Abstract
CCAAT displacement protein (CDP) is a transcriptional repressor that restricts expression of the gp91phox gene to mature myeloid cells. CDP interacts with multiple sites within the −450 to +12 bp human gp91phox promoter, and down-regulation of CDP DNA-binding activity is required for induction of gp91phox transcription in mature phagocytes. Truncation of the gp91phox promoter to −102 to +12 bp removes 4 CDP-binding sites and reveals a promiscuous promoter activity that is active in some nonphagocytic cells. A cis-element at −90 bp is required for derepressed transcription and serves as a binding site for multiple transcriptional activators. We now report that this element also serves as a binding site for CDP. The affinity of CDP for this element is relatively weak compared with upstream CDP-binding sites within the promoter, consistent with the promiscuous transcriptional activity exhibited by the −102 to +12 bp gp91phox promoter fragment. Further analysis of the proximal promoter reveals an additional weak-affinity CDP-binding site centered at approximately −20 bp. Overexpression of cloned CDP represses the −102 to +12 bp gp91phox promoter, indicating that these proximal CDP-binding sites are functionally significant. The constellation of transcriptional activators and a repressor that interacts with the −90 bp cis-element is identical to that observed for a promoter element at −220 bp, reflecting the highly modular organization of the gp91phoxpromoter. These studies illustrate the complex interplay between transcriptional activators and a repressor that contribute to the myeloid-restricted expression of the gp91phox gene.
THE GP91PHOX GENE encodes a component of the NADPH-oxidase complex, which is responsible for the generation of a respiratory burst and microbicidal activity in phagocytic blood cells.1 This gene is highly expressed in mature myeloid cells such as macrophages and granulocytes.2We previously found that the −450 to +12-bp region of the gp91phox promoter directs expression of a linked reporter gene in a subset of monocyte/macrophages (but not granulocytes) in transgenic mice.3 This promoter fragment also responds to interferon-γ stimulation in stably transfected myeloid cell lines.4,5 Extension of the promoter to approximately −2.6 kb does not alter the promoter activity. However, Lien et al6 have reported that inclusion of additional distal elements located 30 to 60 kb upstream of the gp91phox gene results in appropriate transgene expression in the full spectrum of the phagocytic lineage.
The gp91phox gene is under the control of an unusual myeloid cell-restricted promoter, for which transcriptional repression provides an important component of gene regulation.7 We previously reported that binding of the transcriptional repressor CCAAT displacement protein (CDP) to multiple sites in the proximal promoter is necessary to restrict gp91phox expression to mature myeloid cells.8,9 The DNA-binding activity of CDP is down-regulated during phagocyte differentiation, coincident with induction of gp91phox transcription. Furthermore, constitutive overexpression of CDP in a myeloid cell line prevents induction of the gp91phox gene on terminal differentiation.10 Several other lineage-restricted genes are also repressed under these conditions, suggesting that they are also targets of CDP-mediated transcriptional repression. These include the secondary granule proteins neutrophil gelatinase, neutrophil elastase, and lactoferrin.11 12 However, other myeloid cell-restricted components of the NADPH-oxidase complex, such as p47phox and p67phox, are expressed normally, and the cells retain the ability to terminally differentiate in the presence of constitutive CDP.
Removal of CDP-binding sites from the gp91phoxpromoter reveals a strong promiscuous transcriptional activating activity within the −102- to +12-bp promoter. This promoter fragment is active in nonphagocytic [eg, human erythroleukemia (HEL)] cells,9 and other nonphagocytic cell lines, such as HeLa and K562 cells, additionally require the retention of a CCAAT-box at 125 bp for significant promoter activity.9 CDP excludes the binding of widely expressed transcriptional activators to each of 4 CDP-binding sites centered at −350 bp, −220 bp, −150 bp, and −125 bp.5,8,9,13 The CDP-binding site oligonucleotide probes centered at −150 bp and −125 bp (CDP-β and CDP-α) overlap by 25 bp. However, analysis of truncated versions of these binding sites indicates that each represents a distinct CDP-binding site, although portions of the sequence in common are required to provide high-affinity binding sites for CDP.9
The transcriptional activating factors that compete with CDP for overlapping binding sites include previously described proteins such as the CCAAT-box binding factor CP1 and interferon regulatory factors (IRF)-1 and IRF-2, as well as an unidentified factor denoted BID (binding increased duringdifferentiation), which binds to multiple sites. Eklund and Kakar14 reported the cloning of a cDNA that encodes the BID activity (denoted TF1phox in that report). However, Yamit-Hezi et al15 reported this cDNA to be a bacterial transcript that contaminates commercially available libraries. Hence, the identity of the BID DNA-binding factor remains to be determined. The intensity of DNA/protein complexes, which contain these activators, is increased in electrophoretic mobility shift assays (EMSA) on terminal phagocytic differentiation.5,8,9 However, the apparent induction of the activating factor complexes coincides with downregulation of the competing CDP DNA-binding activity. CDP was initially reported as a factor that excludes the binding of a ubiquitous CCAAT-box binding factor (CP1) to an overlapping binding site in the sea urchin sperm histone h2b promoter.16 Hence, we hypothesize that transcriptional activators present in nonphagocytic cells are unable to effectively interact with the gp91phox promoter in the presence of CDP DNA-binding activity.
The −450 to +12 bp gp91phox promoter contains interferon-stimulated response elements (ISRE) and is transcriptionally induced in myeloid cells after stimulation with interferon-γ.17 We have reported that an ISRE at −220 bp of the promoter serves as a binding site for CDP, BID, IRF-1, and IRF-2 and that CDP excludes the binding of these transcriptional activators before terminal phagocytic differentiation (see Fig 3).5,9 We further showed that another ISRE at −90 bp of the gp91phox promoter also serves as a binding site for BID, IRF-1, and IRF-2 and is necessary for activity of the −102 to +12 bp promoter in HEL cells.13 We now report that CDP additionally interacts with a promoter fragment that contains the −90 bp ISRE, as well as with a distinct element at −20 bp of the gp91phox promoter. Furthermore, overexpression of cloned CDP represses the −102 to +12 bp gp91phox promoter. These studies provide new information regarding the significance of CDP function on the regulation of the highly complex gp91phox promoter.
MATERIALS AND METHODS
Oligonucleotides.
