Abstract
The transcriptional mechanisms that drive colony-forming unit granulocyte-macrophage (CFU-GM) myeloid progenitors to differentiate into cells of either the granulocytic or monocytic lineage are not fully understood. We have shown that the c-Maf and c-Myb transcription factors physically interact in myeloid cells to form inhibitory complexes that hinder transactivation of c-Myb target genes through direct binding to Myb consensus sites. These complexes arise in a developmentally regulated pattern, peaking at the promyelocyte stage, or in cell model systems, appearing soon after the induction of monocytic differentiation. We wished to determine if this developmentally related interaction is a consequence of myeloid differentiation or an intrinsic differentiating stimulus. Because the elevated Myb:Maf status seen in differentiating cells can be recapitulated by overexpression of c-Maf in myeloid cell lines, we inducibly expressed the c-Maf cDNA in 2 bipotent human myeloid progenitor cells. Elevated levels of c-Maf protein led to marked increases in Myb:Maf complexes and the accumulation of monocyte/macrophage cells, followed by eventual programmed cell death. Analysis of targets that could mediate these phenotypic changes indicated that c-Maf likely plays a key role in myeloid cell development through dual mechanisms; inhibition of a select set of c-Myb regulated targets, such as Bcl-2 and CD13/APN, coupled with the activation of as yet undefined differentiation-promoting genes.
PRECISE REGULATION OF the c-Myb transcription factor is essential for normal hematopoiesis. Interference with T-cell–specific c-Myb expression in transgenic mice causes growth arrest and reduced proliferative capacity of the T-cell compartment.1 c-Myb null mice die late in gestation and display greatly reduced numbers of hematopoietic progenitors, presumably due to their reduced capacity to establish a normal proliferative program.2 However, these progenitors can still differentiate, suggesting that c-Myb sustains hematopoietic progenitor cells in a proliferative mode, thereby regulating the shift between proliferation and differentiation.2-4 Consequently, inhibition of Myb activity during hematopoiesis could block proliferation, setting the stage for differentiation on receipt of the appropriate inductive signals.
Members of the Maf family of basic region/leucine zipper (bZIP) transcription factors play an essential role in growth and development by regulating tissue-specific gene expression.5-7 These proteins activate or repress transcription depending on their particular protein partner and the context of the target promoter. For example, selective expression of c-Maf dictates the ratio of T-helper cell subsets by positively regulating interleukin-4 (IL-4) expression.5 Similarly, the differentially expressed and highly homologous MafB protein physically binds to and inhibits the activity of the Ets-1 protein in avian myeloid cells.7 Maf family proteins are essential to both avian and mammalian development and differentiation. The tissue restricted avian L-Maf regulates multiple lens-specific genes, and ectopic expression of this protein is sufficient to induce lens differentiation in ectodermal and cultured cells.6 Finally, c-Maf null mice have severe multisystem defects with high mortality rates in late gestation (L. Glimcher, personal communication, July 1998, and Kim et al8), illustrating a fundamental role for c-Maf in mammalian development.
We have recently demonstrated9 that c-Myb and c-Maf physically interact, resulting in the inhibition of Myb-dependent gene transcription in early myeloid cells through a mechanism that requires the binding of c-Myb, but not Maf, to DNA. Although c-Maf mRNA and protein levels remain constant during myeloid development, the formation of inhibitory Myb:Maf-DNA complexes is developmentally regulated, with the highest levels found in promyelocytic cells and in progenitors soon after their induction to differentiate. By contrast, markedly lower levels of these complexes are present in either immature myeloblasts or later developmental stages, and during terminal differentiation. The regulation of complex formation appears to be independent of Myb, as addition of Myb protein does not affect Myb:Maf complex levels, or do levels of free Myb protein correlate with complex levels.9 This developmental pattern of protein interaction suggests that Maf modulation of c-Myb activity may be an important regulatory pathway for the control of transcription and the initiation of myeloid cell differentiation.
Enforced expression of c-Maf in early myeloid cells lacking Myb:Maf complexes results in the formation of inhibitory complexes, thereby recapitulating the regulatory interactions normally occurring in maturing myeloid cells.9 Because downregulation of Myb may prepare cells for differentiation, we postulated that conditional expression of c-Maf would both inhibit obligatory Myb targets and provide the necessary inductive signals to drive cells to differentiate. Here we report that induction of c-Maf in 2 bipotential human myeloid cell lines induces the appearance and accumulation of cells characteristic of the monocytic phenotype. Importantly, levels of Myb:Maf complexes in these induced clones increased in a pattern similar to that seen on phorbol-induced monocytic differentiation, supporting the hypothesis that c-Maf plays a crucial role in myeloid cell differentiation. Finally, prolonged c-Maf induction in these cell lines results in a decrease in Bcl-2 protein levels and subsequent apoptotic cell death, consistent with inhibition of Myb activity and terminal monocytic differentiation.
MATERIALS AND METHODS
Cell lines.
Human cell lines included the myeloid leukemia lines HL-60 (American Type Culture Collection [ATCC], Rockville, MD, CRL 1593) and U937 (ATCC, CRL 1593). Cells were grown in RPMI-1640 medium supplemented with 2 mmol/L L-glutamine and 10% fetal calf serum. HL-60 and U937 cells were induced to differentiate by using 12-0-tetradecanoylphorbol diester (TPA, 5 × 10−6 mol/L) added to the culture medium; cells were harvested at indicated time points after addition. Anti-IL–4 and matched control antibodies (R & D Systems, Minneapolis, MN) were used according to the manufacturer’s instructions.
Expression vectors and reporter plasmids.
The CD13/APN minimal reporter construct −411luc contains genomic sequences from bp −411 through bp +65 of the CD13/APN myeloid promoter in the pGL2basic backbone (Promega, Madison, WI)10. The conditional expression plasmid pMT-CB6-cMaf was made by cloning the full-length murine c-Maf cDNA11 into HindIII/XbaI cut pMT-CB69. Ligation of the Myb-dominant interfering construct (MEnT1, the generous gift of Dr Kathleen Weston, CRC Centre for Cell and Molecular Biology, Chester Beatty Laboratories, London, UK) into XbaI cut pMT-CB6 vector resulted in the conditional Myb dominant negative construct pMT-CB6-MEnT.
Transfection of recombinant plasmids and reporter gene assays.
To analyze inhibition of Myb transcriptional activity, c-Maf stably transfected U937 or HL-60 cloned cell lines were induced with 150 μmol/L ZnSO4 for the indicated time intervals before electroporation with 5 μg of the CD13/APN −411luc promoter construct10 and 2 μg of the control β-actin–secreted alkaline phosphatase (SEAP) plasmid as described.10 The transfection efficiency for each construct was normalized to the control level of SEAP activity12; the reported values were calculated as relative light units (RLU) per unit of SEAP activity. Each point was determined at least 4 times.
