Abstract
The low levels of transduction of human hematopoietic stem cells (HSCs) with Moloney murine leukemia virus (MLV) vectors have been an obstacle to gene therapy for hematopoietic diseases. It has been demonstrated that lentivirus vectors are more efficient than MLV vectors at transducing nondividing cell lines as well as human CD34+ cells and severe combined immunodeficiency disease repopulating cells. We compared transduction of cell lines and Lin− bone marrow cells, using a vesicular stomatitis virus G (VSV-G)-pseudotyped lentivirus or MLV vectors carrying a green fluorescent protein marker gene. As predicted, the lentivirus vector was more efficient at transducing mouse and human growth-inhibited cell lines. The transduction of mouse HSC by lentivirus vectors was compared directly to MLV vectors in a co-transduction assay. In this assay, transduction by ecotropic MLV is a positive internal control for downstream steps in retrovirus transduction, including cell division. Both the VSV-G lentivirus and MLV vectors transduced mouse HSCs maintained in cytokine-free medium at very low frequency, as did the ecotropic control. The lentivirus vector and the MLV vector were equally efficient at transducing bone marrow HSCs cultured in interleukin 3 (IL-3), IL-6, and stem cell factor for 96 hours. In conclusion, although lentivirus vectors are able to transduce growth-inhibited cell lines, the cell cycle status of HSCs render them resistant to lentivirus-mediated transduction, and it is hypothesized that entry into cycle, not necessarily division, may be a requirement for efficient lentivirus-mediated transduction.
Introduction
Hematopoietic stem cells (HSCs) are an attractive target for gene therapy of inherited and acquired blood diseases.1,2 Integration of novel genetic material into the genome of HSCs would result in continuous production of hematopoietic cells with the transferred gene and correction of the disease. The most efficient and most widely used gene transfer system for integrating genes into HSCs is retrovirus-mediated gene transfer. Initial work in mouse model systems using a mouse-specific ecotropic retrovirus vector demonstrated the ability to transduce permanently up to 35% to 40% of mouse long-term repopulating cells (LTRCs).3-6 Gene marking and gene transfer studies in canines,7-9 in nonhuman primates,10-12 and in humans13-17 demonstrated the possibility to mark LTRCs, using an amphotropic retrovirus vector, although the efficiency was much lower, approaching 1%. This low transduction efficiency is a major obstacle to successful treatment of hematopoietic disease by gene therapy.
Transduction of any cell with either a Moloney murine leukemia virus (MLV) or a lentivirus involves at least 3 steps.1,18 With either virus, the first step involves binding of the retrovirus particles to its specific receptor at the cell surface and entry into the cytoplasm. We have shown that low levels of amphotropic retrovirus receptor on HSCs are one important obstacle to transduction.19,20 We and others have shown that retroviruses pseudotyped with a vesicular stomatitis virus G (VSV-G) protein transduce HSCs more efficiently than amphotropic retroviruses.21 22
After entry into the cell, the RNA genome is reverse transcribed into double-stranded DNA. This step requires that the target cell has adequate pools of deoxynucleotides available for DNA synthesis by reverse transcriptase carried in with the RNA genome.23MLV and lentivirus differ at the third step. Double-stranded MLV DNA can only become integrated into the host cell genome after the nuclear membrane breaks down prior to cell division.18,24 One of the major limitations to MLV-mediated gene transfer of HSCs is that HSCs are quiescent, with less than 5% of HSCs in the S, G2, or M phases of the cell cycle.25-31 In contrast, double-stranded lentivirus DNA is transported to the nucleus where it can integrate into the genome without the requirement of cell division.32-34 Numerous studies23,33-37 have demonstrated the superiority of lentivirus over MLV vectors for transduction of human CD34+ cells. Other studies have demonstrated that nondividing mouse, rat, and human liver, brain, or muscle cells38-40 and growth-arrested human and mouse cell lines are efficiently transduced with lentiviral vectors.32,41 42
In this study, we used a co-transduction assay to compare transduction of mouse HSCs with MLV and lentivirus vectors pseudotyped with the VSV-G envelope. This assay makes use of an ecotropic retrovirus as an internal control to demonstrate that HSCs are capable of converting retrovirus RNA to DNA and to assess HSC cycle status through ecotropic retrovirus integration. We report that exposure of HSCs to MLV or lentivirus vectors for 24 hours without cytokines did not result in transduction. In contrast, exposure of HSCs to MLV and lentivirus vectors for 96 hours with cytokines resulted in equivalent transduction. We conclude from this experiment that a large proportion of HSCs are refractory to lentivirus as well as MLV-mediated transduction.
Materials and methods
Mice
All mice were purchased from The Jackson Laboratories (Bar Harbor, ME). Bone marrow (BM) from young adult (5 to 8 weeks old) female C57BL/6J (homozygous Hbbs/Hbbs“single” hemoglobin) mice was used for HSC purification and transduction. Genetically anemic WBB6F1-W/Wv (abbreviatedW/Wv, heterozygous Hbbs/Hbbd “single/diffuse” hemoglobin) mice were used as recipients in all experiments. The National Institutes of Health is fully accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care.
