Using a mouse bone marrow transplantation model, the authors evaluated a Moloney murine leukemia virus (MMLV)-based vector encoding 2 anti-human immunodeficiency virus genes for long-term expression in blood cells. The vector also encoded the human nerve growth factor receptor (NGFR) to serve as a cell-surface marker for in vivo tracking of transduced cells. NGFR+ cells were detected in blood leukocytes of all mice (n=16; range 16%-45%) 4 to 5 weeks after transplantation and were repeatedly detected in blood erythrocytes, platelets, monocytes, granulocytes, T cells, and B cells of all mice for up to 8 months. Transgene expression in individual mice was not blocked in the various cell lineages of the peripheral blood and spleen, in several stages of T-cell maturation in the thymus, or in the Lin−/loSca-1+ and c-kit+Sca-1+ subsets of bone marrow cells highly enriched for long-term multilineage-reconstituting activity. Serial transplantation of purified NGFR+c-kit+Sca-1+bone marrow cells resulted in the reconstitution of multilineage hematopoiesis by donor type NGFR+ cells in all engrafted mice. The authors concluded that MMLV-based vectors were capable of efficient and sustained transgene expression in multiple lineages of peripheral blood cells and hematopoietic organs and in hematopoietic stem cell (HSC) populations. Differentiation of engrafting HSC to peripheral blood cells is not necessarily associated with dramatic suppression of retroviral gene expression. In light of earlier studies showing that vector elements other than the long-terminal repeat enhancer, promoter, and primer binding site can have an impact on long-term transgene expression, these findings accentuate the importance of empirically testing retroviral vectors to determine lasting in vivo expression.
Hematopoietic stem cells (HSC) are key targets for gene therapy approaches to treat inherited hematologic disorders, cancers, and chronic viral diseases such as acquired immunodeficiency syndrome. Successful application of stem cell-based gene therapies to treat blood system disorders will require efficient delivery of the therapeutic gene to engrafting HSC, long-term reconstitution of hematopoiesis from transduced HSC, and stable expression of the therapeutic gene in the affected blood cell lineages. Most clinical trials using retroviral vectors to transfer therapeutic genes into long-term multilineage-reconstituting HSC have relied on derivatives of the Moloney murine leukemia virus (MMLV). Although retroviral marking is routinely observed at high levels in mice reconstituted with transduced HSC, marking is low in clinical trials and in primate models. Additionally, stable expression of therapeutic genes from retroviral vectors remains problematic even in mouse bone marrow transplantation models.
The MMLV long-terminal repeat (LTR) promoter can direct gene expression in blood, spleen, and thymus cells after the transplantation of transduced bone marrow cells to lethally irradiated mice,1,2 and it is active in multiple hematopoietic cell lineages of reconstituted mice, including T cells, B cells, myeloid cells, and erythroid cells.3,4 However, MMLV LTR-driven gene expression generally declines as a function of time after transplantation, and it varies between individual mice and within various tissues analyzed from a single mouse. Moreover, transgene expression from MMLV-based vectors can be drastically reduced by 70% to more than 90% in secondary or tertiary colony-forming units-spleen (CFU-S) derived from serial transplantation of transduced bone marrow cells.5 6
Although the ultimate reasons for the decline and variability in transgene expression after transplantation are unclear, several studies have suggested that the MMLV LTR is transcriptionally silenced in hematopoietic stem/progenitor cells, their progeny, or both after transplantation.5-7 Indeed, the lack of expression from retroviral vectors in CFU-S after serial transplantation was correlated with increased methylation of the LTR.5 6 However, in previous studies RNA levels or enzymatic activity were used to measure expression of the therapeutic gene from cell lysates. Consequently, it is unclear whether the MMLV LTR promoter is active at uniform levels in each cell of the hematopoietic tissues or whether expression varies from cell to cell and between cell types within each tissue. Understanding which is the more accurate picture for retroviral vector expression in HSC and their progeny after transplantation may enable a rational approach toward improved long-term retroviral vector expression. Analysis of MMLV LTR promoter-driven transgene expression at the single cell level would help to clarify whether transgene expression is restricted in HSC, its progeny cells, or both.
For several years we have been developing a stem cell gene therapy approach to deliver genes that blocks replication of the human immunodeficiency virus (HIV) in hematopoietic cells of infected persons.8 9 Success with this approach requires gene transfer to multilineage-reconstituting HSC and long-term expression of anti-HIV genes throughout the hematopoietic system, particularly in progeny cells targeted by HIV, such as CD4+ T cells and macrophages. In this study, we evaluated an MMLV-based vector bearing a combination of 2 anti-HIV genes for long-term expression in a mouse bone marrow transplantation model. Expression of a cell-surface marker gene included in the vector (the human nerve growth factor receptor [NGFR10]) was tracked at the single cell level by flow cytometry as a measure of MMLV LTR promoter activity. We showed that a high frequency of transgene-positive cells and high levels of vector expression can be achieved in multiple lineages of peripheral blood cells for 8 months after transplantation. Transgene expression was evident in all cell lineages examined from the peripheral blood and spleen, in successive stages of T-cell differentiation in the thymus, or in highly enriched bone marrow stem/progenitor cell populations. In addition, we demonstrated that retroviral gene expression is maintained after engraftment and subsequent differentiation of HSC during peripheral blood reconstitution after transplantation in secondary recipients.