Northern and Western blot analysis.
polyA+ RNA was purified from cell lines using the FastTrack 2.0 kit (Invitrogen, Carlsbod, CA). Total RNA was extracted using Tri Reagent (Molecular Research Center, Inc, Cincinnati, OH). A total of 10 μg of poly A+ RNA or 20 μg of total RNA from the indicated cell lines was separated on a 1% agarose-formaldehyde gel, transferred to nylon membranes, and sequentially probed with the BstEII/NcoI fragment containing the 5′ region of murine c-Maf (which excludes the bZIP domain), the 3-kb BamHI fragment of the murine CSF-1R cDNA, and a glyceraldehyde-3-phosphate dehydrogenase (G3PDH) cDNA probe (Clontech, Palo Alto, CA). For Western blot analysis, proteins were separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto nitrocellulose. Membranes were blocked with 5% milk in Tris-buffered saline (TBS) buffer; incubated with the primary antibody (rabbit anti v-Maf and goat anti-actin polyclonal antisera, Santa Cruz Biotechnology, Santa Cruz, CA; monoclonal mouse anti-human Bcl-2, Upstate Biotechnology, Lake Placid, NY; or anti-Myc for detection of Myc-tagged MEnT, Calbiochem, San Diego, CA), followed by horseradish peroxidase (HRP)-linked secondary antibody. Specific HRP-conjugated protein complexes were detected according to the ECL protocol (Amersham, Arlington Heights, IL).
DNA binding assays.
For DNA binding assays, whole-cell lysates from myeloid cells were prepared by resuspending and washing pelleted cells in cold phosphate-buffered saline (PBS) and resuspending the pellet in lysis buffer (20 mmol/L Tris-HCl, pH 7.5, 2 mmol/L dithiothreitol [DTT], 20% glycerol, 50 mmol/L KCl) containing a protease inhibitor cocktail (Boehringer-Mannheim Complete Inhibitor Cocktail Tablets; Boehringer-Mannheim, Indianapolis, IN). After addition of Triton X-100 to 0.5% final concentration (vol/vol), the lysate was incubated on ice for 60 minutes, and the debris pelleted for 15 minutes at 14,000 rpm. Cleared lysate was quantitated and stored at −80°C until use. Cell lysates (20 μg) were preincubated in binding buffer (10 mmol/L HEPES, pH 7.9, 50 mmol/L KCl, 5 mmol/L MgCl2, 10% glycerol, 1 mmol/L DTT, and 1 μg poly d[I-C]) in 25-μL reactions with or without 0.1 μg of the consensus double-stranded oligonucleotide competitors (Myb consensus site: mimAmyb-CTAGGACATTAT AACGGTTTTTTAGT10; Maf consensus site: MARE-TCGAGCTCGGAATTGCTGACT CAGCATTACTC13) for 15 minutes at 4°C before the addition of probe. For supershift experiments, 1 μL of an anti–v-Maf polyclonal antibody (Santa Cruz Biotechnology, cross-reactive with c-Maf) was preincubated with lysate for 10 minutes at 4°C. The 32P-end–labeled genomic fragment probe (CD13/APN XbaI/DdeI fragment; containing sequences bp −433 through −3059) was then added and incubated for an additional 15 minutes at 4°C. Binding reactions were electrophoresed through a 3.5% acrylamide gel containing 10% glycerol in TAE buffer at 4°C. Gels were dried and exposed to x-ray film (Kodak, Rochester, NY).
Apoptosis analysis.
DNA from ZnSO4 induced cells was isolated at the indicated intervals (see Fig 8C) using described methods.14 Briefly, cells were harvested, washed with PBS and resuspended in lysis buffer (1% NP-40 in 20 mmol/L EDTA, 50 mmol/L Tris-HCl, pH 7.5; 10 mL per 1 × 106 cells). After centrifugation, SDS was added to 1% and the supernatants were treated for 2 hours with RNase A (5 mg/mL final concentration) at 56°C followed by digestion with proteinase K (2.5 mg/mL final concentration) for 2 hours at 37°C. The DNA was precipitated, separated by electrophoresis in 1% agarose, and visualized by ethidium bromide staining. TdT-mediated dUTP nick end labeling (TUNEL) assays were performed as described15 on cells treated with zinc for 24 hours (MEnT) or 48 hours (c-Maf).
Reverse transcription-polymerase chain reaction (RT-PCR).
RNA was extracted with TRI-Reagent (Molecular Research Center, Inc) according to the method provided. Fifteen micrograms of total RNA was used as template for first-strand complementary DNA (cDNA) synthesis using Superscript II reverse transcriptase (GIBCO/BRL, Gaithersburg, MD) with 100 U ribonuclease inhibitor, 1 μL random primer (GIBCO/BRL), and nuclease-free water to 36 μL. The reaction was heated to 70°C for 10 minutes and then quick chilled on ice. Second-strand synthesis was performed in 5X first strand buffer (12 μL), 10 mmol/L each dNTP (3 μL), and 3 μL SuperScript II, and the reaction incubated at 25°C for 10 minutes, 42°C for 50 minutes, and at 75°C for 15 minutes. PCR amplification was performed with 1 μL cDNA, 10 μL 10X PCR buffer, 8 μL dNTP (2.5 mmol/L each), 1 μL 3′ primer (100 mmol/L), 1 μL 5′ primer (100 mmol/L), add dH2O up to 99.5 μL, and 0.5 μL AmpliTaq Gold. The cDNA was heat denatured at 94°C for 9 minutes, then amplified through 35 cycles of 94°C, 45 seconds, 58°C, 45 seconds, and 72°C, 45 seconds. For c-Maf RT-PCR, 5′ primer: TGCACTTCGACGACCGCTTCTCGG, 3′ primer: AAGGTGGCTAGCTGGAATCGCG. MafG RT-PCR primers are as published16; IL-4 and b-actin primers were purchased from Stratagene (La Jolla, CA). Positive control plasmid template containing the IL-4 cDNA was purchased from ATCC.
RESULTS
c-Maf promotes monocytic/macrophage differentiation of HL-60 and U937 myeloid cells.
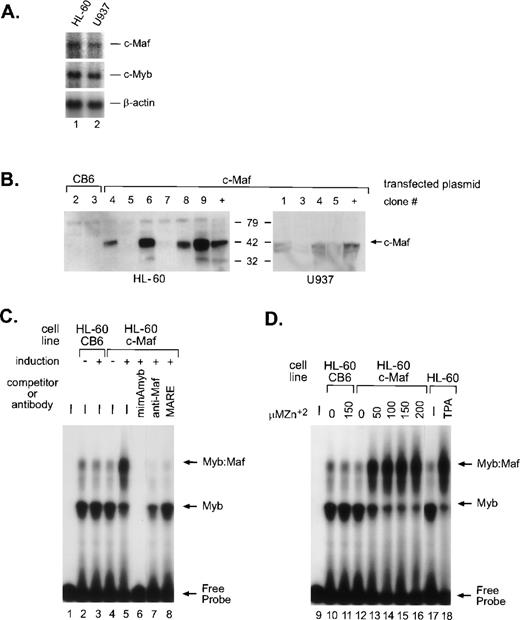
In myeloid cells, the highest levels of Myb:Maf protein complexes are found in those representing early stages of differentiation or in cells soon after the induction of monocytic differentiation. By contrast, markedly lower levels are apparent in either immature myeloblasts or later developmental stages, linking the interaction of these 2 proteins with myeloid differentiation.9 To address whether this interaction is a consequence of myeloid differentiation signals or a differentiating stimulus in and of itself, we engineered HL-60 and U937 cells (containing equivalent, low levels of endogenous c-Maf mRNA, Fig 1A) to inducibly express the full-length murine c-Maf cDNA11 using the metallothionine-based expression vector pMT-CB6.9 Although protein expression from the individual clones varied, extremely high induction of c-Maf was detected in several clonal lines (Fig 1B). The low level of endogenous c-Maf protein in parental cells is not detectable in whole-cell lysates, but is evident on immunoprecipitation followed by Western blot analysis.9 Consistent with our model, electrophoretic mobility shift assay (EMSA) analysis of myeloid cell lysates indicated a dose-dependent increase in the levels of Myb:Maf containing complexes binding to theCD13/APN target promoter fragment on zinc induction of c-Maf-containing clonal lines (Figs 1C, lanes 4 and 5; 1D, lanes 10 to 16), in effect, mirroring the increase observed in cell lysates from TPA-induced (monocytic lineage) parental cell lines (Fig 1D, lanes 17 and 18). These complexes consist of physically interacting Myb and Maf proteins bound to the Myb consensus site in the target promoter fragment as confirmed by competitor oligonucleotide and specific antibody abrogation of the complexes (Fig 1C, lanes 6 to 8 and Hegde et al9). Maf itself does not contact the CD13/APNpromoter probe.9 Therefore, these c-Maf–expressing clones recapitulate the Myb:Maf status of myeloid cells driven to differentiate along the monocyte/macrophage pathway.
