Abstract
Inflammation may contribute to the pathogenesis of atherosclerosis. On the basis of previous reports that human atherosclerotic lesions contain α-defensins, a class of cationic proteins released by activated neutrophils, the study was designed to ask whether defensins modulate the binding and catabolism of low-density lipoprotein (LDL) by human vascular cells. The results of the study demonstrated that defensin stimulated the binding of 125I-LDL to cultured human umbilical vein endothelial cells, smooth muscle cells, and fibroblasts approximately 5-fold in a dose-dependent and saturable manner. Defensin and LDL formed stable complexes in solution and on cell surfaces. Stimulation of LDL binding by defensin was not inhibited by antibodies against the LDL-receptor (LDL-R), or by recombinant receptor-associated protein, which blocks binding of ligands to the α2-macroglobulin receptor/LDL-R–related protein and other LDL-R family members. Furthermore, defensin stimulated the binding, endocytosis, and degradation of LDL by fibroblasts lacking LDL-R. Stimulation of LDL degradation by defensin was inhibited approximately 75% by low concentrations of heparin (0.2 units/mL) and was similarly reduced in CHO cells lacking heparan-sulfate–containing proteoglycans. The effect of defensin was substantially increased in cells overexpressing the core protein of the syndecan-1 heparan sulfate proteoglycan. The α-defensins released from activated neutrophils may provide a link between inflammation and atherosclerosis by changing the pattern of LDL catabolism from LDL-R to the less efficient LDL-R–independent, proteoglycan-dependent pathway.
Introduction
Atherosclerosis is a multifactorial disorder that develops over decades under the influence of both genetic and environmental factors. Retention of low-density lipoproteins (LDL) within the walls of blood vessels is a pathognomonic feature and perhaps the central cause of the disease.1 In accordance with this, elevated plasma levels of LDL resulting from a deficiency in the expression of LDL-receptor (LDL-R),2 mutations in LDL that impair its ability to bind to this receptor,3,4 or mutations in the receptor itself5 are associated with an increased risk of atherosclerotic vascular disease. Failure to endocytose and degrade LDL via the LDL-Rs increases not only its plasma concentration but also its residence time in the vasculature, thereby permitting it to undergo oxidative and nonoxidative (reviewed in Williams and Tabas6) modifications. Modified LDL is taken up by a number of “scavenger” receptors expressed by macrophages and other arterial cell types contributing to the formation of foam cells.3,7 Cytokines and proteolytic enzymes released from foam cells may contribute to smooth muscle cell proliferation, intimal thickening, aneurysm formation, and plaque instability.8
Inflammation within the arterial wall (see Ross8 for review) also contributes to the initiation and propagation of atherosclerotic vascular disease. For example, some atherosclerotic lesions and inflammatory lesions share a number of histologic features.8,9 Persistent vascular infection with certain DNA viruses,10,11 bacteria,12 and chlamydia13; allograft rejection14; and biologically active molecules released from modified LDL6,8may stimulate localized inflammation of the arterial wall, predisposing a person to accelerated lipoprotein retention and atherosclerotic vascular disease. Induction of adhesion receptors for leukocytes on endothelial cells has been implicated in the early stages of the disease.15,16 In addition, elevated levels of acute phase reactants, such as C-reactive protein, fibrinogen, and interleukin-6 correlate with the risk of clinical events and with the response to therapies such as aspirin.17 Furthermore, a correlation between the number of circulating granulocytes and prevalence of atherosclerosis has also been identified in many studies.18-20
Neutrophil-derived (α) defensins may provide one link between inflammation and the development of atherosclerosis. The α-defensins compose a family of closely related peptides that are found primarily in the granules of neutrophils and constitute approximately 5% of the total cellular protein.21 The α-defensins are small (29– to 35–amino acid) cationic peptides characterized by hydrophobic patches that insert in the lipid portion of cell membranes and by 4 arginine residues that are expressed on the opposing molecular surface.22,23 Defensins are incorporated into the cell membrane of prokaryotic organisms during phagocytosis, disrupting ion fluxes and provoking cell lysis.24 During severe infections, defensins are released into the plasma at concentrations approaching 30 μmol/L25,26 and are found at considerably higher concentrations at sites of inflammation.27-29
Importantly, α-defensins promote the accumulation of lipoprotein(a) in the vasculature,30,31 inhibit fibrinolytic activity within clots and on the surface of vascular cells,32,33and are abundant in human atherosclerotic plaques.30,34 Of interest, the overall structure of defensins, with a hydrophobic face and a cationic face, generally resembles many apolipoproteins, such as apoB, apoE, lipoprotein lipase, and hepatic lipase, that mediate the binding of atherogenic lipoproteins to LDL-R family members and to proteoglycans.3-5 35-39 We therefore asked whether α-defensins might also modulate the catabolism of LDL through similar mechanisms.