Materials and methods
Retroviral vectors and producer cell lines
Construction and anti-HIV efficacy of the 1171 vector will be described elsewhere (Veres G, manuscript in preparation). The 1171 vector encoded the RevM1011 and the HIV-1 pol antisense genes cloned in pLN.9 The truncated NGFR10served as a marker to trace expression in vivo. Ecotropic retroviral vector producer cells were derived from the GP-E86 packaging cell line.12
Retrovirus transduction of bone marrow cells
Eight- to 12-week-old (C57BL/Ka.AKR/J)Sys-Ptprca-Thy-1a mice (BA .1; Thy-1.1, Ly-5.2) were used as bone marrow donors. Eight- to 12-week-old congenic (C57BL/6J.SJL/J)Sys-Ptprcb-Thy-1b mice (B6SJL; Thy-1.2, Ly-5.1) were the recipients in our bone marrow transplantation experiments. Mice were bred and maintained at the SyStemix animal facility. B.A1 mice were injected with 5-fluorouracil (150 mg/kg body weight) 5 to 6 days before bone marrow harvest. Cells were flushed from femoral shafts with phosphate-buffered saline (PBS) and 0.2% bovine serum albumin (BSA) and were plated at 106/mL in Whitlock/Witte medium containing 10% fetal bovine serum (FBS; Hyclone Laboratory, Logan, UT), murine stem cell factor (100 ng/mL), murine IL-3 (10 ng/mL), and murine IL-6 (10 ng/mL). Cytokines were from R&D Systems (Minneapolis, MN). Cells were cultured for 24 hours at 37°C, plated onto irradiated (1500 rad) virus producer cells, and co-cultured for an additional 48 hours with protamine sulfate (16 μg/mL) added to the media. Nonadherent bone marrow cells were recovered, resuspended in PBS/0.2% BSA, and 106 cells (200μL) was injected through the tail vein into lethally irradiated (2 × 525 rad) B6SJL recipient mice.
Analysis of transgene expression on peripheral blood cells, thymocytes, and splenocytes
Peripheral blood samples were collected from individual mice by retro-orbital bleeding and analyzed independently. A portion of the sample was stained with a phycoerythrin (PE)-conjugated antibody specific for human NGFR (Boehringer Mannheim, Indianapolis, IN) and a fluorescein isothiocyanate (FITC)-conjugated anti-mouse CD41 antibody (Pharmingen, San Diego, CA) and analyzed for the NGFR expression on erythrocytes and platelets. Red blood cells (RBCs) were defined using characteristic forward/side-scatter properties, and platelets were defined by forward/side-scatter properties and CD41 expression. Both populations were clearly distinguished from leukocytes, which comprised less than 0.5% of the events. The remainder of the sample was diluted in hypotonic lysis buffer to remove erythrocytes, and the remaining cells were stained with anti-Ly-5.2 (CD45.2)-FITC (Pharmingen), anti-NGFR-PE, and a lineage-specific allophycocyanin (APC)-conjugated antibody (either Gr-1, Mac-1 [CD11b], CD3e, or B220 [CD45R], all from Pharmingen). After the final wash, cells were resuspended in PBS/2% FBS with propidium iodide (1 μg/mL), and dead cells were excluded from analysis based on propidium iodide staining and forward-scatter properties. The thymus and spleen from individual mice 8 months after transplantation were made into single-cell suspensions and stained with anti-Ly-5.2-FITC and anti-NGFR-PE. Splenocytes were additionally stained with APC-conjugated anti-Gr-1, anti-Mac-1, anti-B220, anti-CD4, or anti-CD8 (Pharmingen). Thymocytes were stained either with anti-CD8-FITC (Pharmingen), anti-NGFR-PE, and anti-CD4-APC, or with anti-Ly-5.2-FITC and anti-NGFR-PE. Analysis was performed on a FACScalibur instrument (Becton Dickinson Immunochemistry Systems, San Jose, CA). The significance of differences in transgene expression in peripheral blood cells was analyzed using a 1-tailed Student'st test.
Analysis and purification of transgene expressing HSC populations
Total bone marrow cells were collected from the long bones (2 femurs and 2 tibias) of each mouse by flushing with PBS/0.2% BSA. To analyze NGFR expression on Lin−/loSca-1+ cells, bone marrow cells from mice 8 months after transplantation were depleted of RBCs by hypotonic lysis and were stained with anti-Ly-5.2-FITC, anti-NGFR-PE, anti-Sca-1 biotin, APC-conjugated antibodies to lineage-specific antigens (CD4, CD8, Gr-1, Mac-1, and B220), and streptavidin–Texas red. NGFR expression was analyzed on Lin−/loSca-1+ cells from whole bone marrow or from lineage antigen-depleted cells prepared by removal of lineage antigen-stained cells by immunomagnetic bead depletion (Dynal, Great Neck, NJ). Analysis of bone marrow HSC populations from mice 8 months after transplantation with the phenotype c-kit+Thy-1.1loLin−/loSca-1+was performed as previously described.13 Briefly, cells were incubated with a biotinylated anti-Sca-1 antibody and enriched using the MACS system for magnetic bead selection (Miltenyi Biotech, Auburn, CA). Sca-1 enriched cells from individual mice were stained with anti-Thy-1.1-FITC, a cocktail of PE-conjugated antibodies specific for lineage markers, anti-c-kit-APC, and streptavidin–Texas red, and analyzed on a FACStarplus instrument (Becton Dickinson Immunochemistry Systems). For sorting of donor-type NGFR+c-kit+Sca-1+ HSC populations, Sca-1 enriched cells were pooled from 2 mice, stained with anti-NGFR-PE, anti-c-kit-APC, and streptavidin–Texas red, resuspended after the final wash in PBS/2% FBS with propidium iodide, and sorted on a FACStarplus instrument (Becton Dickinson Immunochemistry Systems). Sorted cells were resuspended in PBS/0.2% BSA and injected through the retro-orbital sinus into lethally irradiated (2 × 525 rad) B6SJL recipient mice.