Enrichment of mouse HSCs
BM cells were flushed from the femurs and tibias of the mice into phosphate-buffered saline (PBS), containing 5% fetal calf serum (FCS) (Hyclone). Cells expressing lineage markers were identified, using a mixture of rat monoclonal antibodies directed against the following murine hematopoietic cell markers: B lymphocytes (B220), T lymphocytes (CD4 and CD8), granulocytes (Gr-1), monocytes (Mac-1; all purchased from Caltag Laboratories, San Francisco, CA), and erythroid cells (TER-119; purchased from Pharmingen). Lin+ cells were removed, using goat-anti-rat magnetic immunobeads (Biomag, Framingham, MA). The lineage depletion removed more than 90% of total BM cells.
Lentiviral and retroviral vector construction and production
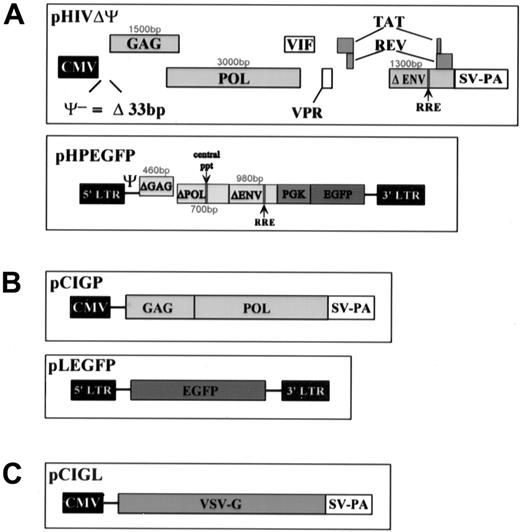
The packaging plasmid, pHIVΔΨ contains the sequence of the human immunodeficiency virus 1 (HIV-1) NL4-3 isolate with deletions of (1) both LTRs, (2) 33 base pairs (bp) of the packaging signal (Ψ) 5′ to the gag gene, (3) 1587 bp of the env gene, (4) the vpu gene, and (5) the nef gene. All the other genes are unaffected. Transcription of the HIV genes is under the control of the cytomegalovirus (CMV)60promoter, derived from the pCI vector (Promega, WI). pHIVΔΨ is a modification of pHIV-PV.43 The transfer vector, pHLPEGFP contains sequences from the HIV-1 NL4-3 isolate, including (1) both LTRs; (2) 1251 bp of the 5′ end of gag; (3) 715 bp of the 3′ end of pol, which contains the central polypurine tract and transcriptional enhancer sequences; and (5) 977 bp of env, containing the REV response element and the second exon of tat. The nef and rev coding sequences are disrupted by the insertion of the PGK promoter and the EGFP marker gene (Clontech, CA). All of the HIV-based vectors are modifications of pHIV-AP G−P−E−F−V−.43The murine retroviral system used for comparison consists of the packaging construct, pCMV-gp,44 that contains the sequence encoding gag and pol from Mo-MLV under the control of the CMV promoter from pCI, and the vector, pLEGFP, that is a derivative of pLN45 with the neo gene replaced by EGFP. The envelope used for pseudotyping in this system is expressed by pCIGL, which contains the VSV-G gene46 47 under the control of the CMV promoter from pCI (Figure 1).
Representation of the VSV-G pseudotyped lentivirus and MLV used in the experiments.
(A) Top: HIV-based packaging construct, bottom: HIV-based vector. (B) Top: MLV-based packaging vector, bottom: MLV-based vector. (C) Envelope construct.
Representation of the VSV-G pseudotyped lentivirus and MLV used in the experiments.
(A) Top: HIV-based packaging construct, bottom: HIV-based vector. (B) Top: MLV-based packaging vector, bottom: MLV-based vector. (C) Envelope construct.
To generate viral supernatants, 293T cells (1 × 107) were plated in a 10-cm dish and transfected with 3 constructs (10 μg packaging construct, 5 μg envelope construct, and 20 μg vector), by Ca2PO4 precipitation (Clontech, CA). The transfection supernatant was collected after 24, 48, and 72 hours, pooled, and filtered through a 0.45-μm filter.
The ecotropic retrovirus was obtained from ψ-CRE MFG-LacZ producer cell lines. The cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% newborn calf serum and heat inactivated for 1 hour at 56°C. For transduction, 90% confluent plates of the producer cell lines were grown overnight in DMEM supplemented with 15% FCS (Hyclone, Logan, UT), heat inactivated 1 hour at 56°C with 4 mmol/L glutamine, 50 mg/mL penicillin, and 50 U/mL streptomycin. The supernatant was then harvested for infection of BM cells. The titer of the producer cell line is known to be approximately 5 × 105 infectious particles/mL.20 The titer of both MMLV VSV-G and lentivirus VSV-G was evaluated by slot blot analysis of RNA extracted from virus-containing medium versus supernatant that contained a known number of infectious particles per milliliter and by transduction of NIH-3T3 cells and 293T cells with serial dilutions of each supernatant.
Transduction of cell lines
Both the MLV-VSV-G and the lentivirus VSV-G supernatant were tested against HeLa and 3T3 cells actively growing or growth inhibited by contact and serum deprivation or aphidicolin (Sigma, St. Louis, MO). Cells were plated in a 75-cm2 tissue culture flask (Corning, Corning, NY) in 10 mL of improved minimum essential medium (IMEM; Gibco, Gaithersburg, MD) supplemented with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) and heat inactivated for 1 hour at 56°C.
