). Luminescence was measured in a MicroLumat LB 96P (Berthold EG & G, Bad Wildbach, Germany).
Northern blot analysis.
HepG2 cells were grown either on petriPERM dishes (confluent cultures) or on conventional cell culture dishes (20% to 30% confluence). Total RNA was isolated according to the method of Chomczynski and Sacchi.23 Samples were subjected to electrophoresis in denaturing 1% agarose gels containing 0.7 mol/L formaldehyde. RNAs were transferred onto nylon membranes (Nytran Plus; Schleicher & Schüll, Dassel, Germany) with a vacuum blotting apparatus (Pharmacia, Uppsala, Sweden). Filters were cross-linked with UV light, dried at 80°C for 2 hours, and prehybridized for 4 hours at 42°C in 45% formamide, 5× SSC, 5× Denhardt solution, 0.1% SDS, and 100 μg/mL sonicated denatured salmon testes DNA. Hybridizations were performed in fresh solution of identical composition supplemented with the radioactive probe (0.5 to 2 × 106 cpm/mL) for 2 days at 42°C. Hybridization probes were polymerase chain reaction (PCR)-generated fragments with primers derived from the published cDNA sequences, with the exception of the 18S rRNA probe, which was an antisense single-stranded DNA oligonucleotide. PCR fragments were [α-32P]dCTP (New England Nuclear)-labeled with a commercially available kit (MBI-Fermentas, St Leon-Rot, Germany). The 18S rRNA probe was labeled with [γ-32P]ATP and T4 kinase. After hybridization, the filters were washed twice for 15 minutes in 2× SSC/0.1% SDS at 50°C and then twice in 0.1× SSC/0.1% SDS at 60°C. Filters were sealed in plastic bags and analyzed by exposure to either Hyperfilm (Amersham) or imaging plates for a Bio Imaging Analyzer (BAS 1000; Fuji, Düsseldorf, Germany).
Assay of EPO and VEGF.
The EPO level was measured in culture supernatants by radioimmunoassay.8 The assay system contained125I-labeled recombinant human EPO (137 TBq/mmol; Amersham), antiserum raised in rabbits against recombinant human EPO, and a human urinary EPO standard calibrated by bioassay against the International Reference Preparation B. After 24 hours of incubation, antibody-bound and free radiolabeled EPO were separated by precipitation with polyethylene glycol (PEG 6000). EPO concentrations were calculated from log-logit plots of the standard curves. The lower detection limit was 5 U/L. Intraassay and interassay coefficients of variation were less than 6% and less than 12% in the relevant range of 20 to 100 U/L.
The VEGF165 level was measured by commercial enzyme-linked immunoassay (Quantikine; R&D Systems, Minneapolis, MN). The intraassay and interassay coefficients of variation were 5% and 7.5%, and the lower detection limit was 9 pg/mL. EPO and VEGF concentrations were related to the total cellular protein measured in lysates of washed cultures using a protein microdetermination kit based on the phenol reagent method (Sigma).
RESULTS
Effect of oligonucleotides on HIF-1 binding.