DNA analysis
High molecular weight genomic DNA was isolated from spleens of individual mice 8 months after transplantation by proteinase K/sodium dodecyl sulfate/RNAase extraction.14 DNA recovered after phenol extraction was ethanol precipitated and resuspended in TE buffer. Proviral integration pattern was analyzed by Southern blot analysis after digestion of 10μg genomic DNA with Hind III, which cleaves the vector 15 nucleotides 5′ to the predicted NGFR translational start site. Proviral copy number was analyzed by digestion with Hind III and Bcl I, which released a 2-kb fragment containing the NGFR and pol/AS sequences. Southern blots were probed with a 1.9-kb fragment (Hind III/Hind II) spanning the NGFR and pol/AS sequences. The probe was generated by the random priming method using [32P]dCTP.
The proviral integrant copy number was quantitated relative to that of the single-copy interferon-° gene (using a 1.2-kb Pst I/Hind III genomic DNA fragment as a probe15) by densitometry using a Molecular Dynamics (Sunnyvale, CA) PhosphorImager system.
Results
Long-term transgene expression on peripheral blood cells
The MMLV-based retroviral vector 1171 (L-M10-IRES-NGFR-pol/AS) encodes 3 genes from 1 polycistronic mRNA transcript: the RevM10 gene,11 the truncated form of the NGFR,10 and an antisense fragment from the HIV-1 pol gene (pol/AS; see Figure4D).9 Translation of the NGFR protein is mediated by the internal ribosomal entry site (IRES) of the human encephalomyocarditis virus16 and thus is linked to the RevM10-pol/AS expression. The expression of RevM10 and a cell surface marker protein (whose translation is mediated by the IRES) are co-linear8,17; therefore, the more easily detectable NGFR surface marker was used to facilitate the tracking of retroviral gene expression by flow cytometry. Ecotropic retroviral vector producer cells were derived from the GP-E86 packaging cell line.12 Transduction of 5-fluorouracil bone marrow and reconstitution of lethally irradiated mice were carried out in 2 independent experiments. In each experiment, groups of 7 to 8 mice (male and female) received either 1171 or mock-transduced bone marrow cells. After 5 months, 4 mice were killed and evaluated for any abnormalities associated with long-term retroviral vector transgene expression in gene-modified cells. Of the remaining mice, 3 from experiment 2 were killed 8 months after transplantation and analyzed in detail for transgene expression in various hematopoietic organs (splenocyte and thymocyte subsets and c-kit+Sca-1+ bone marrow cells). Four mice were analyzed for transgene expression in Lin−/loSca-1+ bone marrow cells. Four mice were killed, and NGFR+c-kit+Sca-1+ bone marrow cells were isolated by cell sorting and were used in secondary transplantation experiments. Chimerism of donor cells was monitored by Ly-5.2 expression in various hematopoietic tissues. Overall, no striking differences were evident in the kinetics of hematopoietic recovery, the extent of donor-type reconstitution of peripheral blood cells, or the proportions of various peripheral blood cell types between the groups of mice receiving 1171 or mock-transduced cells.29
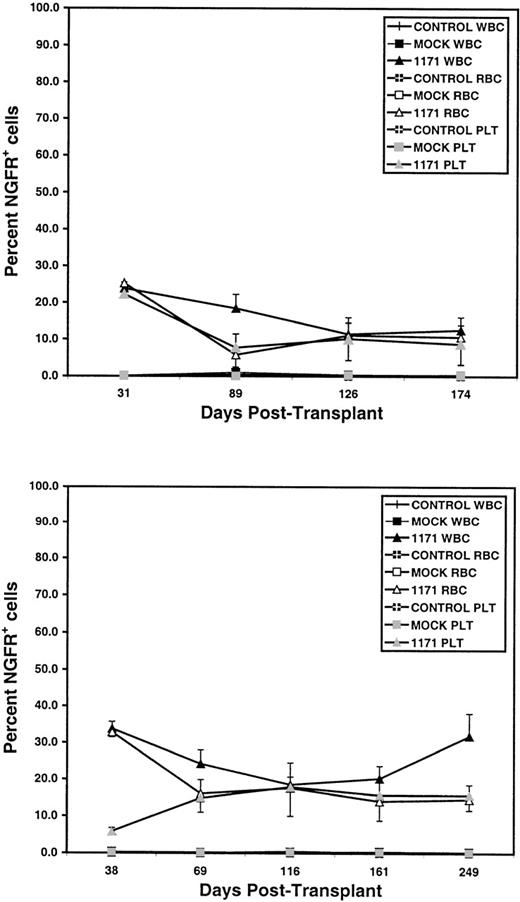
Four to 5 weeks after transplantation, NGFR+ cells were detected in the circulation of all transplanted animals. The frequency of NGFR+ cells ranged from 16.3% to 44.7% for white blood cells, 2.7% to 25.6% for platelets, and 23.6% to 38.7% for erythrocytes. NGFR expression was repeatedly detected in multiple peripheral blood lineages (RBCs, platelets, and donor-type WBCs) for more than 8 months in all mice analyzed (Figures1 and 2). In experiment 1, the percentage of NGFR+ RBCs, platelets, and donor WBCs declined with time and then did not significantly decline further after the second month (Figure 2). Similar results were obtained in experiment 2 for RBCs and donor WBCs, though the percentage of NGFR+ platelets was lowest at day 38 and stabilized at a higher level after the second month. Overall, the frequency of NGFR+ RBCs, platelets, or donor WBCs declined 1- to 2.5-fold by 8 months after transplantation (in experiment 2, NGFR+ platelets increased 2.7-fold overall).