Growth inhibition.
Cells were grown to confluence and serum deprived with 0.1% FCS and ITS liquid media supplement 100× (Sigma, St. Louis, MO) in IMEM for 72 hours before transduction. After transduction, the cells were maintained under the same conditions for 72 hours before analysis.
Aphidicolin.
Cells were grown for 48 hours then cultured in IMEM–10% FCS supplemented with aphidicolin 15 μg/mL for 24 hours before transduction, during transduction, and for 72 hours before analysis.
Actively growing cells.
Cells were plated for 48 hours before transduction and analyzed 72 hours after transduction. All transductions were done by placing a 1:5 dilution (with IMEM ± FCS, ± aphidicolin) of the MLV-GFP or the lentivirus-GFP supernatant over the cells for 24 hours. Transduction efficiency was measured by flow cytometry analysis, using a FACScan (Becton Dickinson, San Jose, CA) for GFP expression and Southern blot analysis for GFP gene integration by standard technique.48Cell cycle analysis was made for each cell line at the moment of transduction under the conditions as cited above, using a NuCycl-PI kit (Exalpha, Boston, MA) according to the manufacturer specifications.
Co-transduction of mouse Lin− BM
Lin− mouse BM cells were plated at a density of 1 to 2 × 105 cells/mL in suspension culture plates (Corning, NY). Transduction medium consisted of lentivirus VSV-G containing supernatant or MMLV VSV-G containing supernatant diluted 1:10 (experiments 1 and 2) and 1:5 in ecotropic MMLV supernatant and 6 μg/mL polybrene (Sigma, St. Louis, MO). One portion of cells was transduced for 24 hours in the absence of cytokines with a supernatant change after 12 hours, the remaining cells were transduced for 96 hours in medium supplemented with cytokines, 100 ng/mL rat stem cell factor (SCF), 50 ng/mL human interleukin 6 (IL-6; both provided by Amgen, Thousand Oaks, CA), and 10 ng/mL murine IL-3 (PeproTech, Rocky Hill, NJ). The medium was changed every 24 hours. After transduction, the cells were transplanted into W/Wvrecipients by tail vein injection. Repopulation of the recipients with donor cells was demonstrated by cellulose acetate electrophoresis analysis of hemoglobin from peripheral blood.49 At 16 weeks posttransplantation, DNA was extracted from 300 μL of peripheral blood collected by retro-orbital venous sinus puncture. Polymerase chain reaction (PCR) was performed on 400 ng of DNA, using reagents and protocols obtained from the manufacturer (Perkin Elmer). Primers were as follows: mouse β-globin (sense primer, 5′GAA GTT GGG TGC TTG GAG AC3′; anti-sense primer, 5′GAG ACT GCT CCC TAG AAT CG3′; fragment size 401 bp; annealing 56°C), LacZ (sense primer, 5′GCC GAC ACC AGA CTA AGA AC3′; anti-sense primer, 5′CCT CTT CGC TAT TAC GCC AG3′; fragment size 289 bp, annealing 66°C), and EGFP (sense primer, 5′CCA TGT GAT CGC GCT GCT TCT CG3′; anti-sense primer, 3′GGC CAC AAG TTC AGC GTG TC3′; fragment size 586 bp, annealing 68°C). Thirty-five cycles were performed for all samples at 94°C for 30 seconds (denaturing), annealing for 30 seconds at primer-specific temperature, and at 72°C for 30 seconds (extension), using a GeneAmp PCR System 9700 (Perkin Elmer). The PCR products were resolved on a 5% polyacrylamide gel.
Transduction was also analyzed by flow cytometry. Peripheral blood (50 μL) was collected by retro-orbital sinus puncture, and red cells were lysed, using ACK Lysing Buffer (BioWhittaker, Walkersville, MD). White cells were resuspended in PBS (Biofluids, Rockville MD) and supplemented with 5% FCS. The cells were then analyzed for green fluorescence, using FACSvantage (Becton Dickinson, San Jose, CA). Dead cells were excluded, using propidium iodide stain, and negative green fluorescence was set at less than 1%, using cells from a naive mouse. Transduction of different lineages was also evaluated by staining the cells with CD3-phycoerythrin (PE), B-220-PE, Mac-1-PE (PharMingen, San Diego, CA). Cells were incubated 30 minutes at 4°C with the antibody at 0.5 μg/106 cells/100 μL. After staining, the cells were washed once and resuspended in DMEM–5% FCS for analysis.
DNA was extracted from the BM, spleen, and thymus of repopulated animals. Southern blot analysis using standard methods48was performed on DNA digested with Sst 1 (Gibco BRL, Gaithersburg, MD) to estimate the number of integrated provirus. Stem cell transduction was demonstrated by insertion-site analysis of DNA digested with Nco1. The filters were probed with a32P-labeled probe for GFP.