Complementary oligonucleotides were synthesized on an Applied Biosystems model 394 synthesizer (Perkin-Elmer, Inc, Foster City, CA). Listed are the upper strands of double-stranded oligonucleotides corresponding to the following regions of the human gp91phox promoter8: CDP-δ (−382 to −313 bp) 5′-gtttaatgtgttttacccagcacgaagtcatgtctagttgagtggcttaaaaattgtgatcaaatagctg-3′; CDP-γ (−261 to −212 bp) 5′-gttatttatctcttagttgtagaaattggtttcattttccactatgttta-3′; CDP-β (−182 to −113 bp) 5′-tttgtagttgttgaggtttaaagatttaagtttgttatggatgcaagcttttcagttgaccaatgattat-3′; CDP-α (−136 to −76 bp) 5′-gcttttcagttgaccaatgattattagccaatttctgataaaagaaaaggaaaccgattgc-3′; 5′CDP-α (−136 to −102 bp) 5′-gcttttcagttga ccaatgatattagccaatttc-3′; CDP-ε (−102 to −65 bp) 5′-ctgataaaagaaaaggaaaccgattgccccaggg- ctgc-3′; 5′CDP-ε (−102 to −81 bp) 5′-ctgataaaagaaaaggaaaccg-3′; 3′CDP-ε (−93 to −65 bp) 5′-gaaaaggaaaccgattgccccagggctgc-3′; and minimal (MIN) (−121 to −94 bp) 5′-aatgattattagcca atttctgataaaa-3′. The following oligonucleotides contain the indicated regions of the human gp91phox promoter: −68 to −30 bp, 5′-ctgctgttttcatttcctcattggaagaagaagcatagt-3′; −39 to −1 bp (CDP-ζ), 5′-gaagcatagtatagaagaaaggcaaacacaacacattca-3′; −13 to +12 bp, 5′-cacaacacattcaacctctgccacc-3′. Additional oligonucleotides include the high-affinity CDP-binding site E3618 5′-cggatccgaattcatcgataatcgattat-3′; a binding site for the factor Spi-B195′-tcgggctcgagtctgaa agaggaacttggttagc- tcgg-3′; and a CCAAT-box element derived from the α-globin gene promoter205′-ggcggcgctcattggctggcgcggagcccg-3′. All oligonucleotides were synthesized with BamHI overhangs (not shown) to facilitate subcloning into plasmid after annealing of complementary sequences.
Cell culture and transfections.
The human cervical carcinoma cell line HeLa and the human erythroleukemic cell line HEL were obtained from the American Type Culture Collection (Rockville, MD). HeLa cells were grown in Dulbecco’s modified Eagle’s medium, supplemented with 10% fetal calf serum, 0.2 mmol/L glutamate, 50 U/mL penicillin, and 50 μg/mL streptomycin. The human myelomonoblastic cell line PLB-985 was a gift of Thomas Rado (Birmingham, AL).21 PLB-985 and HEL cells were grown in similarly supplemented RPMI 1640 medium.
For transfections, HEL cells growing in log phase were resuspended in fresh RPMI media and grown for 18 to 24 hours to a final density of 6 to 8 × 105 cells/mL. After harvesting by centrifugation, cells were resuspended in RPMI media at a concentration of 3 × 107 cells/mL. Three hundred microliters of the cell mixture was aliquoted into cuvettes (0.4 cm diameter; Bio-Rad Laboratories, Richmond, CA) for duplicate electroporations. Each sample contained 1 μg of cytomegalovirus (CMV) promoter/enhancer-beta-galactosidase (β-gal) plasmid, which served as an internal control for transfection efficiency, 5 μg of the target plasmid, −102 to +12 bp of the gp91phoxpromoter linked to a luciferase reporter gene,13 and either 0.5 μg of a CMV-CDP cDNA expression vector, 0.5 μg of CMV-CDP antisense vector, or 0.5 μg of the empty expression vector, which lacks a cDNA insert. Each sample was electroporated by using a Gene Pulser (Bio-Rad) at 220 V, 960 μF. The transfected cells were immediately transferred to 100 mm tissue culture dishes containing 10 mL of fresh RPMI growth media and grown at 37oC under 5% CO2 for 24 hours.
Cells were harvested by centrifugation at 500g for 10 minutes, followed by a single wash with phosphate-buffered saline. Cell pellets were resuspended in 100 μL of lysis buffer (Promega Luciferase Assay Kit; Promega, Madison, WI) and incubated at room temperature for 15 minutes with frequent vortexing. The cells were then pelleted by centrifugation at 500g for 4 minutes at room temperature. Twenty microliters of each extract was assayed for luciferase activity with a Promega Luciferase Kit (Promega) and Lumat 9210 luminometer (Wallac, Inc, Gaithersburg, MD). Thirty microliters of each extract was added to 270 μL of β-gal assay buffer as described,9 followed by incubation at 37°C, and the relative β-gal activity was determined by measuring the OD420 and used to adjust luciferase values to compensate for relative transfection efficiency.
Human foreskin keratinocytes were prepared as described22and transfected as described by Armstrong and Roman.23Briefly, 2.4 × 105 cells were plated in serum free medium (GIBCO-BRL, Gaithersburg, MD) and transfected the next day with 2.5 μg of the −102 to +12 bp gp91phoxpromoter/luciferase reporter gene, 0.1 μg of expression vector containing sense or antisense CDP cDNA (or vector alone), 0.1 μg CMV-β-gal, and 5 μg pUC19 plasmid DNA. Forty-eight hours posttransfection, cells were lysed and luciferase and β-gal activities assayed as described.23 Expression of CDP had no effect on the expression of the β-gal internal control reporter gene.
In vitro DNA-binding protein assays.
Nuclear extracts were prepared by the method of Dignam et al.24 Double-stranded oligonucleotides were labeled by using [γ32P]ATP and T4 polynucleotide kinase, separated by polyacrylamide gel electrophoresis, and eluted by the crush and soak method.25 EMSA was performed as described previously.4 Briefly, 3 to 10 μg of nuclear extract was mixed with 0.1 to 0.5 μg poly [dI-dC] and binding buffer for a final reaction volume of 20 μL. Competitor double-stranded oligonucleotides, CDP antiserum (gift from Ellis Neufeld, Harvard University, Cambridge, MA), or normal guinea pig serum were added as indicated and incubated on ice for 15 to 30 minutes before the addition of 15,000 cpm of probe. After another 15-minute incubation on ice, samples were loaded onto a 0.5× Tris borate/EDTA, 3.5% nondenaturing polyacrylamide gel and electrophoresis was performed at 25 mA at 4oC until the bromophenol blue dye had migrated near the bottom of the gel.