c-Maf in myeloid cell lines and stable cell clones. (A) Endogenous expression of c-Maf mRNA expression levels in human myeloid cell lines as determined by Northern blot analysis of polyA+ RNA from the HL-60 promyelocytic or U937 monoblastic cell lines. A single blot was initially probed with the c-Maf specific cDNA probe, stripped, and reprobed with c-Myb, and finally, β-actin as a control for RNA loading and integrity. Exposure times for optimal detection of c-Maf mRNA were routinely significantly longer than for other probes (1 week v 1 day). (B) Conditional expression c-Maf in stably transfected clones. Individual HL-60 or U937 clonal cell lines containing either CB6 vector alone (left 2 lanes) or c-Maf (right lanes) encoding plasmids were cultured for 24 hours with 150 μmol/L ZnSO4. Cell lysates were analyzed beside control lysates from a previously established c-Maf inducible cell line (+).9 The gels were transferred and probed for c-Maf expression with an antiserum that recognized the c-Maf protein. (C) Myb:Maf complexes increase on c-Maf induction. An end-labeled 129-bp promoter fragment probe containing the functionally definedCD13/APN myeloid promoter elements9 was incubated with uninduced (-) or induced (+) whole-cell lysates from control vector-containing (CB6, lanes 2 and 3), c-Maf–containing (c-Maf clone 6, lanes 4 to 8) stable HL-60 clones. Unlabeled competitor oligonucleotides containing either the consensus Myb site from themim-1 promoter (mimAmyb, lane 6) or the consensus Maf binding site (MARE, lane 8) were added to the assays in 100-fold molar excess before addition of probe. Maf binding to its consensus oligonucleotide interferes with its ability to complex with Myb.9Antibodies recognizing the c-Maf protein (lane 7) were added to binding reactions before probe addition. (D) The increase in Myb:Maf complexes is dose-dependent and comparable to induction on monocytic differentiation. The identical promoter fragment probe used in (C) was incubated with uninduced (0) or induced (indicated concentrations) whole-cell lysates from control vector-containing (CB6, lanes 10 and 11), c-Maf–containing (c-Maf, lanes 12 to 16) stable HL-60 clonal lines or from untreated (lane 17), TPA-treated (lane 18) HL-60 parental cells.
c-Maf in myeloid cell lines and stable cell clones. (A) Endogenous expression of c-Maf mRNA expression levels in human myeloid cell lines as determined by Northern blot analysis of polyA+ RNA from the HL-60 promyelocytic or U937 monoblastic cell lines. A single blot was initially probed with the c-Maf specific cDNA probe, stripped, and reprobed with c-Myb, and finally, β-actin as a control for RNA loading and integrity. Exposure times for optimal detection of c-Maf mRNA were routinely significantly longer than for other probes (1 week v 1 day). (B) Conditional expression c-Maf in stably transfected clones. Individual HL-60 or U937 clonal cell lines containing either CB6 vector alone (left 2 lanes) or c-Maf (right lanes) encoding plasmids were cultured for 24 hours with 150 μmol/L ZnSO4. Cell lysates were analyzed beside control lysates from a previously established c-Maf inducible cell line (+).9 The gels were transferred and probed for c-Maf expression with an antiserum that recognized the c-Maf protein. (C) Myb:Maf complexes increase on c-Maf induction. An end-labeled 129-bp promoter fragment probe containing the functionally definedCD13/APN myeloid promoter elements9 was incubated with uninduced (-) or induced (+) whole-cell lysates from control vector-containing (CB6, lanes 2 and 3), c-Maf–containing (c-Maf clone 6, lanes 4 to 8) stable HL-60 clones. Unlabeled competitor oligonucleotides containing either the consensus Myb site from themim-1 promoter (mimAmyb, lane 6) or the consensus Maf binding site (MARE, lane 8) were added to the assays in 100-fold molar excess before addition of probe. Maf binding to its consensus oligonucleotide interferes with its ability to complex with Myb.9Antibodies recognizing the c-Maf protein (lane 7) were added to binding reactions before probe addition. (D) The increase in Myb:Maf complexes is dose-dependent and comparable to induction on monocytic differentiation. The identical promoter fragment probe used in (C) was incubated with uninduced (0) or induced (indicated concentrations) whole-cell lysates from control vector-containing (CB6, lanes 10 and 11), c-Maf–containing (c-Maf, lanes 12 to 16) stable HL-60 clonal lines or from untreated (lane 17), TPA-treated (lane 18) HL-60 parental cells.
Our initial observations of three different pMT-c-Maf–containing U937 clonal cell lines showed striking morphological changes after 48 hours of zinc induction. These cells proliferated more slowly, were noticeably larger, extensively vacuolated and multinucleated with a decreased nuclear:cytoplasmic ratio (Fig 2, lower left panel), characteristic of macrophage morphology. To verify this phenotypic designation, cells were stained for α-napthyl-butyrate esterase (ANB, Fig 2, right panel), a standard indicator of monocytic specification.17 The development of deep red cytoplasmic staining in 50% to 60% of the c-Maf–overexpressing cells and TPA-treated control cells verified the presence of ANB esterase and confirmed that c-Maf overexpression triggered monocytic differentiation.
Morphologic and lineage analysis of cloned U937 cell lines conditionally expressing c-Maf. Cytospin preparations of parental U937 cells either untreated (top row) or treated with TPA (second row); or U937 clonal lines containing either vector control (U937 CB6, third row) or c-Maf expression constructs (U937 c-Maf, bottom row) incubated for 48 hours with 150 μmol/L ZnSO4. Left column: Wright-Giemsa stain for morphological examination; right column: -napthyl butyrate esterase stain specific for monocytic lineage.
Morphologic and lineage analysis of cloned U937 cell lines conditionally expressing c-Maf. Cytospin preparations of parental U937 cells either untreated (top row) or treated with TPA (second row); or U937 clonal lines containing either vector control (U937 CB6, third row) or c-Maf expression constructs (U937 c-Maf, bottom row) incubated for 48 hours with 150 μmol/L ZnSO4. Left column: Wright-Giemsa stain for morphological examination; right column: -napthyl butyrate esterase stain specific for monocytic lineage.