Materials and methods
Materials
Defensin (human neutrophil peptide 2) and murine monoclonal antidefensin antibodies were prepared, characterized, and iodinated as described;24 our preparation of defensin migrated as a single band of approximately 3 kd on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). LDL was prepared by ultracentrifugation (1.019 < density < 1.63 g/mL) as described.40-42 Aliquots of unlabled LDL were oxidized by incubation for 16 hours at 37°C with 10 μmol/L CuCl2 in the absence of EDTA.43 The extent of oxidation was monitored by measuring thiobarbituric-acid–reactive substances.40 Native (unoxidized) and oxidized LDL were iodinated with the use of iodine monochloride.41Recombinant 39-kd receptor-associated protein (r-RAP) was the kind gift of Dr D. Strickland (Department of Biochemistry, American Red Cross, Rockville, MD).44 Cultures of human umbilical vein endothelial cells (HUVECs) and vascular smooth muscle cells (SMCs) were prepared as described.45 Human skin fibroblasts were obtained from the foreskins of healthy volunteers and patients with homozygous familial hypercholesterolemia having the C660→Stop (single-letter amino acid code) mutation.46 A polyclonal antibody to the human LDL-R was the kind gift of Dr J. Glick (University of Pennsylvania). Chinese hamster ovary (CHO) cells lacking xylosyl transferase (XT−), and therefore incapable of initiating the synthesis of heparan sulfate or chondroitin sulfate chains, and CHO cells lacking glucuronic acid/N-acetylgalactosamine transferase, and therefore selectively deficient in heparan sulfate but overexpressing chondroitin sulfate, were the kind gifts of Dr J. Esko (University of California, San Diego).47 Wild-type CHO cells were from American Type Culture Collection (Rockville, MD). CHO cells overexpressing the human syndecan-1 core protein were generated as described.48
Binding of LDL to cells
Cells grown in 48-well Falcon Multiwell tissue culture plates (Becton Dickinson, Lincoln Park, NJ) in LDL-deficient medium (Bet Haemek, Haemek, Israel) plus 10% fetal calf serum depleted of lipoproteins as described above were studied when they reached approximately 60% to 70% confluency (final density of approximately 5 × 104 cells/well). Cells were chilled to 4°C for 30 minutes and washed twice with cold phosphate-buffered saline (PBS) containing 0.25% bovine serum albumin (BSA). The cells were then incubated for 4 hours at 4°C with 0 to 300 nmol/L125I-LDL (protein Mr approximately 500 kd) alone or in the presence of defensin (0 to 20 μmol/L) in PBS containing 0.25% BSA. Binding was measured in the presence and absence of 10- to 100-fold excess unlabeled LDL added to the incubation medium. Cells were washed 4 times with binding buffer to remove free ligand. Glycine-HCl (50 mmol/L, pH 2.8) was added to elute surface-bound ligand,49 and the cells were solubilized in 1 N NaOH to measure residual cell-associated radioactivity50; the same results were obtained when heparin (10 μg/mL) was used to elute surface-bound LDL in select experiments. Specific (ie, high-affinity, low-capacity) binding was defined as the difference between the binding of LDL/defensin in the presence and the absence of excess LDL. In some experiments, cells were preincubated with defensin (10 μmol/L) for 1 hour at 4°C and washed twice with PBS before incubation with 50 nmol/L 125I-LDL. To examine the involvement of LDL-R, binding of 125I-LDL was measured in the presence of a blocking polyclonal antireceptor or control immunoglobulin or 1 μmol/L r-RAP in the presence of 2 mmol/L Ca++. As a second approach, cells were grown for 48 hours in media containing lipoprotein-depleted serum or the same medium supplemented with LDL to alter the expression of LDL-R and the binding of LDL, and defensin/LDL was measured as above. All experiments were performed in triplicate and were repeated at least 3 times unless otherwise stated. Data are presented as the mean ± standard deviation (SD) of all experiments (n = 9).