RESULTS
Identification of CDP-binding sites within the −102 to +12 bp gp91phox promoter.
Previously, we showed that CDP competes with the binding of transcriptional activating factors such as BID, CP1, IRF-1, and IRF-2 at 4 sites within the human gp91phox promoter, located at approximately −350 bp, −220 bp, −150 bp, and −125 bp.8,9 Subsequently, we showed the presence of an additional binding site for IRF-1, IRF-2, and BID within the −102 to −65 bp promoter region, an element that contains an ISRE (−94 to −82 bp).13 Because CDP and BID binding sites overlap at 3 other locations within the promoter,5 9 we investigated whether CDP binds to the −102 to −65 bp gp91phox promoter region.
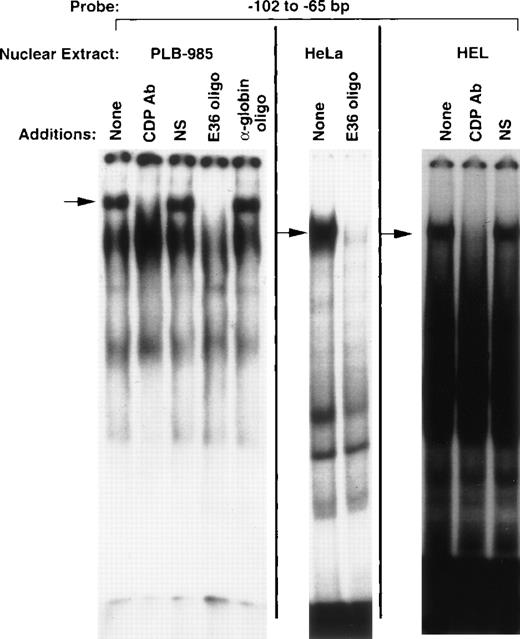
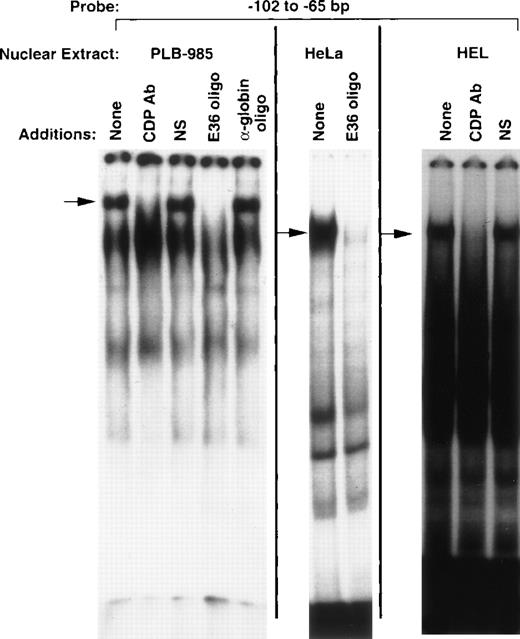
EMSA performed with nuclear extract isolated from PLB-985 myeloid cells and a probe corresponding to the −102 to −65 bp region of the gp91phox promoter reveals a DNA/protein complex of slow mobility (Fig 1) similar to that previously described for CDP.8,9 This complex is specifically disrupted by antiserum raised against CDP, but not by normal serum (Fig 1). Furthermore, this complex contains a sequence-specific DNA-binding activity consistent with CDP, as it is disrupted by addition of a molar excess of an oligonucleotide that contains a high-affinity CDP-binding site (E36),18 but not by an unrelated oligonucleotide (α-globin). This DNA-binding activity is also apparent in nuclear extracts derived from HeLa and HEL cells, as has previously been shown for CDP.8 9 We conclude that the DNA/protein complex of slow mobility contains CDP, and the −102 to −65 bp region of the gp91phoxpromoter is hereafter denoted the CDP-ε binding site.
CDP binds to the −102 to −65 bp region of the gp91phox promoter. EMSA was performed as described in Materials and Methods, with the −102 to −65 bp region of the gp9phox promoter as a probe and nuclear extract isolated from the indicated cell lines. Antiserum directed against CDP, normal guinea pig serum, or 20 ng of double-stranded oligonucleotide competitor was added to the binding reactions where indicated. The E36 oligonucleotide18 is a high affinity CDP-binding site, whereas the -globin oligonucleotide20 contains a CCAAT-box motif from that gene’s promoter and serves as a heterologous competitor. Arrows indicate CDP complexes. Relative DNA/protein complex mobilities cannot be directly compared between different cell lines.
CDP binds to the −102 to −65 bp region of the gp91phox promoter. EMSA was performed as described in Materials and Methods, with the −102 to −65 bp region of the gp9phox promoter as a probe and nuclear extract isolated from the indicated cell lines. Antiserum directed against CDP, normal guinea pig serum, or 20 ng of double-stranded oligonucleotide competitor was added to the binding reactions where indicated. The E36 oligonucleotide18 is a high affinity CDP-binding site, whereas the -globin oligonucleotide20 contains a CCAAT-box motif from that gene’s promoter and serves as a heterologous competitor. Arrows indicate CDP complexes. Relative DNA/protein complex mobilities cannot be directly compared between different cell lines.