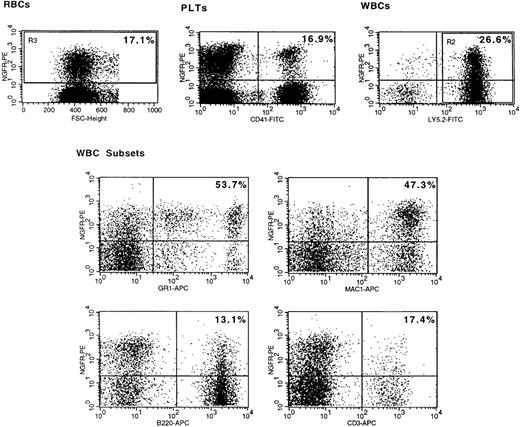
Flow cytometric analysis of NGFR transgene expression in peripheral blood cells on day 249 after transplantation.
Peripheral blood cells were prepared as described in the “Materials and Methods” from a representative mouse (217). (top left) Region 3 (R3) denotes the NGFR+ fraction of red blood cells (RBCs). (top right) R2 is the Ly-5.2+ donor type fraction of live gated white blood cells (WBCs). WBC subsets are donor-type myeloid (Gr-1+ or Mac-1+), B (B220+), and T (CD3e+) cells gated on live cells and R2. PLTs, platelets. The percentage of NGFR+ cells in each is shown in Table1.
Flow cytometric analysis of NGFR transgene expression in peripheral blood cells on day 249 after transplantation.
Peripheral blood cells were prepared as described in the “Materials and Methods” from a representative mouse (217). (top left) Region 3 (R3) denotes the NGFR+ fraction of red blood cells (RBCs). (top right) R2 is the Ly-5.2+ donor type fraction of live gated white blood cells (WBCs). WBC subsets are donor-type myeloid (Gr-1+ or Mac-1+), B (B220+), and T (CD3e+) cells gated on live cells and R2. PLTs, platelets. The percentage of NGFR+ cells in each is shown in Table1.
Sustained multilineage transgene expression from an MMLV-based vector in peripheral blood for more than 8 months after transplantation of transduced bone marrow cells.
Data represented the mean percentage ± SEM of NGFR+cells in peripheral blood of mice at various time-points after transplantation of bone marrow cells transduced with the 1171 vector. Results are shown from 2 independent transductions, experiment 1 (top) and experiment 2 (bottom). NGFR expression was analyzed as in Figure 1with WBCs gated on live donor-type cells. Controls are nonirradiated, age-matched mice from the donor and recipient strains, mocks are mice transplanted with nontransduced cells otherwise prepared as for 1171 transduced cells. Eight mice were analyzed in experiment 1; 7 mice were analyzed in experiment 2 until day 161, and 3 were analyzed on day 249. Note that overall NGFR expression in RBCs, platelets, and donor WBCs declined with time then stabilized after the second month. In experiment 1, the percentage of NGFR+ cells was greater at day 31 than at any subsequent time-point (P < .03, except for donor WBCs at day 31 versus day 89). Similar results were obtained in experiment 2 (P < .05), except that fewer NGFR+ platelets were detected at day 38 than at later time-points. After the second month after transplantation, no other significant declines (P < .05) in the percentage of NGFR+ WBCs, RBCs, or platelets were observed between any of the time-points in either experiment. All other abbreviations are as in Figure 1.
Sustained multilineage transgene expression from an MMLV-based vector in peripheral blood for more than 8 months after transplantation of transduced bone marrow cells.
Data represented the mean percentage ± SEM of NGFR+cells in peripheral blood of mice at various time-points after transplantation of bone marrow cells transduced with the 1171 vector. Results are shown from 2 independent transductions, experiment 1 (top) and experiment 2 (bottom). NGFR expression was analyzed as in Figure 1with WBCs gated on live donor-type cells. Controls are nonirradiated, age-matched mice from the donor and recipient strains, mocks are mice transplanted with nontransduced cells otherwise prepared as for 1171 transduced cells. Eight mice were analyzed in experiment 1; 7 mice were analyzed in experiment 2 until day 161, and 3 were analyzed on day 249. Note that overall NGFR expression in RBCs, platelets, and donor WBCs declined with time then stabilized after the second month. In experiment 1, the percentage of NGFR+ cells was greater at day 31 than at any subsequent time-point (P < .03, except for donor WBCs at day 31 versus day 89). Similar results were obtained in experiment 2 (P < .05), except that fewer NGFR+ platelets were detected at day 38 than at later time-points. After the second month after transplantation, no other significant declines (P < .05) in the percentage of NGFR+ WBCs, RBCs, or platelets were observed between any of the time-points in either experiment. All other abbreviations are as in Figure 1.