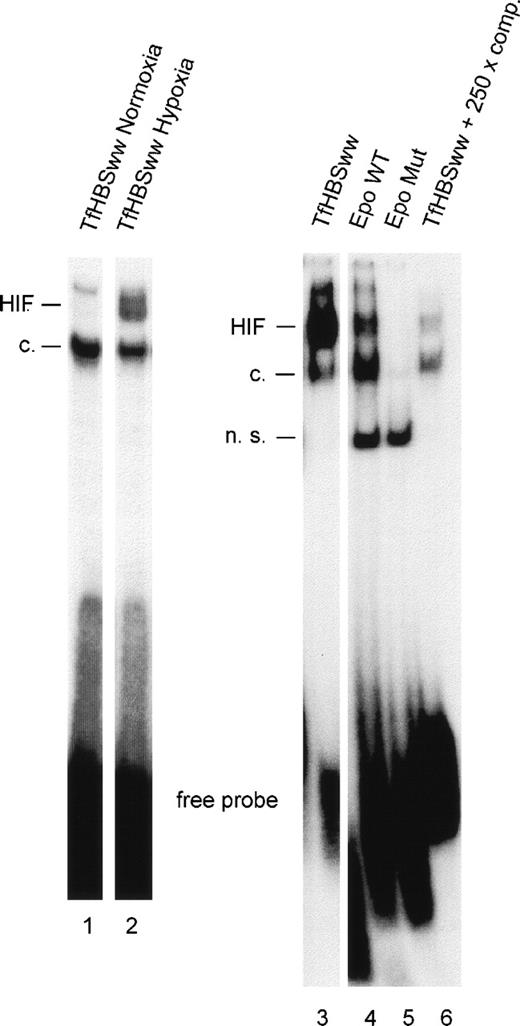
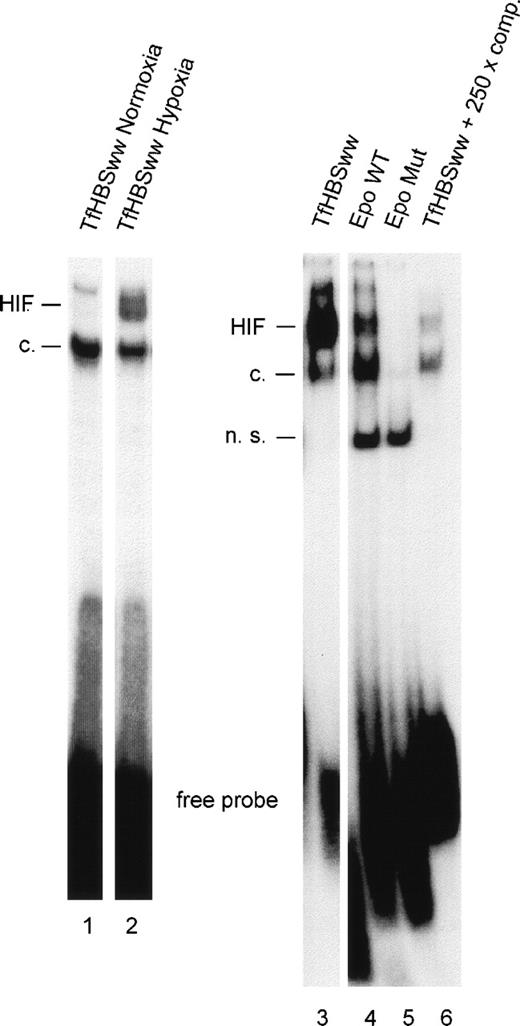
To study the effect of the number of HIF-1 binding sites (HBSs) within the labeled annealed oligonucleotides used for gel mobility shifts, oligonucleotides with 1 (EpoWT) and 2 (TfHBSww) functional HBSs were used, respectively. Equimolar amounts of oligonucleotides were labeled radioactively in parallel, annealed, and used in the binding reactions. In nuclear extracts from Hepa-1 cells, hypoxia led to a clear induction of the HIF-1 DNA binding complex as compared with normoxic conditions (Fig 1, lanes 1 and 2). Signals from hypoxic Hepa-1 cells obtained with TfHBSww (lane 3) were much stronger than signals obtained with EpoWT (lane 4). The mouse hepatoma cell line Hepa-1 was chosen because, upon hypoxic stimulation, Hepa-1 cells accumulate large amounts of activated HIF-1 in the nucleus.22 In nuclear extracts from hypoxic HepG2 cells (human hepatoma cell line), HIF-1 binding to EpoWT was hardly detectable (data not shown).
Gel-shift analysis with nuclear extracts from Hepa-1 cells exposed to normoxia (lane 1) or hypoxia (3% O2 for 4 hours; lanes 2-6). HIF-1 DNA binding activity was examined with 3 different oligonucleotides containing either 2 (TfHBSww) or 1 (EpoWT) consensus HBSs or a mutated HBS (EpoMut). For confirmation of specificity, competition experiments were performed using a 250-fold molar excess of unlabeled annealed TfHBSww oligonucleotide in the binding reaction. HIF, inducible HIF-1 binding; c, constitutive binding; n.s., nonspecific binding.