Experiments were performed to determine whether LDL underwent oxidation in the presence of defensin under the conditions used to measure cell binding and catabolism. To do so, fibroblasts were incubated with 50 nmol/L unlabeled LDL in PBS containing 0.25% BSA in the presence or absence of 10 μmol/L defensin for 4 hours at 4°C, after which the amount of oxidized LDL in the media was measured.40 This experiment was performed twice, each time in triplicate (n = 6).
Binding of defensin to cells and to LDL
Defensin was iodinated and the specific binding to cells was measured as described previously.30 33 In some experiments, binding was measured in the presence of anti–LDL-R or control immunoglobulin. LDL or BSA (10 nm each) was incubated with125I-defensin (0 to 5 μmol/L) for 1 hour at 37°C. The mixture was then applied to a Bio-Gel P-30 polyacrylamide gel (Bio-Rad, Hercules, CA), and the radioactivity in the excluded volume (ie, MW exceeding approximately 40 kd) was measured. Specific binding of defensin to LDL was defined as the difference between the BSA-associated and LDL-associated radioactivities.
Internalization and degradation of LDL
LDL-R–deficient (FH) fibroblasts were incubated with 100 nmol/L 125I-LDL in serum-free medium in the presence or absence of 10 μmol/L defensin for 2 hours at 4°C to permit surface binding without catabolism. The cells were washed 4 times with PBS-BSA; fresh medium without ligands was then added; and the incubations were continued at 37°C for the indicated times followed by chilling to 4°C. Glycine buffer, pH 3.0, at 4°C was added, and the acid-elutable radioactivity was measured as a marker of cell-surface–associated ligand. The difference between the acid eluted and the residual acid-resistant radioactivity was taken as an estimate of internalized ligand that had not been degraded.50 The cell supernatant was removed, and the trichloroacetic acid (TCA) (10% final concentration) soluble radioactivity was measured. Cell-specific degradation was calculated from the difference between cell-associated degradation and spontaneous degradation of ligand in cell-free wells measured in parallel. TCA-precipitable radioactivity in the media was quantified as an indication of retroendocytosis or desorption from the cell surface. To examine the temperature dependence of binding and degradation, HUVECs were incubated with 125I-LDL (100 nmol/L) and defensin (10 μmol/L) for 3 hours at 4°C. The cells were washed 4 times with PBS-BSA and either kept at 4°C or warmed to 37°C for an additional 4 hours. Glycine, pH 3.0, was added, and the residual cell-associated radioactivity was measured. In other experiments, XT− or wild-type CHO cells were incubated for 18 hours at 37°C with 10 nmol/L 125I-LDL in the presence or absence of 10 μmol/L defensin and the presence or absence of 100-fold molar excess unlabeled LDL, and the specific degradation of labeled ligand was measured as described above.