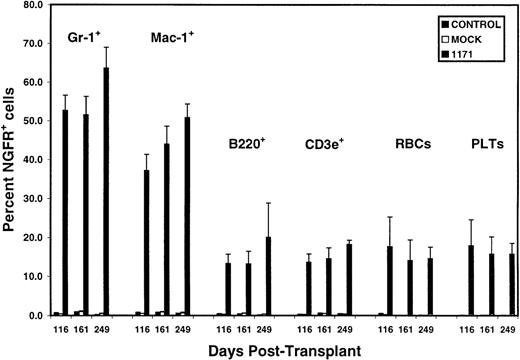
NGFR was detected in all peripheral donor WBC subsets analyzed, including B (CD45R B220+), T (CD3e+), and myeloid (Gr-1+ or CD11b Mac-1+) cells in each mouse (Table 1 and Figures 1 and3). NGFR expression was variable between mice and between different phenotypic populations of peripheral blood cells. Unexpectedly, the percentage of NGFR+ cells was significantly greater in donor type Gr-1+ or Mac-1+ myeloid cells than in RBCs, platelets, or donor lymphoid cells (B220+ or CD3e+) at all time-points (Figure 3). The only exception to this observation was for Mac-1+ versus B220+ cells in mouse 213 at day 249 (Table 1). When both experiments were combined, the frequency of NGFR+ cells was greater in Ly-5.2+Gr-1+ cells than in Ly-5.2+B220+ or Ly-5.2+CD3e+ cells, RBCs, or platelets at all time-points in 13 of 15 mice. These data clearly show that MMLV LTR-driven transgene expression is not blocked in any of the predominant peripheral blood lineages in long-term reconstituted mice. Significantly, because at day 249 NGFR expression was at background levels on host-type WBC subsets (ie, 0.1% to 0.4% of Ly5.2− cells positive for Gr-1, Mac-1, B220, or CD3e were NGFR+), we ruled out the possibility that transgene expression was sustained from the continuous in vivo transfer of the vector or NGFR protein from injected transduced cells to nontransduced cells. It is possible that the greater frequency of NGFR+myeloid cells resulted in part from the lineage-specific enhancement of LTR or IRES activity.
Stable long-term transgene expression in all peripheral donor white blood cell types with preferential representation of NGFR+ cells in Gr-1+ and Mac-1+donor WBCs.
NGFR expression was analyzed by flow cytometry as in Figure 1. Data represented the mean percentage ± SEM of NGFR+ cells in peripheral blood of 7 mice from experiment 2 at various time-points after transplantation of bone marrow cells transduced with the 1171 vector. There were no significant declines (P < .05) in the percentage of NGFR+ cells within a given phenotypic cell population sampled after the second month after transplantation. Note that the frequency of NGFR+ cells was consistently greater in Ly5.2+Gr-1+ or Ly5.2+Mac-1+ myeloid cells than in all other cell types at each time-point. For Gr-1+ versus B220+, CD3e+, RBCs, or platelets,P < .001 (n = 7) at days 116 and 161, andP < 0.03 (n = 3) at day 249. For Mac-1+versus B220+, CD3e+, RBCs, or platelets,P < .009 (n = 7) at days 116 and 161, andP < .02 (n = 3) at day 249, except for Mac-1+versus B220+. All abbreviations are as in Figures 1 and2.
Stable long-term transgene expression in all peripheral donor white blood cell types with preferential representation of NGFR+ cells in Gr-1+ and Mac-1+donor WBCs.
NGFR expression was analyzed by flow cytometry as in Figure 1. Data represented the mean percentage ± SEM of NGFR+ cells in peripheral blood of 7 mice from experiment 2 at various time-points after transplantation of bone marrow cells transduced with the 1171 vector. There were no significant declines (P < .05) in the percentage of NGFR+ cells within a given phenotypic cell population sampled after the second month after transplantation. Note that the frequency of NGFR+ cells was consistently greater in Ly5.2+Gr-1+ or Ly5.2+Mac-1+ myeloid cells than in all other cell types at each time-point. For Gr-1+ versus B220+, CD3e+, RBCs, or platelets,P < .001 (n = 7) at days 116 and 161, andP < 0.03 (n = 3) at day 249. For Mac-1+versus B220+, CD3e+, RBCs, or platelets,P < .009 (n = 7) at days 116 and 161, andP < .02 (n = 3) at day 249, except for Mac-1+versus B220+. All abbreviations are as in Figures 1 and2.
Persistent retroviral gene expression in hematopoietic organs
Long-term retroviral transgene expression was further demonstrated in various hematopoietic organs of 3 mice analyzed 8 months after transplantation. In the thymus, more than 80% of total thymocytes were Ly-5.2+. CD4/CD8 ratios in the thymi of transplanted mice (average, 4.1; n = 4) were not significantly different from those of the age-matched control mice (3.5 and 3.7). On average, 28% of Ly-5.2+ thymocytes expressed the NGFR, and expression varied between mice and between different phenotypic populations of the thymus. However, expression was detected in all mice examined on each of the Ly-5.2+ phenotypic thymocyte subsets, CD4−CD8−, CD4+CD8+, CD4+, and CD8+ cells (Table 2). Hence, it was not restricted at any stage of thymocyte maturation studied. In mouse 213, CD4+CD8+ cells comprised less than 1% of total thymocytes, thereby precluding analysis of NGFR expression in this compartment.
In the spleen, more than 91% of cells were donor Ly-5.2+, and overall 23% of Ly-5.2+ splenocytes expressed the NGFR (Table 3). Again, expression varied from mouse to mouse and between different phenotypic populations. Each Ly-5.2+ lineage subset expressed the NGFR, including Gr-1+ or Mac-1+ myeloid cells, CD4+or CD8+ T cells, and B220+ B cells. Of the various Ly-5.2+ splenocyte populations, NGFR expression was consistently detected on more Gr-1+ myeloid cells than on the other phenotypes. When phenotypic subsets from spleen were compared to peripheral blood of the same mouse, NGFR expression varied though the trend was for expression on a greater proportion of donor Gr-1+ or Mac-1+ cells in peripheral blood than in the spleen. NGFR expression was also variable when compared to donor CD4+ or CD8+ cells in the spleen versus the thymus.