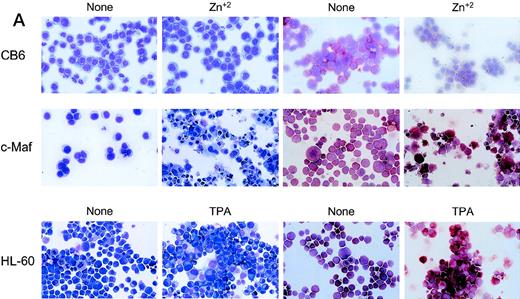
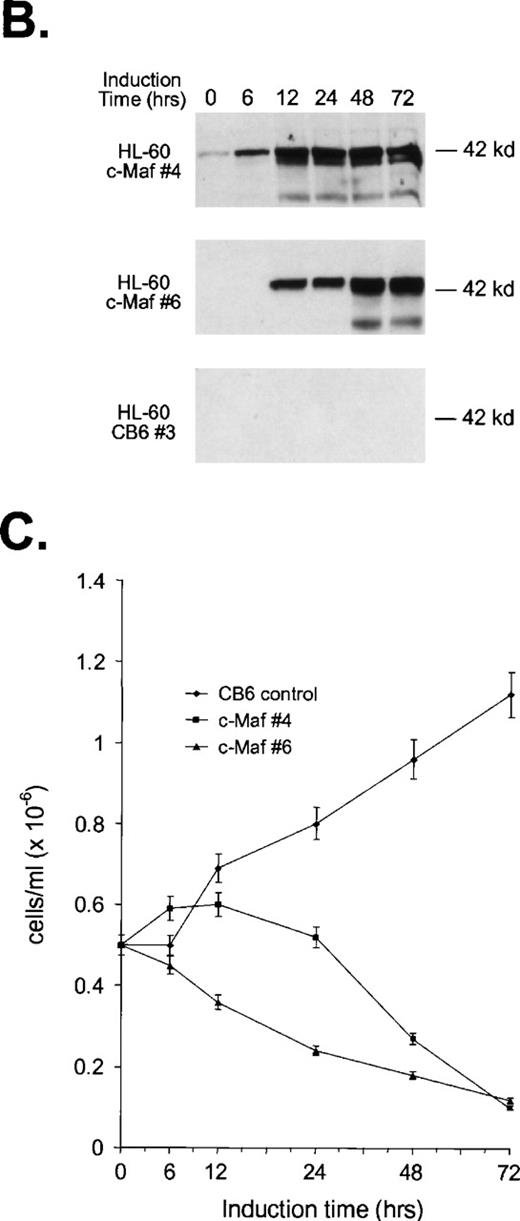
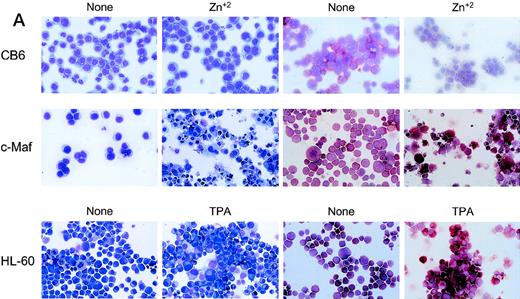
To confirm these observations in other myeloid progenitors, we also examined c-Maf–expressing clones established in HL-60 cells that readily differentiate into monocytes when treated with TPA.18 c-Maf–overexpressing HL-60 cells also displayed characteristics of differentiation along the monocytic lineage, including ANB esterase positive cells with a decreased nuclear:cytoplasmic ratio (Fig 3A). These changes occurred more rapidly in the HL-60 cells than in the U937 lines: early morphologic changes and esterase activity were apparent after 12 hours of zinc treatment (data not shown), which correlated closely with c-Maf protein levels (Fig 3B). By 24 to 48 hours after zinc induction, 70% to 90% of the cells exhibited morphology characteristic of differentiated monocytic cells, comparable to that seen on TPA treatment (89% to 95%). A smaller proportion of multinucleated cells was apparent in c-Maf containing HL-60 as compared with U937 cell cultures (4% v 15% containing 2 to 15 nuclei), and this ratio increased slightly on longer zinc incubation (10% at 48 to 72 hours). ANB esterase staining of cytospin preparations indicated that 45% to 50% of c-Maf–expressing cells displayed this monocyte-specific enzyme activity, identical to ratios obtained in parental cells or HL-60 c-Maf clone 6 cells induced with TPA (≈50%). Similar morphologic and ANB data were obtained with HL-60 c-Maf clone 4, while identical treatment of vector containing control cells showed no analogous changes (data not shown). Finally, the induction of c-Maf in these clones altered their normal growth kinetics, as illustrated by the reduced proliferative capacity of c-Maf containing clones, but not vector control clones after exposure to zinc (Fig 3C).
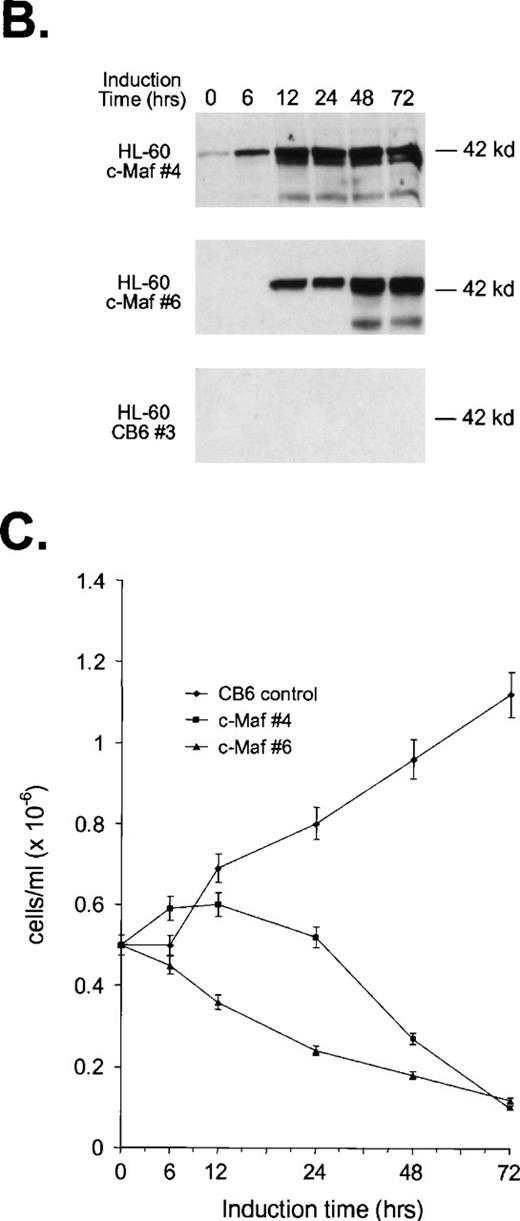
Analysis of cloned HL-60 cell lines conditionally expressing c-Maf. (A) Lineage analysis of stable HL-60 lines expressing c-Maf. Cell lines containing either vector control (CB6, top row) or c-Maf (c-Maf, middle row) were untreated (none) or incubated with 150 μmol/L ZnSO4 (Zn+2), or parental HL-60 cells (bottom row) were untreated (none) or TPA treated (TPA). Cytospin preparations were stained with Wright-Giemsa (left two panels) or -napthyl butyrate esterase monocyte-specific stain (right two panels). (B) Conditional expression of c-Maf over time. Individual HL-60 clonal cell lines containing either CB6 vector (HL-60 CB6 clone 3) or CB6-c-Maf-(HL-60 c-Maf clones 4 and 6) were induced with ZnS04 for the indicated time periods and lysates were probed for c-Maf expression with an antiserum recognizing the c-Maf protein. (C) Induction of c-Maf alters normal growth kinetics. Viable cell numbers were determined for vector control-containing (CB6) or c-Maf–containing (c-Maf clones 4 and 6) stable cell lines by trypan blue dye exclusion at the indicated intervals after zinc exposure. Representative data from 2 individual experiments are presented.