Gel-shift analysis with nuclear extracts from Hepa-1 cells exposed to normoxia (lane 1) or hypoxia (3% O2 for 4 hours; lanes 2-6). HIF-1 DNA binding activity was examined with 3 different oligonucleotides containing either 2 (TfHBSww) or 1 (EpoWT) consensus HBSs or a mutated HBS (EpoMut). For confirmation of specificity, competition experiments were performed using a 250-fold molar excess of unlabeled annealed TfHBSww oligonucleotide in the binding reaction. HIF, inducible HIF-1 binding; c, constitutive binding; n.s., nonspecific binding.
Specificity of HIF-1 binding was confirmed using EpoMut, an oligonucleotide similar to EpoWT except for a mutated HBS. No HIF-1–specific signals, either the inducible band or the constitutive band, were obtained with EpoMut (Fig 1, lane 5). Furthermore, HIF-1 binding to labeled TfHBSww was strongly reduced when a 250-fold excess of unlabeled TfHBSww was added (lane 6).
Influence of proinflammatory cytokines on HIF-1 activation.
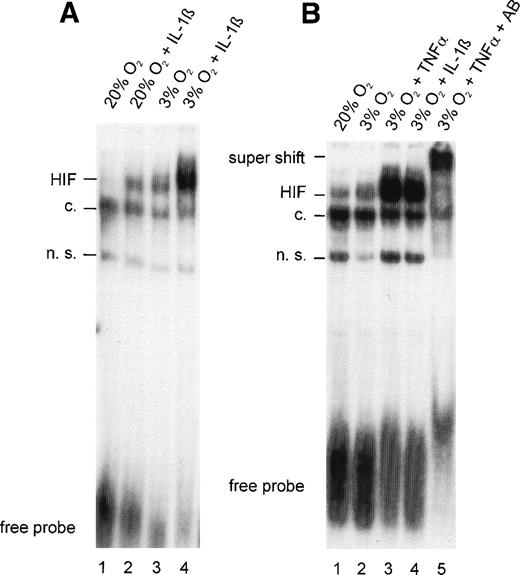
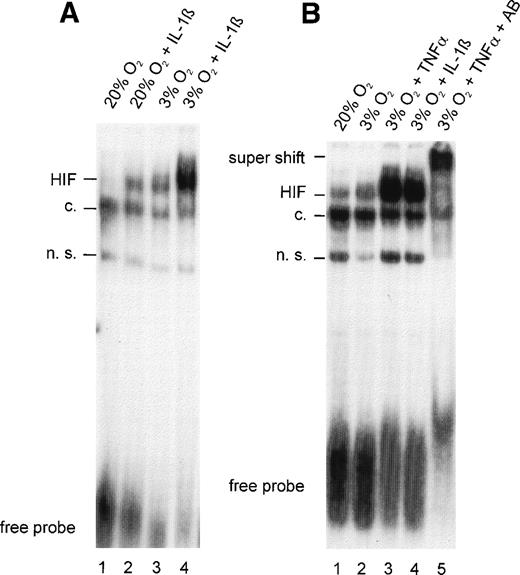
To determine the nature of the inhibitory action of IL-1β and TNF-α on EPO gene expression, the effects of these cytokines on HIF-1 DNA binding activity were analyzed in HepG2 cells. In nuclear extracts from normoxic control cultures, faint inducible bands were observed in slightly overexposed autoradiograms, indicating that some HIF-1α was activated to form the DNA binding complex even under normoxic conditions (Fig 2A and B, lane 1). Because these experiments were performed on regular, gas-impermeable cell culture dishes, strict care was taken to ensure that cultures did not exceed 20% to 30% confluence and incubations were performed with 0.5-mm medium height to ensure short diffusion distances. The effectiveness of this model system was demonstrated by an increase in HIF-1 DNA binding activity after 4 hours of hypoxia (Fig 2A, lane 3). IL-1β treatment resulted in a significant increase in HIF-1 DNA binding both in normoxic cells (Fig 2A, lane 2; compared with the normoxic control) and in hypoxic cells (lane 4; compared with the hypoxic control).
(A) Gel-shift analysis with nuclear extracts from HepG2 cells incubated under normoxic (20% O2) or hypoxic (3% O2) conditions for 4 hours in the absence or presence of IL-1β 300 pg/mL. (B) Gel-shift analysis with nuclear extracts from normoxic HepG2 cells and hypoxic HepG2 cells treated with TNF- 10 ng/mL or IL-1β 300 pg/mL for 4 hours. Specificity was demonstrated in a supershift experiment with anti–HIF-1 antibodies (AB). See Fig 1for abbreviations.