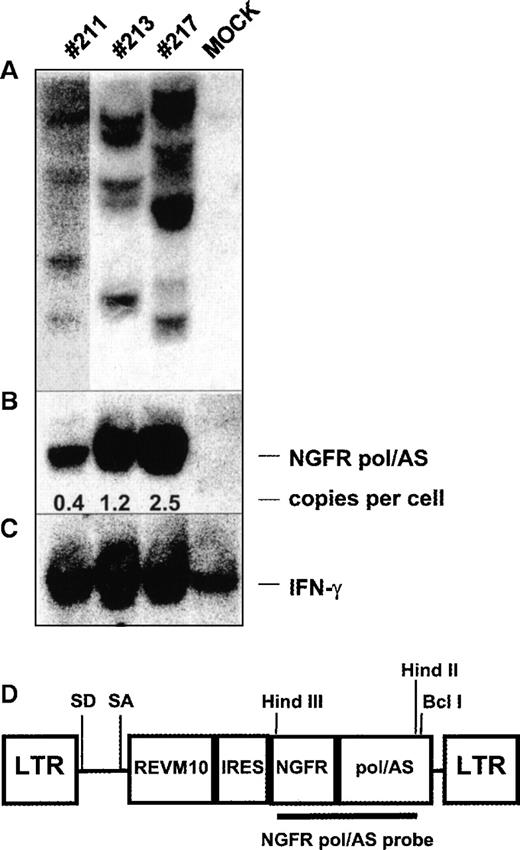
Southern blot analysis was carried out to assess the gene marking frequency and the number of proviral integrants in the spleens of representative mice with long-term transgene expression 8 months after transplantation. We calculated that 0.4, 1.2, and 2.5 proviral copies were detected per genome from mouse 211, 213, and 217, respectively (Figure 4); 4 to 6 proviral integrants were detected in the splenocytes of each mouse. Thus, the NGFR+splenocytes of each mouse were derived from 4 to 6 or fewer retrovirally marked clones.
Southern blot analysis of integration pattern and proviral copy number in splenocytes of mice with long-term multilineage transgene expression.
DNA was isolated from splenocytes of mice whose transgene expression is shown in Table 3 and was digested with Hind III (A, lanes 1-4) or Hind III and Bcl I (B,C, lanes 1-4). (A,B) Southern blot was probed with a 1.9-kb Hind III/Hind II vector fragment encoding the NGFR and pol/AS cDNA. (C) The same blot was reprobed with a genomic DNA fragment from the interferon-γ gene. The blot was exposed to autoradiographic film for 4 days in all mice except mouse 211, which was exposed for 8 days. (D) Schematic diagram of the retroviral vector and NGFR pol/AS DNA probe used in A and B.
Southern blot analysis of integration pattern and proviral copy number in splenocytes of mice with long-term multilineage transgene expression.
DNA was isolated from splenocytes of mice whose transgene expression is shown in Table 3 and was digested with Hind III (A, lanes 1-4) or Hind III and Bcl I (B,C, lanes 1-4). (A,B) Southern blot was probed with a 1.9-kb Hind III/Hind II vector fragment encoding the NGFR and pol/AS cDNA. (C) The same blot was reprobed with a genomic DNA fragment from the interferon-γ gene. The blot was exposed to autoradiographic film for 4 days in all mice except mouse 211, which was exposed for 8 days. (D) Schematic diagram of the retroviral vector and NGFR pol/AS DNA probe used in A and B.
Long-term transgene expression in bone marrow HSC populations
Overall, all blood cell lineages of each mouse examined expressed the NGFR, regardless of whether cells were from peripheral blood or hematopoietic organs. The finding that the NGFR was detected for 8 months on Ly-5.2+Gr-1+ peripheral blood cells suggests continuous replenishment of short-lived granulocytes (half-life, approximately 24 hours)18 by hematopoietic precursors expressing the retroviral transgene. Additionally, retroviral gene expression was detected throughout the various stages of T-cell development in the thymus and in peripheral blood T cells. Based on these data, it is likely that retroviral gene expression is sustained in long-term multilineage-reconstituting cells. This possibility was evaluated in Lin−/loSca-1+ and in c-kit+Sca-1+ bone marrow cells; both populations are highly enriched for long-term multilineage-reconstituting cells in normal19,20 or reconstituted mice.21 Total or Lin+-depleted Lin−/loSca-1+ bone marrow cells from mice 8 months after transplantation were analyzed for Ly-5.2 and NGFR expression. NGFR expression ranged from 0.6% to 22.2% on Lin−/loSca-1+ cells (Table4).