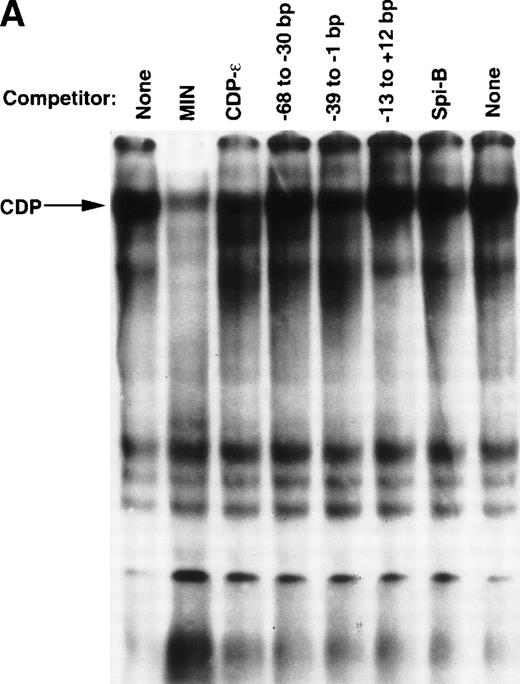
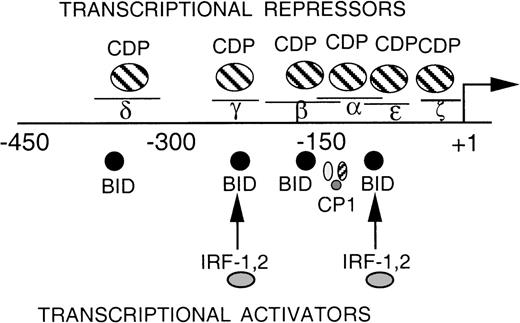
EMSA was performed to assess whether additional CDP-binding sites are present within the proximal gp91phox promoter. A series of overlapping oligonucleotides were synthesized that span the −68 to +12 bp gp91phox promoter, and each was used as a competitor against the CDP-complex formed with the MIN probe. The MIN oligonucleotide contains a portion (−121 to −94 bp) of the CDP-α element and retains a binding site for CDP. Figure 2A illustrates that an oligonucleotide spanning −39 to −1 bp of the promoter partially disrupts the CDP complex, similar to the degree of disruption observed with the CDP-ε oligonucleotide competitor. Additional studies were performed to further characterize this new putative CDP-binding site. A probe containing the −39 to −1 bp region generates an EMSA complex with a mobility indistinguishable from that produced by the CDP-ε probe (Fig 2B, lanes 1 and 2). The putative CDP complex formed with the –39 to –1 bp probe is disrupted by antiserum directed against CDP but not by normal serum (lanes 3 and 4). This complex is partially disrupted by competition with the homologous oligonucleotide or with the CDP-ε element (lanes 6 and 7), suggesting that these 2 promoter elements exhibit a similar affinity for CDP. The EMSA complex formed with the −39 to −1 bp gp91phox promoter probe is also efficiently disrupted by the MIN oligonucleotide for which CDP exhibits a high affinity (lane 8). As expected, the putative CDP complex formed with the −39 to −1 bp probe is not apparent on analysis of nuclear extract derived from PLB-985 cells induced to terminally differentiate (lane 9). We conclude that the −39 to −1 bp probe is bound by CDP, and we hereafter denote this promoter element as the CDP-ζ binding site. Hence, CDP interacts with 2 sites within the −102 to +12 bp gp91phox promoter. Figure 3 illustrates the relative positions of the 6 identified CDP-binding sites within the −450 to +12 bp gp91phox promoter and the transcriptional activating factors previously shown to bind to overlapping sites within these promoter regions.8,9,13 14
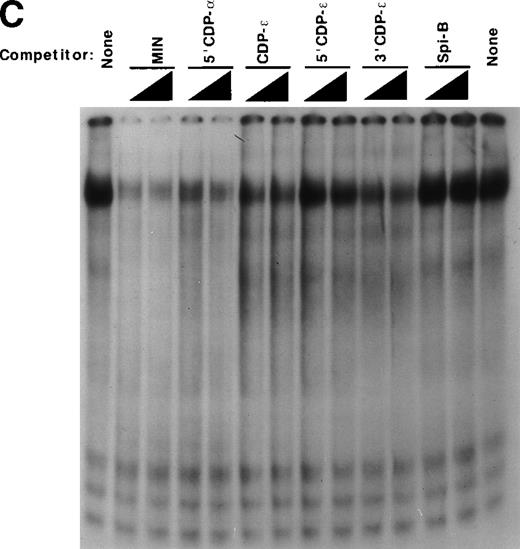
Detection of an additional CDP-binding site in the proximal gp91phox promoter. EMSA was performed as described in Materials and Methods, with the MIN binding site as a probe, nuclear extract derived from HeLa cells, and 10 ng of the homologous competitor oligonucleotide and equimolar amounts of the other competitor oligonucleotides derived from the gp91phox promoter. The Spi-B oligonucleotide is included as a heterologous competitor. EMSA was performed by using the −39 to −1 bp region of the gp91phox promoter as probe. CDP-ɛ is used as a probe in the first lane to provide a size standard for the CDP complex. Reactions contained nuclear extract derived from either PLB-985 cells induced to terminally differentiate into granulocytes (lane 9) or HeLa cells as indicated. Reactions also contain 100 ng of homologous oligonucleotide (or equimolar amounts of other competitor oligonucleotides) (lanes 6-8), antiserum directed against CDP (lane 3), or normal guinea pig serum (lane 4) where indicated. Lanes from a single gel were rearranged during figure preparation for clarity of presentation.
Detection of an additional CDP-binding site in the proximal gp91phox promoter. EMSA was performed as described in Materials and Methods, with the MIN binding site as a probe, nuclear extract derived from HeLa cells, and 10 ng of the homologous competitor oligonucleotide and equimolar amounts of the other competitor oligonucleotides derived from the gp91phox promoter. The Spi-B oligonucleotide is included as a heterologous competitor. EMSA was performed by using the −39 to −1 bp region of the gp91phox promoter as probe. CDP-ɛ is used as a probe in the first lane to provide a size standard for the CDP complex. Reactions contained nuclear extract derived from either PLB-985 cells induced to terminally differentiate into granulocytes (lane 9) or HeLa cells as indicated. Reactions also contain 100 ng of homologous oligonucleotide (or equimolar amounts of other competitor oligonucleotides) (lanes 6-8), antiserum directed against CDP (lane 3), or normal guinea pig serum (lane 4) where indicated. Lanes from a single gel were rearranged during figure preparation for clarity of presentation.
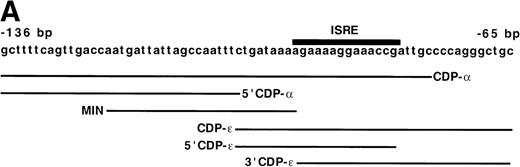
DNA-binding proteins that interact with the proximal gp91phox promoter. The transcriptional repressor CDP competes with the binding of transcriptional activation factors at 6 sites (δ, γ, β, , and ɛ- and ζ-, the proposed fifth and sixth binding sites). The DNA binding activity of CDP is down-regulated during terminal phagocyte development, allowing the transcriptional activators to interact with the gp91phoxpromoter.5,8,9 13 Plus (+)1 indicates the site of transcription initiation. Lines above δ, γ, β, , ɛ, and ζ indicate the positions of each of 5 oligonucleotides used as CDP-binding sites.