Results
Defensins stimulate the binding of 125I-LDL to HUVECs
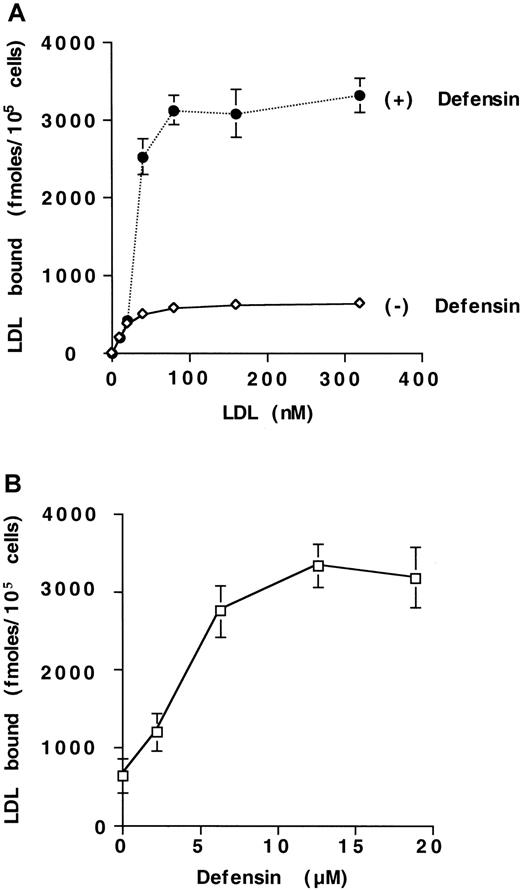
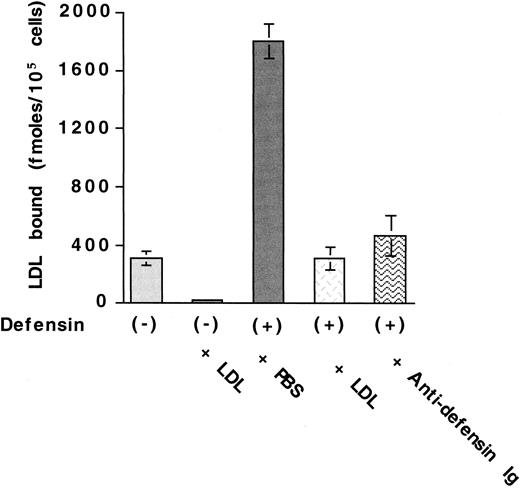
We examined the effect of α-defensin on the interaction of125I-LDL with vascular cells. Defensins stimulate the binding of 125I-LDL to HUVECs (Figure1A). Maximal binding of125I-LDL was increased approximately 5-fold in the presence of 10 μmol/L defensin. with little change in the concentration of125I-LDL required for half maximal binding (22 nmol/L vs 40 nmol/L). Defensin stimulated the binding of 125I-LDL to human SMCs and fibroblasts to a similar extent (not shown). Stimulation of LDL binding by defensin increased approximately 10-fold at 37°C compared with 4°C (not shown) and depended on the concentration of defensin present (Figure 1B). Stimulation was apparent at a defensin concentration of 2.5 μmol/L and reached its half-maximal effect at 4.3 μmol/L, which is within the plasma concentration attained in vivo during severe inflammation.25 26 More than 85% of the binding of 125I-LDL in the presence of defensin was inhibited by excess LDL (Figure2) or by preincubation of125I-LDL/defensin with a monoclonal antidefensin antibody (Figure 2). Incubation of 125I-LDL with defensin under these conditions did not increase lipoprotein oxidation when measured directly.
Defensin stimulates the binding of LDL to HUVECs.
(A) HUVECs were incubated with the indicated concentrations of125I-LDL for 4 hours at 4°C in the presence (+) or absence (−) of 10 μmol/L defensin, and the cell-associated radioactivity was measured. Specific binding was defined as the difference between cell-associated radioactivity in the presence and the absence of 100-fold molar excess unlabeled LDL. (B) HUVECs were incubated with 125I-LDL (50 nmol/L) in the presence of the indicated concentrations of defensin under the experimental conditions described in panel A, and the specific binding was determined. The mean ± SEM of 3 experiments is shown.
Defensin stimulates the binding of LDL to HUVECs.