Results
Retrovirus titering
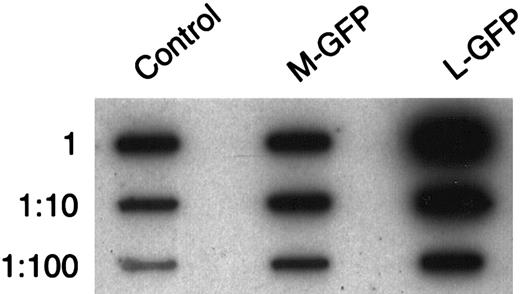
Slot blot analysis of the MLV-VSV-G-EGFP (M-GFP) and lentivirus VSV-G-EGFP (L-GFP) was performed on stocks used in each experiment and compared to slot blot analysis of an ecotropic-GFP retrovirus with a titer at approximately 5 × 106 virus particles/mL (evaluated by NIH-3T3 cell transduction, data not shown). The titers of the MLV and the lentivirus supernatants in each experiment were evaluated to be at least 5 × 107 viral particles/mL, the L-GFP titer being somewhat higher than M-GFP (Figure2). Evaluation of M-GFP and L-GFP titers by transduction of NIH-3T3 cells and 293T cells with serial dilutions of each supernatant gave an estimated titer of more than 107 viral particles/mL for both (data not shown), the titers of L-GFP being slightly higher than M-GFP as demonstrated by the slot blot analysis. In the first 2 murine HSC transduction experiments, the cells were exposed to a multiplicity of infection (MOI) of 5 to 10. In the last experiment, the MOI was 10 to 20.
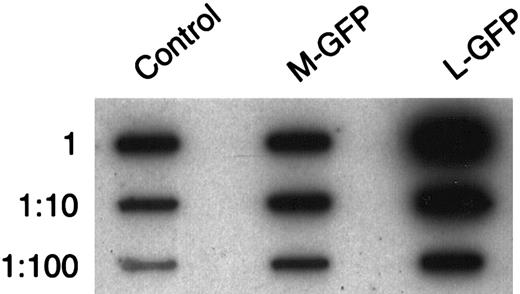
Representative slot blot analysis of the retroviral titers of the supernatant for the M-GFP and L-GFP vectors used in our co-transduction assay.
Equal volumes of each supernatant were taken, viral RNA was extracted, and serial 10-fold dilutions were analyzed by slot blot analysis, using a GFP probe. The titers were compared with a control supernatant from an Ecotropic-GFP-MLV producer cell line of which the titer was known.
Representative slot blot analysis of the retroviral titers of the supernatant for the M-GFP and L-GFP vectors used in our co-transduction assay.
Equal volumes of each supernatant were taken, viral RNA was extracted, and serial 10-fold dilutions were analyzed by slot blot analysis, using a GFP probe. The titers were compared with a control supernatant from an Ecotropic-GFP-MLV producer cell line of which the titer was known.
Transduction of cell lines with MLV and lentivirus vectors
Actively dividing or growth-inhibited HeLa and NIH-3T3 cells were transduced with equivalent amounts of M-GFP and L-GFP retrovirus particles and assayed by fluorescence-activated cell sorter (FACS) and Southern blot analyses for GFP expression and proviral integration. Of actively dividing HeLa and 3T3 cells transduced with the M-GFP, 79% and 34%, respectively, expressed GFP. When transduced with L-GFP, 60% and 24%, respectively, expressed GFP. Of contact-inhibited and serum-deprived HeLa and 3T3 cells transduced with M-GFP, 7.4% and 5.4%, respectively, expressed GFP, whereas 39% and 42% of the same cells transduced with L-GFP, respectively, expressed GFP. Of HeLa and NIH-3T3 cells transduced with M-GFP while growth inhibited with aphidicolin, 1.8% and 0.2% expressed GFP, whereas 13.5% and 11.5% of the same cells transduced with L-GFP expressed GFP. Southern blot analysis of DNA extracted from the cells showed compatible results (Figure 3 and Table1). These results are consistent with previous findings, demonstrating that MLV and lentivirus vectors are equally efficient at transducing dividing cell lines from either mouse or human, but that lentivirus vectors are more efficient at transducing growth-arrested cell lines than MLV vectors. Cell cycle analysis demonstrated that there was at least a 2-fold decrease in the S/G2/M fraction for both 3T3 and HeLa cells under contact inhibition or aphidicolin (data not shown). This incomplete inhibition of the cell cycle may explain why some MLV-mediated transduction in the growth-inhibited cells is observed.
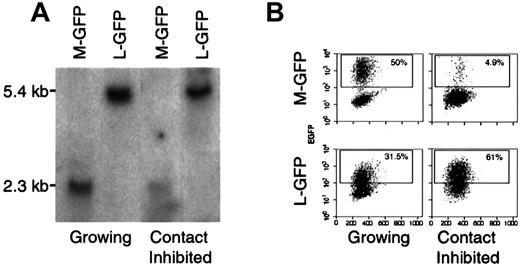
Lentivirus vectors transduce growth-inhibited cell lines more efficiently than MLV vectors.
(A) Representative Southern blot analysis of DNA extracted from 3T3 transduced while growing or while contact inhibited with either a MLV-GFP-VSV-G (M-GFP) or a lentivirus-GFP-VSV-G (L-GFP). After transduction, the cells were either allowed to grow or growth inhibited for 72 hours before analysis. DNA was digested with Sst, which cuts twice in each vector and yields a 2.3-kb fragment for M-GFP and a 5.4-kb fragment for L-GFP. The membrane was probed with a P32-labeled probe. (B) FACS analysis of the same cells as in A. Untransduced 3T3 cells were used as a negative control, the gate was set at less than 0.1% positive. Dead cells were excluded by propidium iodide staining.