DNA-binding proteins that interact with the proximal gp91phox promoter. The transcriptional repressor CDP competes with the binding of transcriptional activation factors at 6 sites (δ, γ, β, , and ɛ- and ζ-, the proposed fifth and sixth binding sites). The DNA binding activity of CDP is down-regulated during terminal phagocyte development, allowing the transcriptional activators to interact with the gp91phoxpromoter.5,8,9 13 Plus (+)1 indicates the site of transcription initiation. Lines above δ, γ, β, , ɛ, and ζ indicate the positions of each of 5 oligonucleotides used as CDP-binding sites.
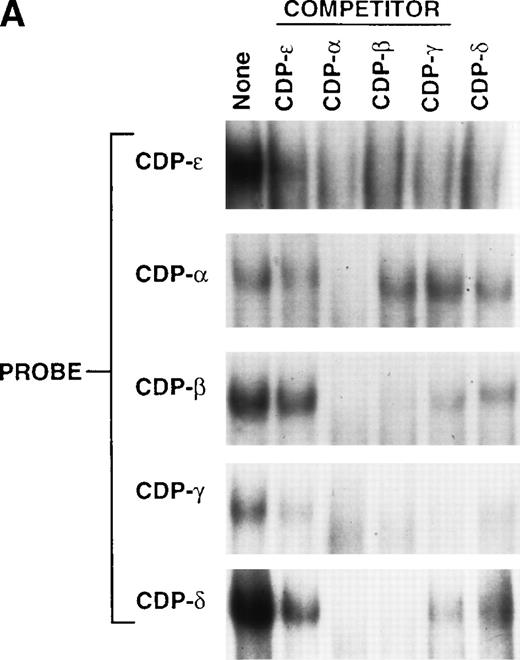
EMSA was performed to examine the relative binding affinity of CDP for the CDP-ε site as compared with the 4 previously identified upstream CDP-binding sites within the gp91phoxpromoter.9 Cross-competition studies were performed with 5 CDP-binding sites derived from the gp91phoxpromoter as both probes and competitors (Fig 4A). The CDP-ε oligonucleotide only partially disrupts the CDP complexes formed with the other 4 CDP-binding site probes, whereas, each of the other 4 CDP-binding sites from the gp91phox promoter completely disrupts the CDP complex formed with the CDP-ε probe. Thus, CDP exhibits a weaker affinity for the CDP-ε binding site than for the other 4 binding sites. As illustrated in the competition studies presented in Fig 2B, CDP exhibits a similar affinity for the CDP-ε and CDP-ζ elements. Hence, the relative affinity of CDP-binding sites is CDP-α > CDP-β > CDP-γ > CDP-δ > CDP-ε = CDP-ζ (Fig4A).9
Comparison of the CDP-ɛ site to upstream CDP-binding sites within the gp91phox promoter. (A) EMSA was performed as described in Materials and Methods, with CDP-binding site probes and nuclear extract isolated from HeLa cells (10 μg for CDP-ɛ; 3 μg for the other 4 probes). Competition is with 10 ng of nonradioactive CDP-ɛ oligonucleotide or an equimolar amount of nonradioactive CDP-, CDP-β, CDP-γ, or CDP-δ oligonucleotides. Because of variations in probe specific activity and film exposure times, the absolute intensity of the CDP complex formed with each probe in the absence of competitor does not reflect the relative affinity of CDP for each binding site. (B) Alignment of 5 CDP-binding sites within the gp91phox promoter against a PCR-selected consensus CDP-binding sequence.18 The lowercase nucleotides do not match the consensus sequence. The percent similarity between each gp91phox promoter site and the PCR selected consensus sequence is indicated.
Comparison of the CDP-ɛ site to upstream CDP-binding sites within the gp91phox promoter. (A) EMSA was performed as described in Materials and Methods, with CDP-binding site probes and nuclear extract isolated from HeLa cells (10 μg for CDP-ɛ; 3 μg for the other 4 probes). Competition is with 10 ng of nonradioactive CDP-ɛ oligonucleotide or an equimolar amount of nonradioactive CDP-, CDP-β, CDP-γ, or CDP-δ oligonucleotides. Because of variations in probe specific activity and film exposure times, the absolute intensity of the CDP complex formed with each probe in the absence of competitor does not reflect the relative affinity of CDP for each binding site. (B) Alignment of 5 CDP-binding sites within the gp91phox promoter against a PCR-selected consensus CDP-binding sequence.18 The lowercase nucleotides do not match the consensus sequence. The percent similarity between each gp91phox promoter site and the PCR selected consensus sequence is indicated.
With the exception of CDP-ζ, each of the identified CDP-binding sites within the gp91phox promoter exhibits significant similarity with a polymerase chain reaction (PCR) selected consensus sequence for CDP binding (Fig 4B).18 However, the CDP-ε–binding site exhibits only 53% similarity to this highly degenerate consensus CDP-binding site sequence, consistent with the relatively low affinity of CDP for this binding site. Although the CDP-ζ element fails to exhibit significant homology to the consensus CDP binding site, the −29 to −13 bp region shares 82% identity with the −105 to −89 bp region of the gp91phox promoter (data not shown), a region in common between the CDP-α and CDP-ε elements. These observations illustrate the difficulty in using the degenerate consensus CDP-binding sequence to identify authentic CDP-binding sites.
The CDP-ε binding site is distinct from the CDP-α binding site.