(A) HUVECs were incubated with the indicated concentrations of125I-LDL for 4 hours at 4°C in the presence (+) or absence (−) of 10 μmol/L defensin, and the cell-associated radioactivity was measured. Specific binding was defined as the difference between cell-associated radioactivity in the presence and the absence of 100-fold molar excess unlabeled LDL. (B) HUVECs were incubated with 125I-LDL (50 nmol/L) in the presence of the indicated concentrations of defensin under the experimental conditions described in panel A, and the specific binding was determined. The mean ± SEM of 3 experiments is shown.
Antidefensin antibody inhibits defensin-induced LDL binding.
125I-LDL (50 nmol/L) was incubated with HUVECs in the presence of buffer, 100-fold molar excess LDL, 10 μmol/L defensin, 10 μmol/L defensin plus 100-fold molar excess unlabeled LDL, or 10 μmol/L defensin plus antidefensin antibody. The mean ± SEM of 3 experiments is shown.
Antidefensin antibody inhibits defensin-induced LDL binding.
125I-LDL (50 nmol/L) was incubated with HUVECs in the presence of buffer, 100-fold molar excess LDL, 10 μmol/L defensin, 10 μmol/L defensin plus 100-fold molar excess unlabeled LDL, or 10 μmol/L defensin plus antidefensin antibody. The mean ± SEM of 3 experiments is shown.
On the basis of these results, we investigated the hypothesis that defensin and LDL form complexes that bind to cells as a unit. To address this possibility, 125I-defensin in varying concentrations was incubated with 10 nmol/L LDL, and the radiolabeled protein that bound to LDL was isolated by size-exclusion chromatography. Defensin bound to LDL in a dose-dependent and saturable manner, first apparent at a defensin concentration of 10 nmol/L (ie, 1:1 molar ratio with LDL) and reaching half-maximal binding at a defensin concentration of approximately 1.2 ± 0.04 μmol/L (mean ± SD; n = 3). Preincubation of HUVECs with saturating concentrations of defensin33 stimulated binding of125I-LDL to the same extent as when both ligands were present simultaneously. This outcome suggests that LDL can bind to cells either as a soluble preformed complex with defensin or directly to cell-associated defensin.
The LDL-R does not mediate defensin-stimulated LDL binding
We next examined the cellular binding sites for defensin and for defensin/LDL complexes. We began by determining if defensin binds to the LDL-R, thereby enhancing the cell-surface binding of LDL through the formation of a bridge between the lipoprotein and the receptor. Consistent with this hypothesis, an anti–LDL-R antibody that blocks LDL binding was able to inhibit the binding of125I-defensin to HUVECs by 43% ± 7% (mean ± SD, n = 3), whereas nonimmune rabbit immunoglobulin (Ig)G did not affect defensin binding. Similarly, the binding of 125I-defensin to FH fibroblasts lacking LDL-R was 39% ± 6% (mean ± SD, n = 3) of binding to wild-type cells. Binding of125I-defensin to FH cells was not inhibited by anti–LDL-R antibody. These data indicate that a substantial fraction of cell-surface binding of defensin depends on LDL-R.
We then directly examined whether defensin enhances the binding of LDL to LDL-R. Several lines of evidence indicated that this was not the case. First, in the presence of defensin, 125I-LDL binds to FH and wild-type fibroblasts to the same extent (Figure3). Second, r-RAP, which inhibits ligand binding of LDL to several LDL-R family members,44inhibited the binding of 125I-LDL to HUVECs by 60 ± 15% (mean ± SD, n = 3) in the absence of defensin, but had no effect on the augmented binding of LDL in the presence of defensin. Third, binding of 125I-LDL to wild-type fibroblasts in the presence of defensin was not inhibited by anti–LDL-R blocking antibody, whereas binding of LDL was inhibited 80% ± 11% (mean ± SD, n = 3); nonimmune rabbit IgG did not affect LDL binding in the presence or absence of defensin. Taken together, these results indicate that neither LDL-R nor LDL-R family members mediate the increased surface binding of LDL in the presence of defensin.