(A) Gel-shift analysis with nuclear extracts from HepG2 cells incubated under normoxic (20% O2) or hypoxic (3% O2) conditions for 4 hours in the absence or presence of IL-1β 300 pg/mL. (B) Gel-shift analysis with nuclear extracts from normoxic HepG2 cells and hypoxic HepG2 cells treated with TNF- 10 ng/mL or IL-1β 300 pg/mL for 4 hours. Specificity was demonstrated in a supershift experiment with anti–HIF-1 antibodies (AB). See Fig 1for abbreviations.
TNF-α was as effective as IL-1β in augmenting hypoxia-induced HIF-1 binding (Fig 2B, lanes 3 and 4). Similar to IL-1β, albeit to a lesser extent, TNF-α increased HIF-1 DNA binding in normoxic cells (not shown). Evidence that HIF-1α was a component of the complex that bound to DNA in TNF-α–treated hypoxic cells was provided by a supershift analysis. The addition of a mixture of OZ12 and OZ15 antibodies (specific for HIF-1α protein) to binding reactions resulted in reduced electrophoretic mobility (Fig 2B, lane 5).
Influence of proinflammatory cytokines on HIF-1α protein level.
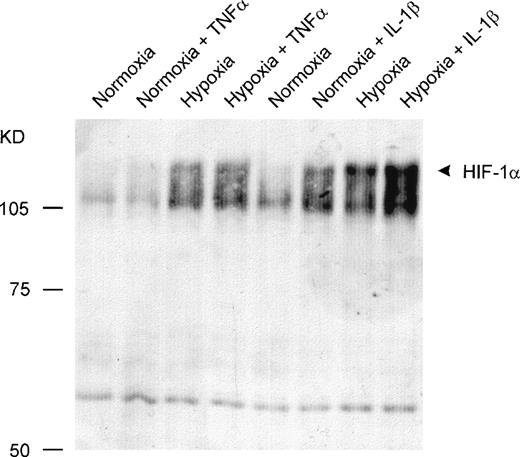
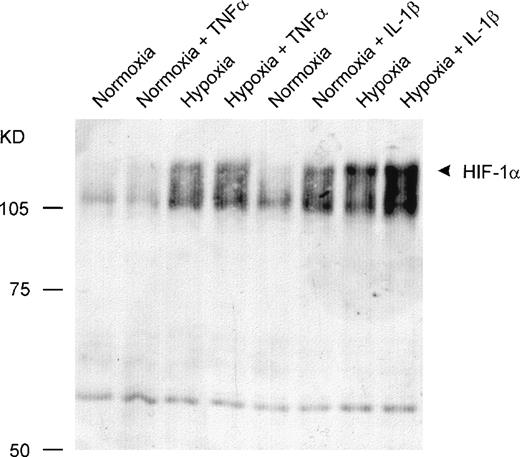
To determine whether the increased DNA binding activity after cytokine treatment was based on elevated amounts of activated HIF-1α or on coactivated proteins’ being part of the DNA binding complex, Western blot assays were performed. Incubation with IL-1β under either normoxic or hypoxic conditions led to increased amounts of HIF-1α protein in the nuclei compared with the normoxic and hypoxic controls. In contrast, TNF-α did not cause a significant accumulation of HIF-1α protein within the nuclei (Fig 3).
Detection of HIF-1 protein by Western blot analysis. Twenty micrograms of nuclear protein per lane was sufficient for detection by a monoclonal anti–HIF-1 antibody. IL-1β caused an accumulation of HIF-1 protein in nuclei from HepG2 cells under normoxic and hypoxic conditions. TNF- was ineffective.
Detection of HIF-1 protein by Western blot analysis. Twenty micrograms of nuclear protein per lane was sufficient for detection by a monoclonal anti–HIF-1 antibody. IL-1β caused an accumulation of HIF-1 protein in nuclei from HepG2 cells under normoxic and hypoxic conditions. TNF- was ineffective.
Action of cytokines on reporter gene expression.