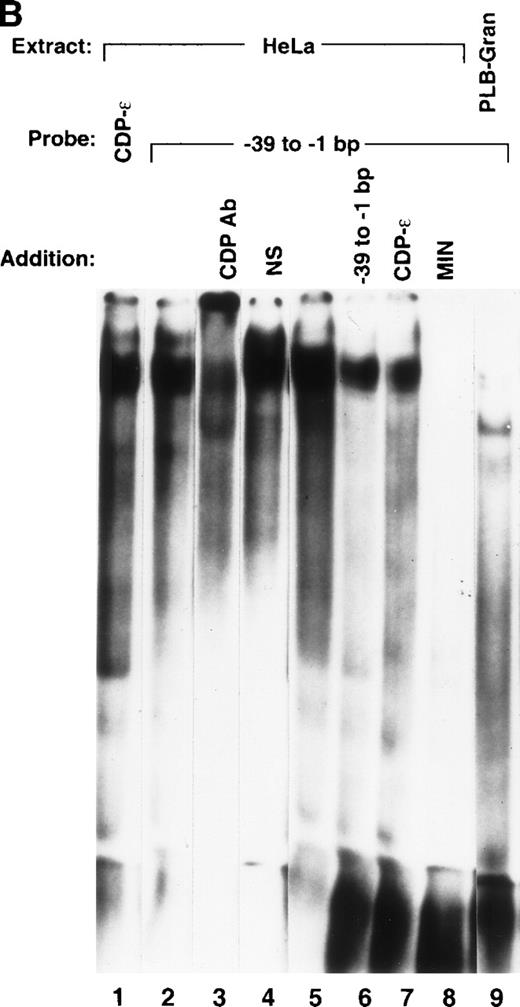
The CDP-ε sequence in the gp91phox promoter overlaps CDP-α by 27 bp (Figs 3 and 5A). However, sequences required for binding of CDP to the CDP-α site have previously been located upstream of this overlapping region.26 Hence, we postulated that binding of CDP to the CDP-ε probe represents a distinct binding event. Additional studies were performed with truncated versions of the CDP-α and CDP-ε sequences to determine if binding of CDP to CDP-ε is because of the overlapping CDP-α sequence. The truncated oligonucleotides include: 5′CDP-α, which lacks the region in common with the CDP-ε site; MIN, which lacks the 5′-end of the CDP-α binding site and extends 9 bp into the overlap region; 5′CDP-ε, which lies entirely within the region in common between CDP-α and CDP-ε and lacks the 3′-end of the CDP-ε sequence; and 3′CDP-ε, which contains the downstream portion of CDP-ε but does not overlap with MIN. Importantly, MIN represents a minimal segment of the CDP-α fragment that retains CDP-binding activity. Mutation of an ATTA motif (−117 to −114 bp) within the MIN element, which lies upstream of the CDP-ε sequence, abolishes the binding of CDP.26
The CDP-ɛ–binding site is distinct from the CDP- binding site. (A) Schematic illustration of the positions of gp91phox promoter regions used in EMSA in comparison to the CDP- site. CDP-, −136 to −76 bp; CDP-ɛ, −102 to −65 bp; 5′CDP-, −136 to −102 bp; MIN, −121 to −94 bp; 5′CDP-ɛ, −100 to −81; 3′CDP-ɛ, −93 to −65 bp. (B) EMSA showing that the binding of CDP to the CDP-ɛ site is distinct from CDP binding to the CDP- site. EMSA was performed as described in Materials and Methods, with nuclear extract isolated from HeLa cells. Antiserum directed against CDP was added to the indicated samples. EMSA was performed by using the MIN oligonucleotide as a probe and nuclear extract derived from HeLa cells. Five or 10 ng of CDP-ɛ (or equimolar amounts of the other oligonucleotides) were added as competitors where indicated. The Spi-B oligonucleotide is included as a heterologous competitor.
The CDP-ɛ–binding site is distinct from the CDP- binding site. (A) Schematic illustration of the positions of gp91phox promoter regions used in EMSA in comparison to the CDP- site. CDP-, −136 to −76 bp; CDP-ɛ, −102 to −65 bp; 5′CDP-, −136 to −102 bp; MIN, −121 to −94 bp; 5′CDP-ɛ, −100 to −81; 3′CDP-ɛ, −93 to −65 bp. (B) EMSA showing that the binding of CDP to the CDP-ɛ site is distinct from CDP binding to the CDP- site. EMSA was performed as described in Materials and Methods, with nuclear extract isolated from HeLa cells. Antiserum directed against CDP was added to the indicated samples. EMSA was performed by using the MIN oligonucleotide as a probe and nuclear extract derived from HeLa cells. Five or 10 ng of CDP-ɛ (or equimolar amounts of the other oligonucleotides) were added as competitors where indicated. The Spi-B oligonucleotide is included as a heterologous competitor.
Each of these oligonucleotides serves as a binding site for CDP when used as a probe in EMSA (Fig 5B), as a slow-mobility complex is formed with each that is disrupted by the addition of antiserum directed against CDP. Importantly, the 5′CDP-α and CDP-ε elements overlap by only 1 nucleotide, and the MIN and 3′CDP-ε elements do not overlap. The observation that each of these 4 promoter regions form CDP EMSA complexes shows that at least 2 distinct CDP-binding sites are present within the −136 to −65 bp region of the gp91phox promoter.
EMSA competition studies were performed by using the MIN probe to assess the relative affinity of CDP for each oligonucleotide derived from the −136 to −65 bp region of the gp91phox promoter. Competition with a molar excess of the homologous oligonucleotide (MIN) completely disrupts the CDP complex (Fig 5C). The residual band visible after competition migrates slightly above the position of the CDP complex and is also apparent after disruption by CDP antiserum (Fig 5B). A similar degree of disruption is apparent with the 5′CDP-α competitor. CDP-ε is a less efficient competitor, as disruption is not complete at the lower oligonucleotide concentration tested. This affinity is similar to that produced by the 3′CDP-ε oligonucleotide. The 5′CDP-ε oligonucleotide exhibits an even lower affinity for CDP, as complete disruption of the CDP complex is not observed for either concentration of competitor tested. Addition of an unrelated oligonucleotide (Spi-B) had no significant effect on the CDP complex. We conclude that the CDP-ε element contains a distinct binding site for CDP, but sequences in common between the CDP-α and CDP-ε binding sites are necessary for high-affinity binding of CDP to the CDP-ε promoter region.
CDP represses the −102 to +12 bp gp91phox promoter.