Lentivirus vectors transduce growth-inhibited cell lines more efficiently than MLV vectors.
(A) Representative Southern blot analysis of DNA extracted from 3T3 transduced while growing or while contact inhibited with either a MLV-GFP-VSV-G (M-GFP) or a lentivirus-GFP-VSV-G (L-GFP). After transduction, the cells were either allowed to grow or growth inhibited for 72 hours before analysis. DNA was digested with Sst, which cuts twice in each vector and yields a 2.3-kb fragment for M-GFP and a 5.4-kb fragment for L-GFP. The membrane was probed with a P32-labeled probe. (B) FACS analysis of the same cells as in A. Untransduced 3T3 cells were used as a negative control, the gate was set at less than 0.1% positive. Dead cells were excluded by propidium iodide staining.
Transduction of mouse HSCs with MLV and lentiviral vectors
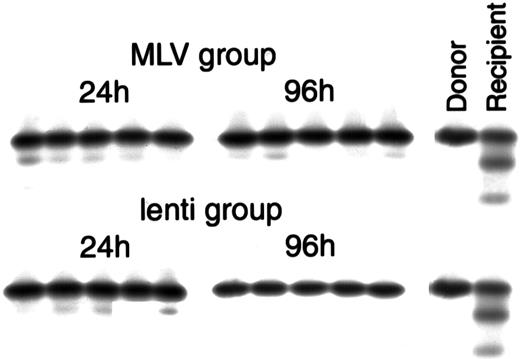
To compare transduction of mouse HSCs by lentivirus vectors to transduction with MLV vectors, we used a co-transduction assay in which each group of cells was transduced with a LacZ ecotropic MLV vector as a control. This internal control is used to establish that the HSCs can convert viral RNA to DNA. Furthermore, because the ecotropic virus particles contain an MLV vector, ecotropic transduction indicates that some HSCs are dividing. The co-transduction was performed with the MLV eco-LACZ and either the VSV-G M-GFP (MLV group) or VSV-G L-GFP (lentivirus group). Two different transduction protocols were tested on lineage-negative BM cells. In the first protocol, Lin−cells were cultured for 24 hours in virus-containing medium without cytokines with a medium change after 12 hours. In the second protocol, Lin− cells were cultured for 96 hours in virus-containing medium supplemented with IL-3, IL-6, and SCF, with a medium change every 24 hours. Following transduction, the cells were collected and injected in W/Wv recipients. Cellulose acetate hemoglobin electrophoresis performed 5 to 6 weeks after transplant demonstrated conversion of all the transplanted animals (heterozygous Hbbs/Hbbd“single/diffuse” hemoglobin) to the phenotype of the donor animals (homozygous Hbbs/Hbbs “single” hemoglobin), indicating repopulation by donor cells (Figure4).
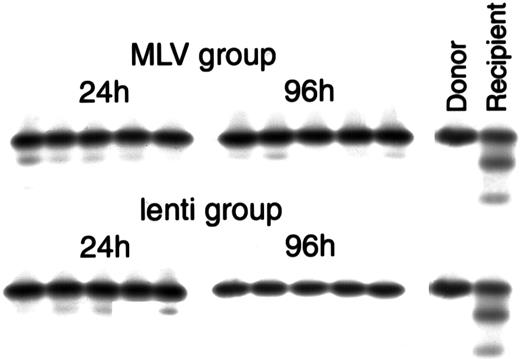
Cellulose acetate hemoglobin electrophoresis of peripheral blood collected from the recipients 5 to 6 weeks after transplant, showing repopulation with donor cells.
The MLV group represents animals transplanted with cells co-transduced with eco-MLV-LacZ and M-GFP, the lenti group represents animals transplanted with cells co-transduced with eco-MLV-LacZ and L-GFP. Twenty-four hours and 96 hours represent the 2 different transduction protocols. The two left lanes represent controls: donor C57BL/6J (homozygous Hbbs/Hbbs “single” hemoglobin) and untransplanted W/Wv(heterozygous Hbbs/Hbbd “single/diffuse” hemoglobin).
Cellulose acetate hemoglobin electrophoresis of peripheral blood collected from the recipients 5 to 6 weeks after transplant, showing repopulation with donor cells.
The MLV group represents animals transplanted with cells co-transduced with eco-MLV-LacZ and M-GFP, the lenti group represents animals transplanted with cells co-transduced with eco-MLV-LacZ and L-GFP. Twenty-four hours and 96 hours represent the 2 different transduction protocols. The two left lanes represent controls: donor C57BL/6J (homozygous Hbbs/Hbbs “single” hemoglobin) and untransplanted W/Wv(heterozygous Hbbs/Hbbd “single/diffuse” hemoglobin).