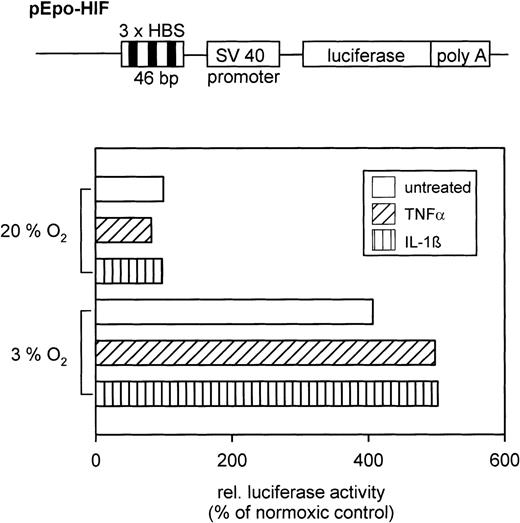
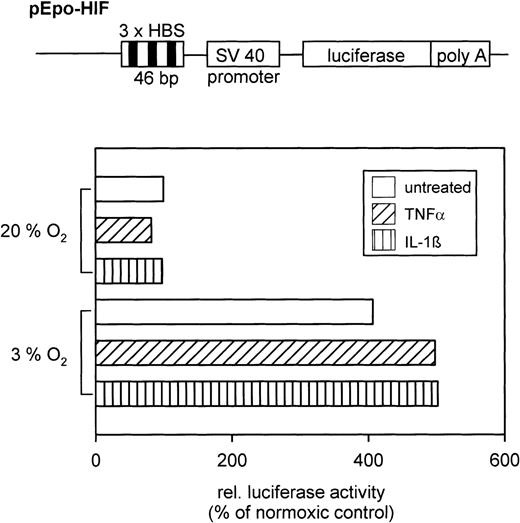
To prove that the augmented HIF-1 DNA binding activity after cytokine treatment is functional, reporter gene assays were performed. After transfection of HepG2 cells with the pEpo-HIF plasmid, luciferase activity in cell extracts was determined. Hypoxia increased luciferase activity compared with normoxia (Fig 4). Furthermore, cytokines amplified luciferase activity by 25% in hypoxic cells (compared with hypoxia alone), whereas the cytokines had no effect in normoxic cells.
(Top) Schematic drawing of the relevant parts of the pEpo-HIF plasmid (not in scale). (Bottom) Typical result of a reporter gene expression assay in HepG2 cells treated with either TNF- or IL-1β under normoxic or hypoxic conditions. Relative light units were normalized to the protein level per dish, and values are shown in relation to the normoxic control (=100%).
(Top) Schematic drawing of the relevant parts of the pEpo-HIF plasmid (not in scale). (Bottom) Typical result of a reporter gene expression assay in HepG2 cells treated with either TNF- or IL-1β under normoxic or hypoxic conditions. Relative light units were normalized to the protein level per dish, and values are shown in relation to the normoxic control (=100%).
Effect of cytokines on steady-state HIF-1α and VEGF mRNA content.
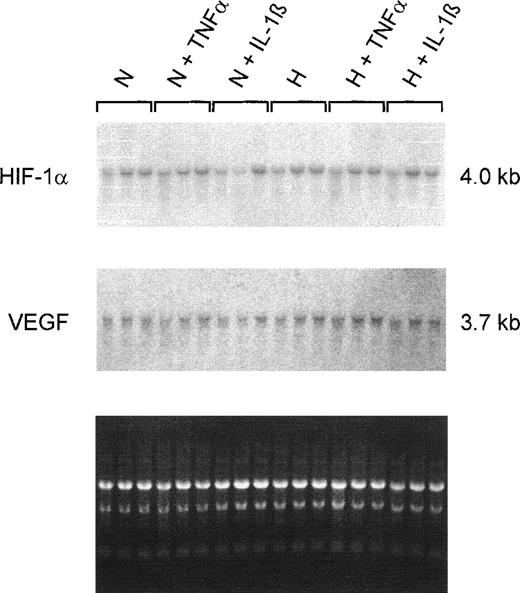
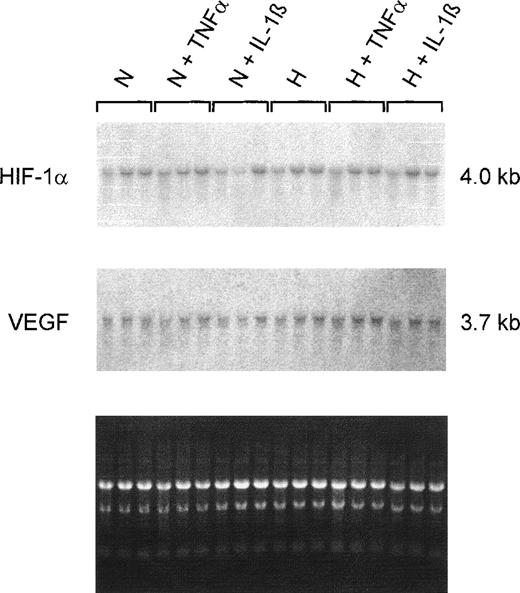
Northern blot analyses on total RNAs from 20% to 30% confluent HepG2 cultures showed no changes in the HIF-1α steady-state mRNA content during normoxic or hypoxic incubation. Additionally, no differences were detectable when cells were treated with TNF-α or IL-1β (Fig5). Because IL-1β and TNF-α affect VEGF gene expression in