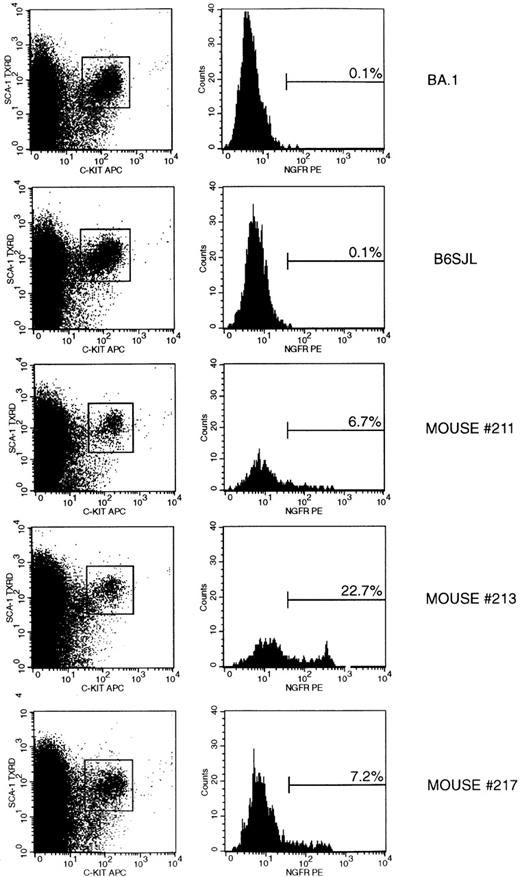
Several studies have shown that the Lin−/loSca-1+ cells are a heterogeneous population of long-term and transiently reconstituting multipotent cells.20,22,23 Expression of the c-kit gene is a key marker of long-term reconstituting cells, and Lin−/loSca-1+c-kit−cells lack detectable long-term reconstituting potential.20,24 25 Retroviral gene expression was further examined in c-kit+Sca-1+ cells enriched from bone marrow of individual mice by positive selection for Sca-1+ cells. As shown in Figure5, total bone marrow or Sca-1+-enriched cells expressing relatively high levels of the c-kit+ and Sca-1+ antigens are a distinct population on dual-parameter dot plots. Stringent gating of this c-kit+Sca-1+ population showed that more than 96% of these cells were Ly-5.2+ and more than 94% were Lin−/lo in 3 reconstituted mice 8 months after transplantation and in age-matched control mice (data not shown). Using this gating strategy, 20% to 22% of Lin−/loSca-1+ cells were c-kit+, and Thy-1.1loLin−/lo cells comprised 42% to 53% of c-kit+Sca-1+ cells from the 3 reconstituted mice or control mice. NGFR expression in the c-kit+Sca-1+ population was limited to the Ly-5.2+ donor-type cells and was distributed evenly between the Thy-1.1− and Thy-1.1lo cells (data not shown). In addition to these 3 mice, bone marrow cells were pooled in 2 independent experiments from 2 mice 8 months after transplantation. Overall, 6.7% to 22.7% of c-kit+Sca-1+ cells from all long-term reconstituted mice analyzed were NGFR+ (Table 4). The mean fluorescence intensity of NGFR staining was reduced in Lin−/loSca-1+ or c-kit+Sca-1+ cells relative to more mature bone marrow or peripheral blood cells. Consistently fewer NGFR+ cells were detected in the HSC populations than in the donor WBC of each mouse.
FACs analysis of transgene expression in bone marrow c-kit+Sca-1+ stem/progenitor cells 8 months after transplantation.
Bone marrow from 4 long bones of each mouse was recovered, Sca-1 selected, and analyzed by flow cytometry as described in “Materials and Methods.” (left) c-kit versus Sca-1 staining of live-gated cells. (boxes) Regions used to define c-kit+Sca-1+ cells. (right) Histograms of NGFR expression on live-gated c-kit+Sca-1+ cells. (markers) Proportion of NGFR+ cells. Two age-matched control mice (BA.1 and B6SJL) and 3 mice transplanted with bone marrow cells transduced with the 1171 vector are shown.
FACs analysis of transgene expression in bone marrow c-kit+Sca-1+ stem/progenitor cells 8 months after transplantation.
Bone marrow from 4 long bones of each mouse was recovered, Sca-1 selected, and analyzed by flow cytometry as described in “Materials and Methods.” (left) c-kit versus Sca-1 staining of live-gated cells. (boxes) Regions used to define c-kit+Sca-1+ cells. (right) Histograms of NGFR expression on live-gated c-kit+Sca-1+ cells. (markers) Proportion of NGFR+ cells. Two age-matched control mice (BA.1 and B6SJL) and 3 mice transplanted with bone marrow cells transduced with the 1171 vector are shown.
Transgene expression in peripheral blood of secondary transplant recipients
The potential of phenotypically defined bone marrow populations for multilineage reconstitution and continued retroviral gene expression was evaluated in 2 experiments. In each experiment, NGFR+c-kit+Sca-1+ cells were purified from pooled bone marrow of 2 mice 8 months after transplantation by cell sorting and were transferred to lethally irradiated recipients. In the first experiment, recipient mice received approximately 1900 NGFR+c-kit+Sca-1+ cells, and, after 2 months, the RBCs, platelets, and donor WBCs were reconstituted in 5 of 5 mice with Ly-5.2+NGFR+cells. Two mice received 200,000 total bone marrow cells pooled from these donors for comparison with the HSC-enriched grafts. Injection of approximately 100 NGFR+c-kit+Sca-1+ cells in the second experiment resulted in the multilineage reconstitution of Ly-5.2+NGFR+ cells in 3 of 5 mice. Donor reconstitution of sorted cells measured 2 months after transplantation ranged from 12% to 52%, and NGFR expression was detected on 13% to 60% of Ly-5.2+ WBCs, 4% to 40% of RBCs, and 4% to 35% of platelets (Table 5). The mean percentage NGFR+ cells in the peripheral blood of the secondary transplant recipient mice was approximately twice that of the primary recipients measured at 2 to 3 months after transplantation. The variance in NGFR expression (SEM) was greater in the secondary transplant recipients, perhaps because fewer mice were used here than in the primary transplant groups.
Discussion
In this study, we analyzed the potential of the MMLV promoter to direct long-term transgene expression in the peripheral blood, hematopoietic organs, and bone marrow stem/progenitor compartment of reconstituted mice. FACS analysis of peripheral blood showed that multilineage reconstitution with NGFR+ donor-type cells early (4 to 5 weeks) after transplantation was sustained for up to 8 months in all the mice studied from 2 independent transduction experiments. Similar data were obtained in 2 experiments with related vectors in which the RevM10 translation start site was abolished (unpublished results). Careful hematologic and histologic analyses did not reveal any abnormalities associated with long-term transgene expression in gene-modified cells.29
Previous studies have shown that MMLV-based vectors are expressed in the peripheral blood, spleen, thymus, or bone marrow after transplantation.1-4 However, transgene expression levels generally declined with time and varied between individual mice and between different tissues of the mouse. Although we observed an overall decline in transgene expression during the first 2 months after transplantation, long-term expression beyond the second month was remarkably stable in all lineages examined. Several experimental differences in the current study might have contributed favorably to our observation of consistent, long-term, multilineage transgene expression. We evaluated transgene expression frequency on a per cell basis in donor-derived cells; many earlier studies scored transgene expression from cell lysates. The NGFR marker gene used here had not been, to our knowledge, evaluated previously in the mouse bone marrow transplantation setting. It is possible that the combination of detection reagents and expression level at the cell surface results in a greater detection frequency for transgene-expressing cells. It is also possible that the immunogenicity or negative influence on hematopoiesis of this gene product is less pronounced or negligible than other marker genes (ie, CD24, CD8, mPrP, or the neomycin phosphotransferase gene).2 Although the level of therapeutic transgene expression in target cells required for clinical efficacy varies according to each disease indication, the data presented here show that stable, long-term transgene expression can be achieved from MMLV-based retroviral vectors in a variety of blood cell lineages found in the periphery or in hematopoietic organs.