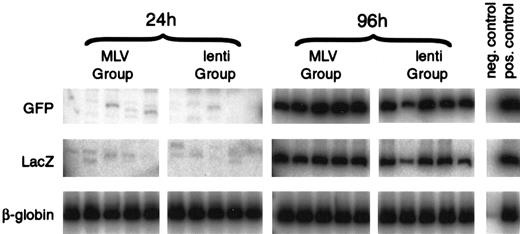
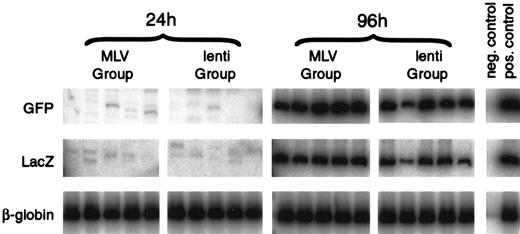
Transduction was analyzed using a PCR assay of DNA extracted from peripheral blood of the transplanted animals 13 to 16 weeks after transplant. Analysis of animals injected with cells transduced for 24 hours without cytokines revealed that the ecotropic proviral sequences were not detectable in any of the animals (0 of 30) from either the MLV or lentivirus groups. The M-GFP proviral sequences were detected in 1 of 15 animals and the L-GFP sequences were detected in 1 of 15 animals, both at low levels (Figure 5 and Table2). Analysis of animals injected with cells transduced for 96 hours in the presence of cytokines revealed that the ecotropic proviral sequences were detectable in 12 of 15 animals from the MLV group and in 14 of 15 animals from the lentivirus group, indicating that some of the HSCs were capable of converting viral RNA to DNA and were dividing. M-GFP proviral sequences were detected in 10 of 15 animals, and L-GFP proviral sequences were detected in 10 of 15 animals (Figure 5 and Table 2), indicating equivalent transduction with both MLV and lentivirus vectors under these conditions. The PCR reaction was validated by analyzing serial dilution of DNA extracted from both a LacZ retrovirus producer cell line and a GFP with a known copy number. Both were sensitive to 1/1000 copy number and in a linear range up to at least 1/10 copy number. Each analysis was run in parallel with DNA samples, containing 0.001 or 0.005, 0.05, and 0.1 copies/cell. The negative animals all had less than 0.001 copies/cell. In the first 2 experiments, the positive animals contained approximately 0.01 copies/cell. In the third experiment, the positive animals contained approximately 0.1 copies/cell for M-GFP–mediated transduction and approximately 0.05 copies/cell for L-GFP–mediated transduction.
Lentivirus and MLV vectors transduce HSCs with similar efficiency.
Representative co-transduction assay of Lin− mouse BM cells co-transduced with MLV-GFP-VSV-G (M-GFP) and ecotropic-MLV-LacZ (MLV group) or lentivirus-GFP-VSV-G (L-GFP) and ecotropic-MLV-LacZ (lenti group). Lin− mouse BM cells were transduced for 24 hours without cytokines with medium changed at 0 and 12 hours or for 96 hours in IL-3, IL-6, and SCF with medium changed every 24 hours. After transduction, the cells were injected intoW/Wv recipients. Peripheral blood DNA from the recipients was analyzed 16 weeks after transplant for the presence of the proviruses by PCR, using GFP- and LacZ-specific primers. PCR of the VSV-G-EGFP provirus generates a 586-bp product, and PCR of the ecotropic-lacZ provirus generates a 289-bp product. DNA from the producer cell lines was used as a positive control, and DNA from an untransplanted mouse was used as a negative control. The mouse adult β-globin gene was amplified as an internal control. Phosphor Imager analysis confirmed that each sample had equivalent amounts of DNA.
Lentivirus and MLV vectors transduce HSCs with similar efficiency.
Representative co-transduction assay of Lin− mouse BM cells co-transduced with MLV-GFP-VSV-G (M-GFP) and ecotropic-MLV-LacZ (MLV group) or lentivirus-GFP-VSV-G (L-GFP) and ecotropic-MLV-LacZ (lenti group). Lin− mouse BM cells were transduced for 24 hours without cytokines with medium changed at 0 and 12 hours or for 96 hours in IL-3, IL-6, and SCF with medium changed every 24 hours. After transduction, the cells were injected intoW/Wv recipients. Peripheral blood DNA from the recipients was analyzed 16 weeks after transplant for the presence of the proviruses by PCR, using GFP- and LacZ-specific primers. PCR of the VSV-G-EGFP provirus generates a 586-bp product, and PCR of the ecotropic-lacZ provirus generates a 289-bp product. DNA from the producer cell lines was used as a positive control, and DNA from an untransplanted mouse was used as a negative control. The mouse adult β-globin gene was amplified as an internal control. Phosphor Imager analysis confirmed that each sample had equivalent amounts of DNA.
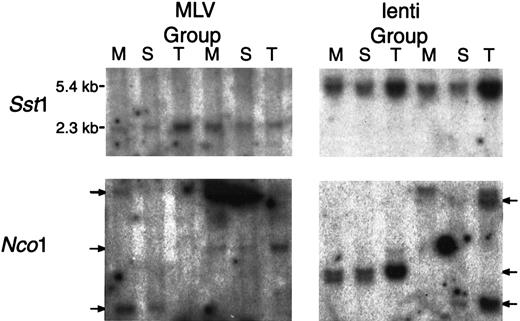
DNA was extracted from BM, spleen, and thymus of 2 mice in both the 96-hour M-GFP–transduced and L-GFP–transduced groups. Southern blot analysis was performed on DNA digested with Sst1 and probed with a GFP probe. The M-GFP provirus yields a 2.3-kilobase (kb) fragment and the L-GFP provirus yields a 5.4-kb fragment. The GFP gene was detectable in the BM, spleen, and thymus of all the animals analyzed (Figure 6). For insertion-site analysis, Southern blot analyses were performed also on DNA digested with Nco1, which cuts once in both proviruses, and probed with a GFP probe. Identical cell-virus junction fragments were detected in the bone, spleen, and thymus of animals in both groups. Although the cell population obtained from the BM, spleen, and thymus do not represent pure populations, this result indicates a likely transduction of stem cells by both M-GFP and L-GFP (Figure 6).