other cell types,12 the filters were reprobed with a VEGF-specific probe. No obvious changes in VEGF steady-state mRNA content induced by cytokines were detected. Furthermore, the 4-hour hypoxic incubation was apparently not sufficient to increase VEGF mRNA levels in HepG2 cells under our culture conditions.
Northern hybridizations with HIF-1– and VEGF-specific probes on total RNAs from HepG2 cells. Cultures were grown in conventional culture dishes to 20%-30% confluence. Normoxic cells (N) were incubated in 20% O2 and hypoxic cells (H) in 3% O2 for 4 hours with TNF 10 ng/mL or IL-1β 300 pg/mL. The ethidium bromide fluorescence of the RNA is shown at the bottom. Neither of the 2 cytokines had an effect on HIF-1 or VEGF mRNA levels after 4 hours of incubation.
Northern hybridizations with HIF-1– and VEGF-specific probes on total RNAs from HepG2 cells. Cultures were grown in conventional culture dishes to 20%-30% confluence. Normoxic cells (N) were incubated in 20% O2 and hypoxic cells (H) in 3% O2 for 4 hours with TNF 10 ng/mL or IL-1β 300 pg/mL. The ethidium bromide fluorescence of the RNA is shown at the bottom. Neither of the 2 cytokines had an effect on HIF-1 or VEGF mRNA levels after 4 hours of incubation.
Effect of IL-1β and TNF-α on EPO and VEGF production.
Confluent HepG2 cultures grown under conventional conditions in gas-impermeable polystyrene dishes produced EPO and VEGF in large amounts. Previous microelectrode measurements of the pericellular pO2 indicate that such cultures are hypoxic due to the diffusion-limited O2 supply.19 20 The 24-hour rate of production of immunoreactive EPO was decreased when either IL-1β or TNF-α were added to the cultures. In contrast, these cytokines did not alter the rate of production of VEGF (Table1).