Defensin stimulates the binding of LDL to FH cells.
Wild-type (WT) or LDL-R–deficient fibroblasts (FH) were incubated with 50 nmol/L 125I-LDL in the absence (left side) or presence (right side) of 10 μmol/L defensin and the specific binding was determined as above. The mean ± SEM of 3 experiments is shown.
Defensin stimulates the binding of LDL to FH cells.
Wild-type (WT) or LDL-R–deficient fibroblasts (FH) were incubated with 50 nmol/L 125I-LDL in the absence (left side) or presence (right side) of 10 μmol/L defensin and the specific binding was determined as above. The mean ± SEM of 3 experiments is shown.
Defensin increases the binding and degradation of LDL
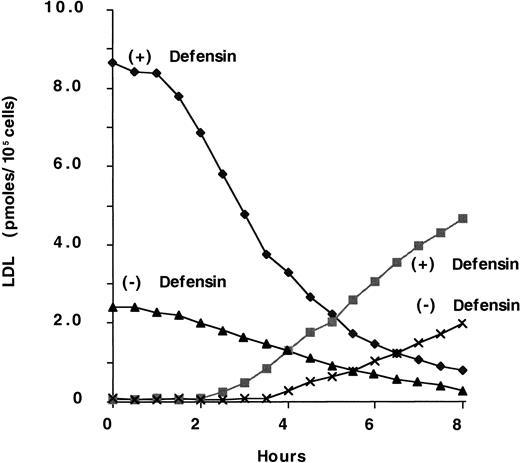
We then examined the effect of defensin on the binding and degradation of 125I-LDL. To do so, we first examined FH cells in view of the finding that defensins stimulate the binding of125I-LDL in an LDL-R–independent manner. Consistent with this finding, defensin augmented the degradation of125I-LDL by FH fibroblasts (Figure4). The t1/2 for disappearance of LDL from the surface in the presence of defensin was approximately 2 to 2.5 hours. For comparison, we examined the internalization of 125I-LDL by wild-type fibroblasts in the presence or absence of defensin. In the presence of defensin, the t1/2 of 125I-LDL internalization was the same as that obtained with FH cells, whereas in the absence of defensin the t1/2 was 9 to 13 minutes (not shown). Thus, the time course of LDL internalization by wild-type and FH fibroblasts was the same in the presence of defensin and was too slow for internalization by LDL-R, which, like other coated-pit receptors, mediates internalization with a t1/2 of 5 to 15 minutes. Rather, the internalization of LDL in the presence of defensin is more consistent with the behavior of ligands bound to syndecan proteoglycans.48 Furthermore, cells grown in the presence or absence of LDL to modulate their expression of LDL-R bound and degraded similar amounts of defensin/LDL (not shown).
Defensin stimulates the binding and degradation of LDL.
FH fibroblasts were incubated with 100 nmol/L 125I-LDL in the presence (+) or absence (−) of 10 μmol/L defensin for 2 hours at 4°C. The cells were washed 4 times with PBS-BSA and then warmed to 37°C for the indicated times. Glycine buffer, pH 3.0, was added, and the cell surface–associated radioactivity in the presence (♦) or absence (▴) of defensin was measured. The cell supernatant was removed and the TCA-soluble radioactivity in the presence (▪) or absence (×) of defensin was measured. The experiment shown is representative of 3 experiments so performed.
Defensin stimulates the binding and degradation of LDL.
FH fibroblasts were incubated with 100 nmol/L 125I-LDL in the presence (+) or absence (−) of 10 μmol/L defensin for 2 hours at 4°C. The cells were washed 4 times with PBS-BSA and then warmed to 37°C for the indicated times. Glycine buffer, pH 3.0, was added, and the cell surface–associated radioactivity in the presence (♦) or absence (▴) of defensin was measured. The cell supernatant was removed and the TCA-soluble radioactivity in the presence (▪) or absence (×) of defensin was measured. The experiment shown is representative of 3 experiments so performed.