Analysis of cloned HL-60 cell lines conditionally expressing c-Maf. (A) Lineage analysis of stable HL-60 lines expressing c-Maf. Cell lines containing either vector control (CB6, top row) or c-Maf (c-Maf, middle row) were untreated (none) or incubated with 150 μmol/L ZnSO4 (Zn+2), or parental HL-60 cells (bottom row) were untreated (none) or TPA treated (TPA). Cytospin preparations were stained with Wright-Giemsa (left two panels) or -napthyl butyrate esterase monocyte-specific stain (right two panels). (B) Conditional expression of c-Maf over time. Individual HL-60 clonal cell lines containing either CB6 vector (HL-60 CB6 clone 3) or CB6-c-Maf-(HL-60 c-Maf clones 4 and 6) were induced with ZnS04 for the indicated time periods and lysates were probed for c-Maf expression with an antiserum recognizing the c-Maf protein. (C) Induction of c-Maf alters normal growth kinetics. Viable cell numbers were determined for vector control-containing (CB6) or c-Maf–containing (c-Maf clones 4 and 6) stable cell lines by trypan blue dye exclusion at the indicated intervals after zinc exposure. Representative data from 2 individual experiments are presented.
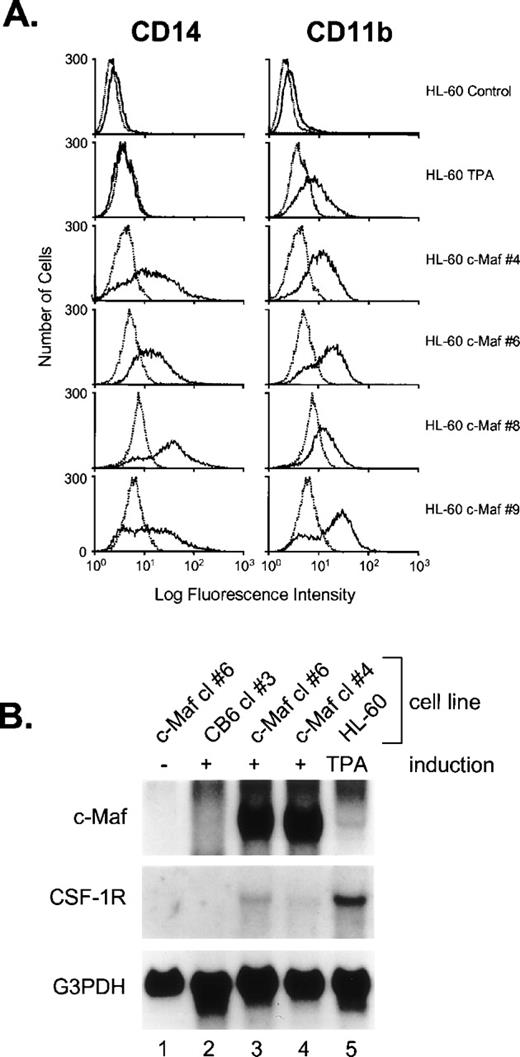
The induction of monocytic differentiation by c-Maf was further confirmed in individual HL-60 clones cultured in zinc (protein levels shown in Fig 1B) by flow cytometry and Northern blot analysis (Fig 4). Elevated cell-surface levels of the myeloid maturation marker CD11b to levels equivalent to those seen on TPA induction and the monocyte-restricted CD14 protein to levels higher than TPA induced cells (Fig 4A), and the appearance of low, but significant, levels of CSF-1 receptor mRNA (Fig 4B) confirms the monocytic phenotype of the c-Maf–containing cells. Thus, the enforced expression of c-Maf in multiple independent clones in two different cell lines induces phenotypic changes consistent with monocytic differentiation.
c-Maf induces the expression of maturation markers in HL-60 cell lines. (A) Fluorescence-activated cell sorting (FACS) analysis of the expression of the CD14 and CD11b myeloid maturation markers on untreated HL-60 cells, HL-60 cells treated with TPA for 24 hours, and 4 individual c-Maf expressing HL-60 clones treated with zinc for 24 hours. Negative control antibody binding is indicated by dashed lines. (B) Expression of c-Maf mRNA (top panel), CSF-1 receptor (CSF-1R, center panel), or control G3PDH (lower panel), as determined by Northern blot analysis of total RNA from uninduced (−, lane 1) or zinc-induced (+) control vector-containing (CB6, lane 2), c-Maf–containing (lanes 1, 3, and 4) stable HL-60 clones; or TPA-treated HL-60 parental cells (lane 5). A single blot was initially probed with the c-Maf–specific cDNA probe, stripped, and reprobed with CSF-1R and G3PDH; G3PDH served as a control for RNA loading and integrity.
c-Maf induces the expression of maturation markers in HL-60 cell lines. (A) Fluorescence-activated cell sorting (FACS) analysis of the expression of the CD14 and CD11b myeloid maturation markers on untreated HL-60 cells, HL-60 cells treated with TPA for 24 hours, and 4 individual c-Maf expressing HL-60 clones treated with zinc for 24 hours. Negative control antibody binding is indicated by dashed lines. (B) Expression of c-Maf mRNA (top panel), CSF-1 receptor (CSF-1R, center panel), or control G3PDH (lower panel), as determined by Northern blot analysis of total RNA from uninduced (−, lane 1) or zinc-induced (+) control vector-containing (CB6, lane 2), c-Maf–containing (lanes 1, 3, and 4) stable HL-60 clones; or TPA-treated HL-60 parental cells (lane 5). A single blot was initially probed with the c-Maf–specific cDNA probe, stripped, and reprobed with CSF-1R and G3PDH; G3PDH served as a control for RNA loading and integrity.
IL-4 does not play a role in inducing monocytic differentiation.