DISCUSSION
HIF-1 was initially identified as a dimeric hypoxia-inducible transcription factor that interacts with the hypoxia-response element of the hepatic EPO gene enhancer.13,14,21 Subsequently, HIF-1 has been shown to initiate transcription of several other pO2-controlled genes, including the VEGF gene. Because IL-1β and TNF-α inhibit the synthesis of EPO but not VEGF, we considered it of interest to study the effects of these proinflammatory cytokines on HIF-1α mRNA and protein levels, as well as HIF-1 DNA binding activity and transcriptional activation. The finding of increased HIF-1 DNA binding following IL-1β or TNF-α treatment in human hepatoma cells (HepG2) was unexpected. This may not only be relevant with respect to O2 and energy homeostasis, because recent studies have shown that HIF-1 is involved in hypoxia-mediated apoptosis.24
Several mechanisms can contribute to the regulation of gene expression. IL-1β and TNF-α have been found to reduce EPO mRNA levels in hypoxic human hepatoma cells.7,9 The cytokines are thought to act at the transcriptional level because they do not reduce the half-life of EPO mRNA, which is 1.5 hours in hypoxic HepG2 cells.9 Since HIF-1 DNA binding was increased (Fig 2), the inhibition of EPO gene expression by cytokines appears to be mediated by other cis-acting elements. The augmented HIF-1 DNA binding activity after IL-1β treatment can be attributed, at least in part, to the increased HIF-1α protein level in the nuclei (Fig 3). On the other hand, the effect of TNF-α on HIF-1 DNA binding is probably mainly due to concomitant activation of other proteins that are part of the activated complex. Cyclic adenosine monophosphate (cAMP)-responsive element (CRE) binding-1 (CREB-1) protein has been shown to augment hypoxia-induced activity of the HIF-1 site in the EPO gene enhancer25,26 and to increase HIF-1–dependent lactate dehydrogenase A gene transcription.27 HIF-1 binding element and CRE are thought to overlap in the EPO 3′ enhancer.26Whether IL-1β and TNF-α interfere with CREB-1 binding is presently unknown. Previous attempts have failed to demonstrate effects of these cytokines on cAMP levels and protein kinase A activity in HepG2 cells, although the addition of cAMP analog largely prevents the in vitro inhibition of EPO production by IL-1β or TNFα.9
Transfection studies with HIF-1α and HIF-1β DNA constructs have shown that overexpression of the HIF-1 protein alone does not increase EPO production in either normoxic or hypoxic Hep3B cells.28Moreover, other transcription factors are important for hypoxic gene expression. The role of hepatic nuclear factor-4 (HNF-4), which binds long-chain acyl-coenzyme A thioesters,29 in hypoxically induced gene expression is only partly elucidated. But for full hypoxic induction of the hepatic EPO gene, the binding of HNF-4 is required.30 However, studies of the effects of IL-1β or TNF-α on HNF-4 DNA binding have not been reported. Thus, it is not surprising that our reporter gene assays have shown that increased HIF-1 DNA binding activity is functional under hypoxic conditions only.
The 5′ flanking region of the EPO gene contains a number of elements potentially responsive to cytokines, including nuclear factor Kappa B (NFκB) and GATA transcription factors binding sites.1,31 NFκB is activated in HepG2 cells upon treatment with IL-1β or TNF-α,32 but a suppressive role in EPO gene expression has not been reported. GATA-2 binding represses EPO gene transcription.33 Hydrogen peroxide, a putative intracellular messenger between the O2 sensor and the EPO gene,34 inhibits EPO production by increasing GATA-2 levels in Hep3B cells.35 Thus, it must be considered that IL-1β and TNF-α may act by a similar mechanism.
In contrast to their effects on EPO production, IL-1β and TNF-α did not decrease VEGF mRNA levels and VEGF production in HepG2 cells. Others have found that IL-1 increases VEGF mRNA levels in smooth muscle36 and fibroblasts.37,38 TNF-α is effective in keratinocytes39 and the epidermoid carcinoma line A-431.40 HIF-1 is considered the maintrans-acting factor in controlling the rate of transcription of the VEGF gene.12,41 We propose that HIF-1 activation is induced not only by hypoxia but also by the proinflammatory cytokines IL-1β and TNF-α. If so, HIF-1 may play an important role not only in oxygen and energy homeostasis but also in immune responses. For instance, hepatic genes that can be activated by HIF-1 encode acute-phase proteins42 and inducible nitric oxide synthase.43 Our observation that cytokines can activate HIF-1 in normoxic conditions emphasizes its possible role as atrans-acting factor in inflammatory processes.
ACKNOWLEDGMENT
We are grateful to Dr O. Hankinson (Los Angeles, CA) and Dr D. Livingston (Boston, MA) for generously providing Hepa-1 cells and anti–HIF-1α antibodies, respectively. We thank Dr A. Rolfs (Zürich, Switzerland) for assistance in setting up the HIF-1 electrophoretic mobility shift assay and Dr T. Kietzmann for providing the pEpo-HIF plasmid.
Supported by a grant from the Deutsche Forschungsgemeinschaft (SFB 367-C8).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Thomas Hellwig-Bürgel, Dipl-Biol, Institut für Physiologie, Medizinische Universität zu Lübeck, Ratzeburger Allee 160, D-23538 Lübeck, Germany; e-mail: Hellwig@physio.mu-luebeck.de.