Heparan sulfate–containing proteoglycans mediate defensin-stimulated LDL degradation
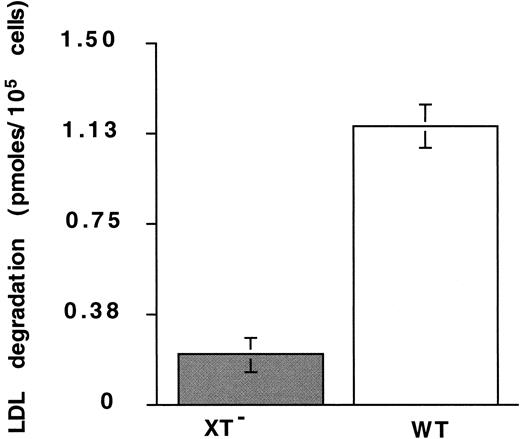
Heparan sulfate–containing proteoglycans (HSPGs) have recently been implicated in the binding and degradation of LDL.6 48Therefore, we investigated the possibility that defensin and/or defensin/LDL complexes bind to and are internalized through HSPGs. Binding of defensin/125I-LDL complexes to HUVECs was inhibited approximately 75% by 0.2 U/mL heparin (data not shown), a concentration well below that required to inhibit LDL binding to LDL-R and that had no effect on LDL binding in absence of defensin. Binding of 125I-defensin to XT−CHO cells, which lack heparan- and chondroitin-sulfate–containing proteoglycans, was reduced 33 ± 11% (mean ± SD; n = 3) relative to wild-type cells. Furthermore, degradation of125I-LDL in the presence of defensin (10 μmol/L) by XT− cells was reduced more than 80% at 4 hours compared with wild-type cells (Figure 5). In addition, degradation of 125I-LDL in the absence of defensin was reduced more than 4-fold in the WT cells, whereas degradation by XT− cells was essentially the same in the presence and absence of defensin (not shown).
Degradation of defensin/LDL: role of proteoglycans.
The degradation of 125I-LDL (10 nm) by XT−cells in the presence of defensin (10 μmol/L) was measured at 4 hours as described in the legend to Figure 4. The mean ± SEM of 3 experiments is shown.
Degradation of defensin/LDL: role of proteoglycans.
The degradation of 125I-LDL (10 nm) by XT−cells in the presence of defensin (10 μmol/L) was measured at 4 hours as described in the legend to Figure 4. The mean ± SEM of 3 experiments is shown.
XT− cells are deficient in chondroitin, dermatan, and heparan sulfate. To identify the specific role of heparan sulfate, we first studied cells selectively deficient in this glycosaminoglycan. Cells lacking heparan sulfate exhibited approximately 70% less degradation of LDL in the presence of defensin than wild-type CHO cells (Figure 6) and a proportionate reduction in cell surface-bound and endocytosed ligand (not shown). Furthermore, cells that overexpress syndecan-1 HSPG exhibited a 30% increase in binding (not shown) and an approximately 50% and 250% increase in the degradation of LDL in the presence of defensin above wild-type cells and heparan sulfate–deficient cells, respectively (Figure 6), indicating a role for this proteoglycan.
Degradation of defensin/LDL: role of heparan sulfate–containing proteoglycans.
Experiments were performed as described in Figure 4. The 4-hour time point is shown. HS (−) refers to CHO cells selectively deficient in heparan sulfate. HS (+) refers to syndecan-1–expressing CHO cells. The mean ± SEM of 3 experiments is shown.
Degradation of defensin/LDL: role of heparan sulfate–containing proteoglycans.
Experiments were performed as described in Figure 4. The 4-hour time point is shown. HS (−) refers to CHO cells selectively deficient in heparan sulfate. HS (+) refers to syndecan-1–expressing CHO cells. The mean ± SEM of 3 experiments is shown.