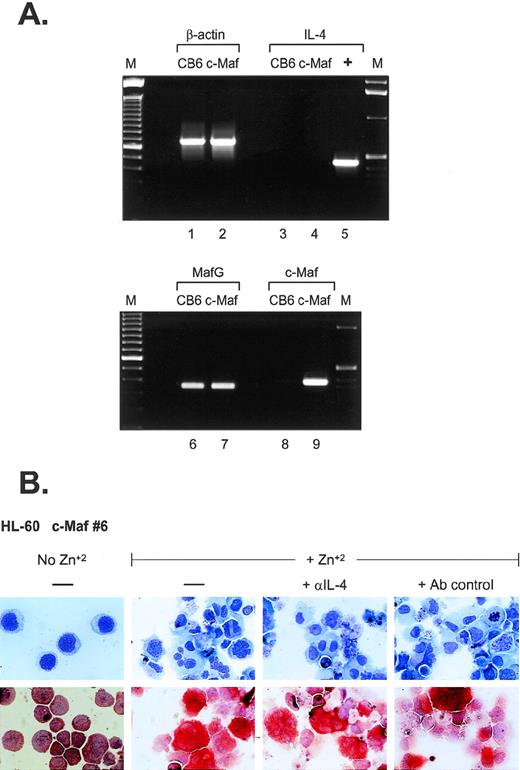
What are the possible mechanisms of monocytic differentiation induced by c-Maf? Relatively few hematopoietic target genes have been identified that are directly regulated by c-Maf through its activity as a DNA-binding transcription factor. Recent data have indicated that c-Maf directly activates the expression of IL-4 in T-helper cells and can induce the expression of the endogenous IL-4 gene in IL-4–negative B-cell lines.5 In several models of experimental inflammation, exogenous IL-4 can initiate the formation of multinucleated giant cells (MGC) through macrophage fusion.19-23 MGC contain multiple nuclei within extensively spread cytoplasm,24 similar to the phenotype of a percentage of c-Maf–induced monocytes, raising the possibility that overexpression of c-Maf in myeloid cell lines could also induce ectopic IL-4 production. To address this issue, we performed RT-PCR analysis (Fig 5A). Although c-Maf mRNA is highly induced in our myeloid clones (lanes 8 and 9), its upregulation does not initiate IL-4 mRNA synthesis (lanes 3 and 4). In addition, we added neutralizing antibodies to IL-4 to cultures of zinc-induced c-Maf clonal cell lines; these antibodies had no discernable effect on the previously observed morphologic changes or ANB esterase positivity (Fig5B). Similarly, addition of inhibitory concentrations of cyclosporin A (a potent and specific inhibitor of the calcineurin-dependent transcription factors required for IL-4 transcription25 26) had no effect on the c-Maf–induced phenotypic changes (data not shown). These results indicate that in our system, c-Maf induction does not initiate endogenous IL-4 expression and, therefore, IL-4 does not contribute to the observed monocytic differentiation.
IL-4 expression is not induced in c-Maf–expressing cell lines. (A) IL-4 mRNA is not induced on c-Maf protein expression. RT-PCR analysis of cDNA templates from induced CB6 vector control (lanes 1, 3, 6, and 8) or c-Maf-containing clone 6 (c-Maf, lanes 2, 4, 7, and 9) using primer pairs detecting β-actin control (lanes 1 and 2), IL-4 (lanes 3 to 5), MafG (lanes 6 and 7), or c-Maf (lanes 8 and 9). M, marker lanes; +, IL-4 positive control DNA template. (B) IL-4 neutralizing antibodies do not affect c-Maf–induced monocytic differentiation. Neutralizing antibodies directed against IL-4 or isotype-matched control antibodies were added to cultures of HL-60 c-Maf clone 6 cells at the time of zinc induction. Top row: Wright-Giemsa stain; bottom row: monocyte-specific ANB esterase stain of cytospin preparations.
IL-4 expression is not induced in c-Maf–expressing cell lines. (A) IL-4 mRNA is not induced on c-Maf protein expression. RT-PCR analysis of cDNA templates from induced CB6 vector control (lanes 1, 3, 6, and 8) or c-Maf-containing clone 6 (c-Maf, lanes 2, 4, 7, and 9) using primer pairs detecting β-actin control (lanes 1 and 2), IL-4 (lanes 3 to 5), MafG (lanes 6 and 7), or c-Maf (lanes 8 and 9). M, marker lanes; +, IL-4 positive control DNA template. (B) IL-4 neutralizing antibodies do not affect c-Maf–induced monocytic differentiation. Neutralizing antibodies directed against IL-4 or isotype-matched control antibodies were added to cultures of HL-60 c-Maf clone 6 cells at the time of zinc induction. Top row: Wright-Giemsa stain; bottom row: monocyte-specific ANB esterase stain of cytospin preparations.
Inhibition of Myb activity is not sufficient to drive differentiation.
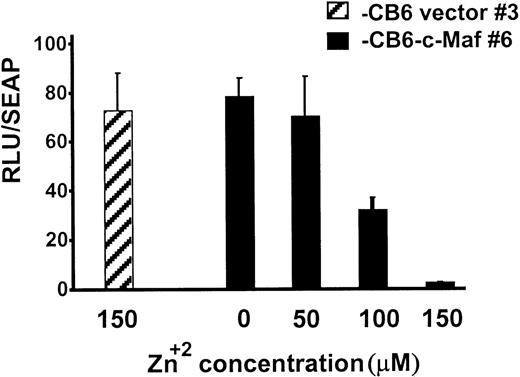
During myeloid differentiation, c-Maf forms inhibitory complexes with c-Myb, which function independently of Maf’s binding to DNA.9 Because the c-Maf protein controls gene expression through DNA-dependent, as well as DNA-independent, mechanisms, Maf-induced differentiation could be triggered by either or both of these mechanisms.5-7,9,11,27 To confirm that c-Maf induction altered Myb transactivation in our stable HL-60 clones, we transiently transfected these cells with Myb-dependent CD13/APNpromoter-driven reporter constructs.9 10 c-Maf expression inhibited luciferase activity of the reporter gene in these cells in a dose-dependent fashion with 90% inhibition at the highest dose tested (Fig 6). Therefore, Myb-dependent transcription is functionally impaired in these cells and in the U937 clonal lines as well (data not shown).
Myb-dependent transcription is functionally impaired in clonal lines of HL-60 cells expressing c-Maf. HL-60 c-Maf clone 6 (▪) or CB6 vector control clone 3 (▨) were incubated at the indicated ZnSO4 concentrations for 12 hours to induce c-Maf protein expression, and then transiently transfected with 5 μg of the −411luc reporter construct containing sequences sufficient for wild-type level, tissue-appropriate expression from theCD13/APN myeloid promoter. Luciferase activities were assayed at 6 hours and normalized for differences in transfection efficiency with SEAP activity produced by the control β-actin-SEAP plasmid.
Myb-dependent transcription is functionally impaired in clonal lines of HL-60 cells expressing c-Maf. HL-60 c-Maf clone 6 (▪) or CB6 vector control clone 3 (▨) were incubated at the indicated ZnSO4 concentrations for 12 hours to induce c-Maf protein expression, and then transiently transfected with 5 μg of the −411luc reporter construct containing sequences sufficient for wild-type level, tissue-appropriate expression from theCD13/APN myeloid promoter. Luciferase activities were assayed at 6 hours and normalized for differences in transfection efficiency with SEAP activity produced by the control β-actin-SEAP plasmid.
Myb mRNA and protein levels normally decrease during hematopoietic cell differentiation.28-30 Downregulation is obligatory for differentiation to proceed, as enforced expression of Myb blocks differentiation of hematopoietic cell lines.31-34 Given the functional relationship between Myb levels and hematopoietic cell differentiation, it was possible that inhibition of Myb activity alone was sufficient to produce the commitment and phenotypic changes observed in the c-Maf–expressing cell lines. To test this possibility, we established stable lines that conditionally express a dominant interfering Myb containing the Drosophila Engrailed repressor domain fused to the Myb DNA binding domain. Ectopic expression of this chimeric protein has been shown to actively inhibit transcription from multiple promoters containing Myb consensus sites.1,10 35Its conditional expression in multiple stably transfected HL-60 clones failed to induce the cell-surface markers (Fig 7) or cytosolic ANB esterase activity alterations (data not shown) seen during parallel induction of c-Maf–expressing clones over a range of time points. Therefore, inhibition of Myb activity alone is not sufficient to initiate c-Maf–like monocytic differentiation in these cells, suggesting that c-Maf subserves other functions during monocytic commitment.