Both lentivirus and MLV vectors transduce stem cells.
Southern blot analysis of DNA extracted from the BM (M), spleen (S), and thymus (T) of animals was transplanted with cells transduced for 96 hours in cytokines with M-GFP (MLV group) and L-GFP (lenti group). DNA was collected 16 weeks after transplant and digested withSst1, which cuts twice in each vector, producing a 2.3-kb fragment for M-GFP and a 5.4-kb fragment for L-GFP, andNco1, which cuts once in each vector, for insertion-site analysis. The membranes were probed with a P32-labeled GFP probe.
Both lentivirus and MLV vectors transduce stem cells.
Southern blot analysis of DNA extracted from the BM (M), spleen (S), and thymus (T) of animals was transplanted with cells transduced for 96 hours in cytokines with M-GFP (MLV group) and L-GFP (lenti group). DNA was collected 16 weeks after transplant and digested withSst1, which cuts twice in each vector, producing a 2.3-kb fragment for M-GFP and a 5.4-kb fragment for L-GFP, andNco1, which cuts once in each vector, for insertion-site analysis. The membranes were probed with a P32-labeled GFP probe.
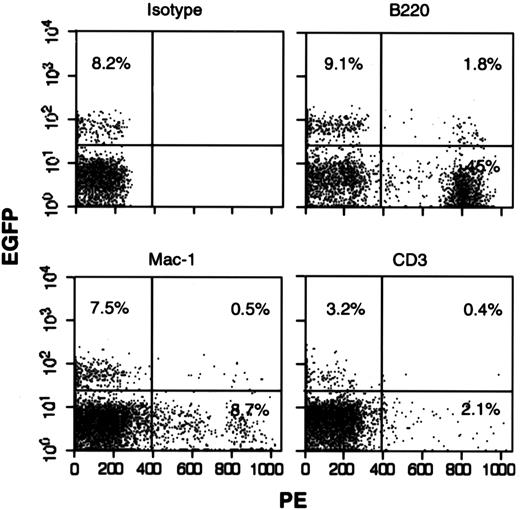
GFP expression in peripheral blood of the animals was analyzed by FACS. A naive mouse was used as a negative control, and dead cells were excluded by propidium iodide staining. In 2 experiments, 6 weeks after transplant, GFP could not be detected in the peripheral blood of either M-GFP– or L-GFP–transduced mice, both in the 24-hour or in the 96-hour protocol. In the third experiments, GFP could not be detected in the animals transplanted with cells transduced at 24 hours. In the animals transplanted with cells transduced at 96 hours with M-GFP, 6 weeks after transplant, GFP was detectable in all animals at an average of 3.4% of positive cells (range 1.17-6.09; data not shown). Sixteen weeks after transplant, GFP was no longer detectable. In the animals transplanted with cells transduced at 96 hours with L-GFP, 6 weeks after transplant, GFP was detectable in all the animals at an average of 2.11% (range 1.09-2.99). Sixteen weeks after transplant, GFP expression was still detectable in 2 animals (levels 3.4% and 7.9%). Lineage-specific expression was analyzed by staining the cells with CD3-PE, B220-PE, and Mac-1–PE. Double-positive cells (GFP and PE) were observed for all lineages analyzed (Figure7). For animals transduced with M-GFP, although most have approximately 0.1 copies/cell in the peripheral blood, the fact that no GFP is detectable by FACS implies silencing of the GFP gene. There was also some silencing in the animals transduced with L-GFP, although at a lower level, which leads us to believe that a gene driven by a retroviral LTR is more prone to silencing in mouse HSCs than a gene driven by a pgk promoter.
GFP expression in multiple lineages.
FACS analysis of 1 mouse transplanted with cells transduced with the lentivirus-GFP in our 96-hour with cytokines protocol. Peripheral blood was collected 16 weeks after transplant and stained with an isotype-PE, B-220-PE, Mac-1-PE, or CD3-PE. Gates were set for GFP, using an untransplanted mouse and for lineage by isotype staining. Dead cells were excluded with propidium iodide.
GFP expression in multiple lineages.
FACS analysis of 1 mouse transplanted with cells transduced with the lentivirus-GFP in our 96-hour with cytokines protocol. Peripheral blood was collected 16 weeks after transplant and stained with an isotype-PE, B-220-PE, Mac-1-PE, or CD3-PE. Gates were set for GFP, using an untransplanted mouse and for lineage by isotype staining. Dead cells were excluded with propidium iodide.
Discussion
A consistent observation in human and large-animal gene transfer studies has been the inefficient transduction of hematopoietic stem cells with MLV vectors10,11,14 50 The low transduction efficiency has been attributed to different factors: low retrovirus titer, lack of integration of the retrovirus into the genome because of quiescence of the stem cells, or insufficient number of retrovirus receptors on the cell surface. Lentivirus vectors have been shown to transduce nondividing cells, leading to the hypothesis that lentivirus vectors should transduce HSCs at higher frequencies than MLV vectors.