Discussion
This study demonstrates that α-defensins stimulate the binding of 125I-LDL to vascular endothelial cells, SMCs, and fibroblasts. Defensin increased the maximal binding of125I-LDL, consistent with the generation of new binding sites on the cell surface. Binding of LDL was increased to the same extent when defensin and LDL were added to the cells simultaneously as when the cells were preincubated with defensin followed by addition of125I-LDL. The effect of defensin was dose-dependent and saturable, and its biologic activity was observed at concentrations shown previously to bind to endothelial cells and SMCs30,32,33 and to circulate in plasma during severe inflammation.25
Several lines of evidence indicate that the binding of defensin/LDL to the vascular cells is not mediated via the LDL-R or LDL-R family members.2 First, binding of defensin/LDL to fibroblasts was not inhibited by anti–LDL-R antibodies or by r-RAP.44Second, defensins stimulate the endocytosis and degradation of125I-LDL by LDL-R–deficient FH fibroblasts and wild-type cells to the same extent. Rather, our data indicate that defensin forms stable complexes with LDL in solution and on cell surfaces. The resultant defensin/LDL complexes bind to heparan sulfate–containing proteoglycans including syndecan-1.6,48,51,52 The time course of internalization of defensin/LDL, which is measured in hours, is also inconsistent with the kinetics of LDL-R–mediated internalization, but is consistent with the rate of internalization and degradation of LDL by syndecan HSPGs.48 Binding of defensin/LDL to cells is also inhibited by low concentrations (0.2 U/mL) of heparin and is markedly reduced on cells lacking HSPGs. Taken together, these data indicate that defensin promotes surface binding, endocytosis, and degradation of LDL via HSPGs.
Our data indicate that defensin, which contains 4 arginine residues on its molecular surface, binds to LDL-R and therefore would be predicted to competitively inhibit the binding of LDL.23 Binding of defensin to fibroblasts is itself partially inhibited by anti–LDL-R antibody, and binding of defensin to LDL-R–deficient fibroblasts is reduced compared with wild-type cells. This interpretation is consistent with the finding that contact between cationic residues in apoB of LDL is required for optimal binding to LDL-R.38,53Defensin bound to LDL-R appears unavailable to bind LDL, in contrast to defensin bound to HSPGs, which binds LDL quite well. Thus, in the presence of defensin, binding of LDL to the LDL-R is likely to be competitively inhibited, whereas binding to HSPGs is promoted. Diversion of LDL from the LDL-R to HSPGs by defensin may slow the degradation of the lipoprotein31 and subject it to oxidation and other modifications. Thus, the studies reported here provide insight into one mechanism by which leukocyte defensins may modulate LDL metabolism and thereby the development of atherosclerosis.
We have previously observed that defensin is readily detected in atherosclerotic human coronary and cerebral arteries.30,34During systemic infection, α-defensins are released into plasma in micromolar concentrations.25,26 Defensins are found at considerably higher concentrations at sites of inflammation27-29 that are likely to simulate concentrations within the vessel wall that develop as result of neutrophil senescence or neutrophil activation. The studies presented here indicate that the binding of LDL by vascular cells is promoted by α-defensin at these same concentrations. These studies also suggest that α-defensins should be added to the list of bridging molecules that link atherogenic lipoproteins to HSPGs, such as lipoprotein lipase, apoE, and hepatic lipase.54 55 The α-defensins also have the interesting and distinctive property of being released during inflammation, which sets them apart from the other 3. Thus, α-defensins may be considered as representing a potential new class of “inflammatory lipoprotein-binding apoproteins.” However, additional studies will be required to define the effect of defensin on LDL metabolism in vivo.
Supported in part by grants HL58107 and HL60169 from the National Institutes of Health, grant 960105000 from the American Heart Association, and a grant from the Joint Research Fund of the Hebrew University and Hadassah University Hospital. During part of this work, K.J.W. was an Established Investigator of the American Heart Association and Genentech.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Abd Al-Roof Higazi, Department of Pathology and Laboratory Medicine, 513A Stellar-Chance, 422 Curie Blvd, Philadelphia, PA 19104; e-mail: higazi@mail.med.upenn.edu.