MEnT does not induce the expression of monocyte specific markers in HL-60 cell lines. Flow cytometric analysis of the expression of the CD14 and CD11b maturation markers on untreated, zinc treated (18 hours), or TPA treated (18 hours) HL-60 MEnT-containing cells. Negative control antibody binding is indicated by dashed lines. Data shown are a representative tracing of clonal line, HL-60 MEnT clone 1 (Western blot showing protein expression Fig 9A).
MEnT does not induce the expression of monocyte specific markers in HL-60 cell lines. Flow cytometric analysis of the expression of the CD14 and CD11b maturation markers on untreated, zinc treated (18 hours), or TPA treated (18 hours) HL-60 MEnT-containing cells. Negative control antibody binding is indicated by dashed lines. Data shown are a representative tracing of clonal line, HL-60 MEnT clone 1 (Western blot showing protein expression Fig 9A).
Maf-induced differentiation terminates in apoptosis.
Active inhibition of Myb in avian myeloid and mammalian T cells results in the downregulation of the antiapoptotic effector, Bcl-2, a direct transcriptional target of Myb, followed by induction of apoptosis.35 36 We observed that induction of either c-Maf or the dominant interfering Myb construct (MEnT) in HL-60 clonal lines generates substantial cell death when compared with zinc-treated control cells (Fig 3C and data not shown). Flow cytometric analysis of the DNA content of propidium iodide stained cells showed the appearance of a significant percentage of c-Maf, and a higher ratio of positive control MEnT expressing lines, present in a sub-G1 peak by 24 hours and as shown at 72 hours (Fig 8A), indicative of apoptotic cell death. Untreated cells (Fig 8A) or vector control cells treated with zinc (data not shown) did not result in similar DNA content profiles. We confirmed that c-Maf containing cells were dying by apoptosis by TUNEL assay (Fig 8B) and DNA fragmentation analysis (Fig 8C). This latter assay also indicated that c-Maf–induced apoptosis is delayed relative to that induced by the positive control MEnT fusion protein (24 to 48 hours v 6 hours), probably reflecting differences in the range of Myb targets inhibited by Maf versus MEnT.
c-Maf–expressing cells die by apoptosis. (A) c-Maf– and MEnT-containing cells show a sub-G1 peak indicative of apoptotic cell death. Positive control MEnT-containing (MEnT, top two panels) or c-Maf–containing (c-Maf, lower panels) that had been untreated or treated with ZnSO4 for 72 hours, stained with propidium iodide, and analyzed for DNA content by flow cytometry. Representative data is presented. (B) c-Maf– and MEnT-expressing cells show increased TUNEL staining. HL-60 clones overexpressing MEnT, CB6 vector, or c-Maf were treated with zinc for 48 hours (c-Maf, CB6) or 24 hours (MEnT), stained, and assessed by flow cytometry. Representative data are presented. (C) c-Maf– and MEnT-containing cells show DNA “laddering” characteristic of apoptosis. DNA was extracted from negative control CB6 (lanes 1 and 2), c-Maf–containing (c-Maf, lanes 3 to 6), or positive control MEnT-containing cells (lanes 7 to 9) that had been treated with ZnSO4 for the indicated time periods and separated by agarose gel electrophoresis.
c-Maf–expressing cells die by apoptosis. (A) c-Maf– and MEnT-containing cells show a sub-G1 peak indicative of apoptotic cell death. Positive control MEnT-containing (MEnT, top two panels) or c-Maf–containing (c-Maf, lower panels) that had been untreated or treated with ZnSO4 for 72 hours, stained with propidium iodide, and analyzed for DNA content by flow cytometry. Representative data is presented. (B) c-Maf– and MEnT-expressing cells show increased TUNEL staining. HL-60 clones overexpressing MEnT, CB6 vector, or c-Maf were treated with zinc for 48 hours (c-Maf, CB6) or 24 hours (MEnT), stained, and assessed by flow cytometry. Representative data are presented. (C) c-Maf– and MEnT-containing cells show DNA “laddering” characteristic of apoptosis. DNA was extracted from negative control CB6 (lanes 1 and 2), c-Maf–containing (c-Maf, lanes 3 to 6), or positive control MEnT-containing cells (lanes 7 to 9) that had been treated with ZnSO4 for the indicated time periods and separated by agarose gel electrophoresis.
Because it was possible that apoptosis was triggered by Maf downregulation of Bcl-2 through inhibition of Myb activity, we determined Bcl-2 protein levels in the induced cells by Western blot analysis (Fig 9). As expected, conditional expression of the MEnT Myb interfering protein in 2 HL-60 clonal cell lines resulted in decreased Bcl-2 expression (Fig 9A). Induction of c-Maf in HL-60 cells also reduced Bcl-2 protein levels between 12 and 24 hours of induction (Fig 9B), directly corresponding with the upregulation of Maf protein and the formation of inhibitory Myb:Maf complexes (Fig 1C). By contrast, addition of zinc to vector containing control cell lines showed no change in Bcl-2 levels (Fig 9B). Overall, these data suggest that Maf inhibits Myb transregulatory activity, which in turn, downregulates Bcl-2, resulting in apoptotic cell death.
c-Maf–expressing cells downregulate Bcl-2 protein levels. (A) Cells expressing MEnT have decreased Bcl-2 protein levels. Western blot analysis of MEnT and Bcl-2 protein expression in uninduced or induced HL-60 clones (clones 1 and 2) containing the ZnSO4 inducible MEnT Myb dominant-interfering construct. (B) Bcl-2 protein levels decrease coordinately with c-Maf expression. Western blot analysis of c-Maf, Bcl-2, and actin control protein levels in HL-60 c-Maf–containing clone 6 incubated with ZnSO4 for the indicated time periods. Vector control cells were treated identically and assayed for Bcl-2 and actin protein levels (CB6 control).
c-Maf–expressing cells downregulate Bcl-2 protein levels. (A) Cells expressing MEnT have decreased Bcl-2 protein levels. Western blot analysis of MEnT and Bcl-2 protein expression in uninduced or induced HL-60 clones (clones 1 and 2) containing the ZnSO4 inducible MEnT Myb dominant-interfering construct. (B) Bcl-2 protein levels decrease coordinately with c-Maf expression. Western blot analysis of c-Maf, Bcl-2, and actin control protein levels in HL-60 c-Maf–containing clone 6 incubated with ZnSO4 for the indicated time periods. Vector control cells were treated identically and assayed for Bcl-2 and actin protein levels (CB6 control).
DISCUSSION
Here we show that elevated, conditional expression of c-Maf in bipotent myeloid cell lines is sufficient to promote differentiation to monocytes. Elevation of c-Maf causes an increase in the levels of Myb:Maf complexes, recapitulating the increase seen when these lines are induced to monocytic differentiation. Enforced expression of the c-Maf protein correlated precisely with the induction of morphological changes, the appearance of cell-surface markers, and the induction of cytosolic enzyme activities characteristic of monocytic differentiation. Maf’s induction of differentiation was independent of the expression of IL-4, a direct transcriptional target of c-Maf, but was associated with the inhibition of c-Myb activity, including the Myb targets CD13/APN and Bcl-2. This inhibition of Bcl-2 correlated with the induction of apoptosis following the differentiation of c-Maf–expressing cells and was consistent with Bcl-2 downregulation seen during normal terminal myeloid differentiation.37-39 However, inhibition of c-Myb alone does not appear to account for all of the effects of c-Maf, as conditional expression of dominant interfering Myb proteins in HL-60 cells did not induce c-Maf–like differentiation. Hence, c-Maf likely plays a key role in myeloid cell development through dual mechanisms; inhibition of selected c-Myb regulated targets coupled with the activation of as yet undefined differentiation-promoting genes.