The decline in retroviral vector expression after transplantation of transduced bone marrow cells has been attributed to restricted expression in long-term multilineage-reconstituting HSC and their progeny compared to transient multilineage-reconstituting cells or more mature cells.5,6 Support for this proposal comes from the finding that serial transfer of bone marrow cells can lead to undetectable MMLV-LTR-driven transgene expression in 70% to 90% of secondary or tertiary CFU-S.5,26 Decreased expression over time has also been observed with vectors having LTRs and additional elements from other murine retroviruses.27,4 However, the issue of how transgene expression in phenotypically or functionally defined HSC populations relates to that in peripheral blood cells remains controversial. Analysis of a murine Friend spleen focus-forming virus vector showed that though transgene expression was detected in bone marrow Lin−/loSca-1+stem/progenitor cells 2 to 4 months after transplantation, expression declined in WBCs from 100% at 4 weeks to 23% of mice positive at week 28.28 In contrast, preselection of cells expressing a CD24 transgene from a murine stem cell-based vector resulted in high-level, long-term transgene expression (81%) in bone marrow Lin−/loSca-1+ cells after transplantation, yet only 28% of peripheral WBCs expressed the transgene.7 Clearly, the observation of a greater frequency of transgene expression in bone marrow stem/progenitor cells than in peripheral blood is at odds with the view that retroviral gene expression is suppressed in HSC but not in more mature blood cells.
We demonstrated that NGFR expression was sustained 8 months after transplantation in a significant fraction of bone marrow Lin−/loSca-1+ stem/progenitor cells, and in at least some of the primitive c-kit+Sca-1+ subset enriched for long-term multilineage-reconstituting HSC. Transgene expression was consistently detected in fewer Lin−/loSca-1+ or c-kit+Sca-1+ bone marrow cells than in peripheral WBCs. However, long-term expression was stable in peripheral blood cells and was clearly demonstrated in multiple lineages of the peripheral blood and spleen cells 8 months after transplantation. Moreover, transgene expression was detected throughout the various stages of T-cell lineage differentiation—from the bone marrow stem/progenitor compartment and immature CD4+CD8+ thymocytes to the more mature CD4+ or CD8+ thymocytes and peripheral CD3e+ T cells. Additionally, NGFR was expressed in splenic and peripheral B220+ B cells. In the peripheral blood, granulocytes have a relatively short half-life (approximately 24 hours)18 and are continually replenished from progenitor cells in the bone marrow. The consistent detection of the NGFR on donor type Gr-1+ cells for up to 8 months suggested continuous production of NGFR+ granulocytes from NGFR+progenitors. Similarly, sustained production of NGFR+platelets was likely supported by NGFR+megakaryocytes in the bone marrow.
Transplantation of NGFR+c-kit+Sca-1+ cells into secondary recipients demonstrated that these HSC populations are sufficient for hematopoietic recovery and multilineage reconstitution of lethally irradiated mice. Significantly, all secondary recipients were repopulated with NGFR+ RBCs, platelets, and donor-type WBCs, directly demonstrated for the first time that retroviral gene expression in multilineage-reconstituting HSC was sustained in the peripheral blood progeny cells after serial transplantation. The serial transplantation data, in conjunction with the analysis of primary transplant recipients, provided strong evidence that silencing of MMLV LTR-driven transgene expression is most pronounced in long-term, multilineage-reconstituting HSC during the hematopoietic recovery phase after myeloablation and transplantation. Once durable chimerism of transgene-expressing HSC is established, progeny cells expressing the transgene are continuously generated and contribute to the various peripheral blood cell lineages.
Other investigators5,6 have reported that pronounced silencing of MMLV expression at the mRNA level is accompanied by extensive methylation of the viral LTR in secondary CFU-S. Although long-term expression in the peripheral blood of secondary recipients was not studied, it is likely that the MMLV vector would be silenced there as well. Multiple modifications of the vector LTR and primer binding site were shown to improve the frequency of transgene expression from proviral integrants.6 In our study, relatively few retrovirally marked clones contributed to spleen reconstitution (4 to 6 or fewer). Because the proviral copy number per cell was greater than the percentage of NGFR+ cells, it is likely that not all proviral integrants were transcriptionally active. Nonetheless, the finding that transgene expression persisted for at least 8 months demonstrated that some of the proviral integrants were not silenced. Because vector elements other than the LTR enhancer, promoter, and primer binding site can have an impact on long-term transgene expression (eg, addition of the neomycin phosphotransferase gene),2 our findings emphasized the critical role for in vivo analysis when evaluating the potential of retroviral vectors for long-term expression in the hematopoietic system.
Acknowledgments
The authors thank Nobuko Uchida and Stan Tamaki for help with KTLS cell isolation.
Reprints:Timothy W. Austin, SyStemix Inc, 3155 Porter Drive, Palo Alto, CA 94304; email: tim.austin@pharma.novartis.com.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.