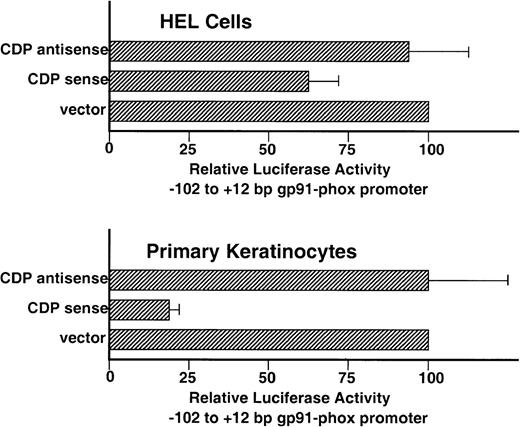
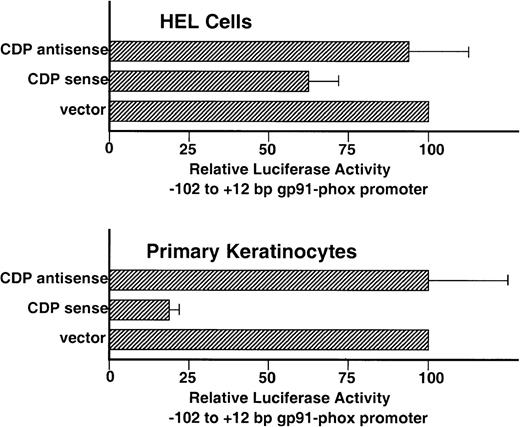
Although the −102 to +12 bp gp91phox promoter contains 2 weak-affinity binding sites for CDP (CDP-ε and CDP-ζ), it exhibits significant transcriptional activity after transient transfection into HEL cells, which contain endogenous CDP DNA-binding activity (Fig1).9 This promoter activity requires binding sites for the transcriptional activating factors BID and IRF-1/IRF-2 that lie within an ISRE in the CDP-ε element (Fig 5A).13 We hypothesized that the persistence of promoter activity in the presence of the CDP-ε and CDP-ζ binding sites may reflect the relatively weak binding affinity of CDP for these elements and reasoned that the −102 to +12 bp gp91phox promoter might be repressed if the amount of CDP DNA-binding activity was further elevated in HEL cells. To test this, cotransfection experiments were performed in which expression vectors containing either CDP cDNA or antisense CDP were transiently transfected into HEL cells along with the −102 to +12 bp gp91phox/luciferase reporter gene construct. Figure 6illustrates that the proximal gp91phox promoter is repressed approximately 30% after cotransfection with the CDP overexpression vector, but is unaffected by cotransfection with the antisense vector. Although statistically significant (P < .006), the magnitude of the CDP-mediated repression in HEL cells is small. This is presumably because of the relatively weak affinity of CDP for the CDP-ε and CDP-ζ elements and the strong basal transcriptional activity exhibited by the −102 to +12 bp gp91phox promoter in HEL cells (approximately as strong as the SV40 early promoter).9 To further assess the functional significance of the CDP-binding sites within the −102 to +12 bp gp91phox promoter, similar cotransfection experiments were performed by using primary keratinocyte cultures. We previously showed that CDP represses human papillomavirus gene expression in proliferating keratinocytes.22 Cotransfection of the −102 to +12 bp gp91phoxpromoter/luciferase plasmid with the CDP-overexpression vector into keratinocytes results in an 80% reduction in reporter gene expression, whereas cotransfection with the vector containing the CDP antisense cDNA has no significant effect on reporter gene expression (Fig 6).
Overexpression of CDP represses the −102 to +12 bp gp91phox promoter. HEL and primary keratinocyte cultures were cotransfected with the −102 to +12 bp gp91phox promoter/luciferase vector and either a CDP expression vector (CDP sense), the expression vector containing the CDP cDNA in the antisense orientation (CDP antisense), or the expression vector lacking a cDNA insert (vector). Transfections were performed as described in Materials and Methods. Data represent the mean ± standard error of at least 3 transfections, with at least 2 distinct plasmid preparations. The level of luciferase expression generated in cotransfections containing the expression vector lacking an insert is defined as 100%.
Overexpression of CDP represses the −102 to +12 bp gp91phox promoter. HEL and primary keratinocyte cultures were cotransfected with the −102 to +12 bp gp91phox promoter/luciferase vector and either a CDP expression vector (CDP sense), the expression vector containing the CDP cDNA in the antisense orientation (CDP antisense), or the expression vector lacking a cDNA insert (vector). Transfections were performed as described in Materials and Methods. Data represent the mean ± standard error of at least 3 transfections, with at least 2 distinct plasmid preparations. The level of luciferase expression generated in cotransfections containing the expression vector lacking an insert is defined as 100%.
DISCUSSION
Restriction of gp91phox expression to terminally differentiating myeloid cells is due, in part, to the repression of this gene’s transcription in undifferentiated cells by the DNA-binding protein CDP.8-10 CDP-binding sites have been reported in a large number of promoters and enhancers, many of which exhibit lineage-restricted activity in a diverse spectrum of cell types. These include the neural cell adhesion molecule, γ-globin, neutrophil collagenase, lactoferrin, histone, tyrosine hydroxylase, CD8a, β-myosin, cystic fibrosis transmembrane conductance regulator, and c-myc gene promoters, the mouse mammary tumor virus long terminal repeat, heavy-chain immunoglobulin enhancer, human papillomavirus type 6 long control region, herpes simplex virus thymidine kinase promoter, and c-mos enhancer.11,12,16,22,27-38 In addition, CDP has recently been implicated in tumor progression and has been found to interact with the polyomavirus large T antigen.39,40 CDP also represses expression of the cyclin-dependent kinase inhibitor p21WAF1 in a cell-cycle–dependent manner.41 Hence, a better understanding of the mechanisms of CDP function and regulation is of general interest. Our laboratory previously reported that CDP binds to 4 sites within the proximal promoter region (−382 to −76 bp) of the gp91phox gene.8,9 Truncation of the promoter to −102 to +12 bp removes 4 CDP-binding sites and reveals a latent promoter activity that is active in some nonphagocytic cell lines. Studies were undertaken to identify elements within the −102 to +12 bp gp91phox promoter that are required for transcriptional activity in the absence of repression mediated by CDP-binding to upstream elements. We previously reported that an element between −102 and −86 bp is necessary for de-repressed gp91phox promoter activity. This element contains an ISRE, which serves as a binding site for IRF-1, IRF-2, and BID.13 We now report that this promoter region is also bound by CDP, similar to the constellation of DNA-binding factors interacting with an ISRE at −220 bp of the gp91phox promoter.9,13 An additional CDP-binding site is also detected at approximately −20 bp of the promoter. Hence, the gp91phox promoter appears to be highly modular and contains multiple binding sites for a number of transcriptional activators (CP1, BID, IRF-1, and IRF-2) and for the repressor CDP. CDP excludes the binding of transcriptional activators at each of 5 binding sites within the gp91phoxpromoter (the presence of activators binding to the CDP-ζ element has not been assessed).8,9,13 This is consistent with previous descriptions of CDP as a factor that “displaces” activators from a promoter.8,9,16,33 In addition, however, CDP also contains an active repression domain that can function after addition to an unrelated DNA-binding protein.33 This region of CDP has recently been shown to associate with the histone deacetylase HDAC1.38 The ability of CDP to actively repress transcription mediated by adjacent promoter elements is apparent within the gp91phox promoter. Truncation of the gp91phox promoter from −138 to −102 bp results in the removal of the CDP-α site, including the binding sites for both CDP and the competing transcriptional activating factor CP1. Despite the ablation of the activator binding site, this truncation leads to a 10-fold increase in promoter activity after transient transfection into HEL cells.9 Hence, CDP represses gp91phox promoter activity both through physical exclusion of transcriptional activators as well as repression of adjacent promoter elements.