Our results, comparing MLV and lentivirus transduction of growth-arrested mouse and human cells lines, are consistent with a large body of published work, demonstrating superior transduction of nondividing cells by lentivirus vectors as demonstrated, for example, by Mochizuki et al39 and by Naldini et al.41Mochizuki et al39 found that on contact-inhibited human skin fibroblast and rat cerebellar neurons, lentivirus vectors transduced 20% to 30% of the cells while MLV transduction was nearly undetectable. Naldini et al41 found that on growth-arrested HeLa cells, lentivirus vectors transduced 70% to 90% of the cells and MLV vectors only of 5% to 8%. On rat fibroblast lentivirus vectors transduced 17% to 50% of the cells and MLV vectors 1% to 11%.
Our results, comparing MLV- and lentivirus-transducing mouse HSCs, indicate that HSC transduction with lentivirus occurs under conditions that also promote MLV transduction. In contrast to growth-arrested cells lines that are mostly in G1/S (in aphidicolin)51 or G2(radiation),52 HSCs are primarily in G0.31,53,54 We have omitted 5-fluorouracil treatment of donor cells and fibronectin from our transduction protocol because these transduction conditions are postulated to recruit or signal HSCs into cell cycle. We hypothesize that resting G0HSCs are resistant to both MLV and lentivirus transduction. Support for this hypothesis can be found in related work with other systems. Naldini et al41 observed that cells lines maintained in G0 for longer periods were transduced by lentivirus vectors with a lower efficiency. Park et al,55 in an in vivo experiment of mouse liver cell transduction, observed decreased lentivirus-mediated transduction of noncycling hepatocytes. Our results are consistent with failure of G0 cells to either reverse transcribe viral RNA23,56,57 or to allow integration into the genome.58
Other studies33-35,37,43 have examined transduction of CD34+ cells, whether in vitro or in severe combined immunodeficiency (NOD/SCID) repopulation assay. All of these studies have demonstrated better transduction with lentivirus vectors compared with MLV. Our results differ from those results. One explanation may be that we have focused on the transduction of LTRCs, as opposed to the transduction of a mixture of LTRCs and progenitor cells. Our model of mouse transplantation may be identifying only a subset of stem and progenitor cells capable of long-term repopulation. We believe that our mouse model is a closer model to human HSCs than the NOD/SCID model. It is well demonstrated that lentiviruses transduce SCID repopulating cells (SRCs) efficiently (up to 30%),34 but there is no data regarding human or nonhuman primate HSC transduction using a lentivirus vector. However, some investigators have demonstrated high transduction efficiency of SRCs in NOD/SCID models using an amphotropic MLV (up to 30%).59 In our mouse model, the transduction efficiency of mouse HSCs using an amphotropic retrovirus was very low,19,22 just like the one observed in human trials.14
We have previously demonstrated that a VSV-G–pseudotyped MLV was as efficient at transducing mouse HSCs as an ecotropic retrovirus.22 In the present study, both MLV and lentivirus are almost as efficient as the ecotropic MLV control for transduction of mouse HSCs in dividing cells, which is an improvement over what was seen with an amphotropic-pseudotyped MLV in the same model. It is possible that lentivirus vectors may perform better in human HSCs than in mouse HSCs. However, on the basis of similarity of MLV and lentivirus transduction of mouse and human cell lines, as well as the transduction of primary mouse and rat liver and neuronal cells, we favor the explanation that integration of lentivirus vectors into mouse and human HSCs occurs under the same conditions.
It has been shown that the transduction efficiency of human CD34+ cells using a VSV-G–pseudotyped lentivirus vector increased significantly with the MOI, up to an MOI of 1000.23 In contrast, Miyoshi et al34 did not see a significant increase in transduction efficiency when increasing the MOI from 60 to 300. Also, Rebel et al60 were able to transduce 25% of the SRCs, using a VSV-G MLV with a high MOI (130-260). The MOI used here was low (5-10). Doubling the MOI increased the transduction efficiency for both the MLV and the lentivirus vectors. On the basis of these results and the literature findings, we have no reason to believe that a further increase in the MOI would advantage the lentivirus vector over the MLV vector.
Our packaging plasmid is capable of expressing the HIV-1 vpr accessory gene, which has been shown to be cytostatic, or even of inducing apoptosis in target cells.61 Because the lentivirus vector was able to transduce prestimulated mouse HSCs, we do not believe this is the reason for the low transduction efficiency of unstimulated HSCs.
In summary, lentivirus vectors were not able to transduce mouse HSCs without cytokine stimulation, and both lentivirus and MLV vectors performed equally in our mouse co-transduction assay when cells were transduced in cytokine-containing medium. Although cell division may not be necessary for lentivirus vector integration, progression to the G1 stage may be required. Our data suggest that protocols for HSC transduction may continue to depend on cell cycle progression to allow efficient integration of either MLV or lentivirus vectors.
J.L.D. is an employee of Systemix Inc.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David M. Bodine, Hematopoiesis Section, GMBB, NHGRI, 49 Convent Dr, Rm 3A11, NIH, Bethesda, MD 20892-4442; e-mail:tedyaz@nhgri.nih.gov.