The morphology of the c-Maf–induced cells differs to some extent from that of cells resulting from standard effectors of monocytic or granulocytic differentiation, precluding characterization based on morphology alone. While the monocyte specific indicator, ANB esterase activity, is readily detected in 50% of the cells in cultures induced to express c-Maf, characterization of granulocytic differentiation in the remaining cells is less clear. However, expression of the CAAT enhancer binding protein (CEBP)α transcription factor is rapidly downregulated to undetectable levels in our c-Maf containing clones following induction of c-Maf expression,40 a pattern compatible with that seen in mature peripheral blood monocytes and bipotent cell lines induced toward monocytic differentiation.41 By contrast, high levels of CEBPα expression are seen in mature neutrophils and bipotent cell lines induced to differentiate toward granulocytes,41suggesting that the effects of c-Maf are selective for promoting monocytic differentiation of myeloid cells.
An additional striking morphological change observed on induction of c-Maf expression is the appearance of large, multinucleated, intensely ANB esterase positive cells. These cells strongly resemble MGC, which are characterized by randomly arranged nuclei within extensively spread cytoplasm.24 MGC are a primary histologic feature of chronic inflammatory processes such as tubercular granulomas19,23 and result from macrophage fusion rather than endomitosis. MGC are formed through a 2-step process; progenitor cells must first mature to macrophages before membrane fusion can occur.20 21 Therefore, the presence of cells that resemble MGC in c-Maf–induced clonal lines is consistent with our assertion that c-Maf drives cells toward monocytic differentiation.
Surprisingly, although c-Maf can initiate IL-4 expression in IL-4 negative B cells,5 and IL-4 promotes MGC formation,20-22 attempts to implicate IL-4 in the formation of the large cells in our cultures have been unsuccessful. Endogenous IL-4 mRNA was not upregulated in c-Maf expressing myeloid cells and blocking of IL-4 activity with neutralizing antibodies had no effect on c-Maf–induced phenotypic alterations or differentiation (Fig 5). It is thus possible that other effectors shown to promote MGC formation, such as IL-13 or interferon-γ (IFN-γ),42 are responsible for formation of MGC-like cells in our clones.
Downregulation of Myb expression is strictly required for the progression of hematopoietic cell differentiation,31-34 but the question of whether Myb plays a role in inducing cells to differentiate is still unresolved. In our system, inhibition of Myb alone by the MEnT construct does not induce monocytic differentiation. Although the arrest of Myb-driven transcription by the MEnT dominant interfering protein results in alterations in cell morphology, we see no evidence of differentiation as determined by production of alpha-napthyl-butyrate esterase or induction of monocytic cell-surface markers over a wide range of time points. By contrast, these indicators of monocytic differentiation are readily detectable in parallel cultures of c-Maf inducible lines. Therefore, our observations support a hypothesis enjoining a broader role for c-Maf in inducing monocytic differentiation, as inhibiting Myb activity alone does not appear to recapitulate c-Maf–induced differentiation when assayed at early (6 to 18 hours) or later (72 hours) time points.40 Furthermore, the fact that c-Maf–expressing cells do not immediately undergo programmed cell death, even though Myb is inhibited, reinforces this observation and may suggest that other antiapoptotic proteins either continue to function or are activated by Maf expression, thereby delaying apoptosis until after differentiation. Conversely, because the MEnT construct used in this study inhibits transcription from any promoter containing Myb binding sites and because Maf does not inhibit all Myb-regulated genes,9 the MEnT encoded protein undoubtedly inhibits a larger array of genes than does Myb:Maf alone. It is possible that physiologic Myb downregulation does induce differentiation, but the global repression of Myb transcription by MEnT forces cells to undergo an accelerated apoptosis, bypassing or surpassing the normal differentiation phase induced by the less abrupt physiologic Myb downregulation. Cell lines expressing functionally defined mutant Maf constructs will help to clarify this question.
In the hematopoietic system, terminal differentiation of myeloid cells is tightly associated with programmed cell death.37,38,43 Levels of Bcl-2 mRNA and its encoded protein coordinately decrease during normal myelomonocytic maturation and terminal monocytic differentiation of HL-60 cells,37-39providing a mechanistic link between normal differentiation and programmed cell death. The gene encoding the antiapoptotic protein Bcl-2 is a direct transcriptional target of Myb, and inhibition of Myb activity leads to apoptosis in hematopoietic cells.35 36 In agreement, we observed apoptotic cell death in c-Maf expressing HL-60 clones that paralleled c-Maf expression, Myb:Maf inhibitory complex formation, cell differentiation, and a decline in Bcl-2 protein levels. Whether this Bcl-2 decrease is the result of Maf’s inhibition of Myb transcriptional activity or is a consequence of the physiologic Bcl-2 decline seen during myeloid cell differentiation remains to be determined.
If Myb mRNA levels normally decline during development, would it be necessary to invoke a second developmental strategy to inhibit Myb-regulated genes? It is possible that to prepare cells for differentiation, it is necessary to selectively refine Myb’s positive effects on a subset of responder genes before its physiologic downregulation. Myb:Maf complex levels do not strictly correlate with Myb mRNA and protein levels in myeloid cell lines: despite low levels of Myb:Maf in complex, U937 cells contain Myb mRNA levels equivalent to those in HL-60 cells, where concentrations of Myb:Maf in complex with DNA are significantly higher (Fig 1A, and Hegde et al9). This suggests that Myb levels alone do not dictate Myb:Maf complex formation, so that high Myb levels could transactivate some target genes even as Myb:Maf complexes are blocking the expression of others. Interestingly, in promoter assays using fibroblasts that do not express c-Myb and thus lack Myb:Maf complexes, c-Myb positively autoregulates its own transcription through binding to Myb consensus sites located in its 5′-flanking region.44 However, Myb transcription is negatively regulated through these same Myb sites when assays are performed in Myb-expressing T cells.45 While we have not studied Myb:Maf complex formation in T cells, it is intriguing to speculate that the negative regulation observed in these cells results from endogenous Myb:Maf inhibition of Myb transcription as an initial step in the differentiation-linked decline of Myb mRNA. Studies to address these questions during myeloid cell development are important to our further understanding of Maf’s role in myelopoiesis.
ACKNOWLEDGMENT
We thank Drs Rick Bram, David Shapiro, John Cleveland, and Paul Brindle for helpful comments, Dr Kathy Weston for her generous gift of plasmids, John Zacher for photomicrography, Liz Mann for technical help, and John Gilbert for editorial assistance.
Supported by Grant No. CA-70909 from the National Institutes of Health (to L.H.S.), by Grant No. CA-21765 from the National Cancer Institute Cancer Center Support (CORE), and by the American Lebanese Syrian Associated Charities (ALSAC), St Jude Children’s Research Hospital.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Linda H. Shapiro, PhD, Department of Pathology and Laboratory Medicine, St Jude Children’s Research Hospital, Memphis, TN 38105; e-mail:linda.shapiro@stjude.org.