Although the CDP-α and CDP-ε sites within the gp91phox promoter overlap, analysis of truncated binding site probes in EMSA indicates that these binding sites are distinct. However, overlapping sequence is required for high affinity binding of CDP to the CDP-ε element. A similar relationship was previously reported for the overlapping CDP-β and CDP-α sites within the gp91phox promoter.9 Whether CDP simultaneously binds to adjacent sites has not been determined, although previous studies show a footprint that extends to the 3′-end of the CDP-α element.8 This footprint is strongest at the 5′-end of CDP-α (−130 to −103 bp) and weaker at the 3′-end (−102 to −76 bp). Given the data presented in this report, the extended CDP footprint visible with the CDP-α probe could include low-affinity interaction of CDP with the region in common between CDP-α and CDP-ε (ie, 5′CDP-ε in Fig 5).
The significant transcriptional activity directed by the −102 to +12 bp gp91phox promoter indicates that the presence of 2 weak-affinity CDP-binding sites within this DNA fragment is insufficient to fully repress promoter function. However, the magnitude of CDP function via these 2 proximal binding sites is difficult to establish, because it has not been possible to specifically ablate these CDP-binding sites and retain overlapping binding sites for transcriptional activators. Hence, the strength of the proximal promoter in the absence of CDP-mediated repression is unknown. Furthermore, the presence of multiple CDP-binding sites raises the possibility that CDP binds cooperatively to the gp91phox promoter. Hence, the occupancy of CDP at the CDP-ε and CDP-ζ sites may be higher in vivo in the context of an intact promoter than is suggested by the low affinity that CDP exhibits for these sites in in vitro EMSA studies by using isolated binding site probes.
CDP contains 4 distinct DNA-binding domains, including a homeodomain and 3 cut-repeats.18 42-44 Each of these 4 domains exhibits DNA-binding activity when expressed as glutathioneS-transferase fusion proteins and each may possibly make contact with a discrete region of a CDP-binding site. It is possible, then, that CDP may also bind to “chimeric” binding sites comprised of portions of the adjacent elements depicted in Fig 3. In this context, it is interesting that all 6 oligonucleotide probes that cover various regions within the −136 to −65 bp gp91phox promoter form CDP EMSA complexes. Importantly, overexpression of CDP results in repression of the −102 to +12 bp gp91phox promoter in the absence of higher-affinity upstream CDP-binding sites. This repression is likely mediated via the 2 weak-affinity CDP-binding sites within the −102 to +12 bp promoter, although it remains a formal possibility that the observed repression represents an indirect effect of CDP overexpression.
The difference in the magnitude of CDP-mediated repression of the −102 to +12 gp91phox promoter in HEL cells and keratinocyte cultures may reflect differences in the complement or concentration of transcriptional activators that counteract the activity of CDP, or differences in the level of CDP expression driven by the CMV promoter/enhancer. Cell-specific differences in gp91phox promoter behavior have previously been described. For example, the −102 to +12 bp gp91phox promoter is highly active in HEL cells, but is much weaker in K562 and HeLa cell lines.9 Similarly, the −450 to +12 bp gp91phox promoter is fully active in a subset of monocyte/macrophages after introduction into transgenic mice, but the majority of phagocytic cells are unable to express this transgene.3 Mutations within the gp91phox promoter have also been described that lead to a variant form of chronic granulomatous disease,45,46 in which the vast majority of phagocytic cells are unable to express gp91phox and generate a respiratory burst, yet a subset of the lineage appears to function normally.47 48 Further studies will be required to determine the differences between HEL cells and primary keratinocytes that result in variable effectiveness of CDP-mediated repression.
Importantly, the down-regulation of CDP DNA-binding activity, although necessary for gp91phox induction, is not a myeloid cell-specific event and is not sufficient for gp91phox induction. CDP is down-regulated more generally on terminal differentiation and withdrawal from the cell cycle and occurs in cells that do not express gp91phox.49,50 We hypothesize that down-regulation of CDP DNA-binding activity poises a promoter for expression and that restriction of gp91phoxtranscription to mature myeloid cells requires the combinatorial interaction and coordinate regulation of numerous transcription factors. Lineage-restricted members of the Ets family of transcription factors (PU.1 and Elf-1) have recently been found to interact with the gp91phox promoter element that is mutated in some cases of chronic granulomatous disease.46,51 52 These activators presumably play a role in the induction of gp91phox expression on down-regulation of CDP-mediated transcriptional repression during terminal phagocytic differentiation. The studies reported here contribute to our understanding of the network of transcriptional regulators that direct myeloid cell-restricted expression of the gp91phox gene.
ACKNOWLEDGMENT
We are grateful to Jean Bang and Grova Mae Lewis for excellent technical assistance with the keratinocyte experiments and to Ellis Neufeld for providing us with CDP overexpression vectors and antiserum directed against CDP.
Supported by National Institutes of Health (NIH) Grants No. CA58947 and AI31494, which were awarded to D.G.S. and A.R., respectively, and an Arthritis Foundation Biomedical Science Grant awarded to D.G.S. S.H. was supported by a Department of Education training grant for Graduate Assistance in Areas of National Need (GAANN).
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to David G. Skalnik, PhD, Wells Center for Pediatric Research, Cancer Research Building, Room 472, 1044 West Walnut St, Indianapolis, IN 46202; e-mail: dskalnik@iupui.edu.