Abstract
Lineage-specific transcription factors play crucial roles in the development of hematopoietic cells. In a previous study, it was demonstrated that Ras activation was involved in thrombopoietin-induced megakaryocytic differentiation. In this study, constitutive Ras activation by H-rasG12V evoked megakaryocytic maturation of erythroleukemia cell lines F-36P and K562, but not of myeloid cell line 32D cl3 that lacks GATA-1. However, the introduction of GATA-1 led to reprogramming of 32D cl3 toward erythrocytic/megakaryocytic lineage and enabled it to undergo megakaryocytic differentiation in response to H-rasG12V. In contrast, the overexpression of PU.1 and c-Myb changed the phenotype of K562 from erythroid to myeloid/monocytic lineage and rendered K562 to differentiate into granulocytes and macrophages in response to H-rasG12V, respectively. In GATA-1–transfected 32D cl3, the endogenous expression of PU.1 and c-Myb was easily detectable, but their activities were reduced severely. Endogenous GATA-1 activities were markedly suppressed in PU.1-transfected and c-myb–transfected K562. As for the mechanisms of these reciprocal inhibitions, GATA-1 and PU.1 were found to associate through their DNA-binding domains and to inhibit the respective DNA-binding activities of each other. In addition, c-Myb bound to GATA-1 and inhibited its DNA-binding activities. Mutant GATA-1 and PU.1 that retained their own transcriptional activities but could not inhibit the reciprocal partner were less effective in changing the lineage phenotype of 32D cl3 and K562. These results suggested that GATA-1 activities may be crucial for Ras-mediated megakaryocytic differentiation and that its activities may be regulated by the direct interaction with other lineage-specific transcription factors such as PU.1 and c-Myb.
Introduction
Transcription factors play a key role during the development of hematopoietic cells through a series of gene transcriptions necessary for their growth, differentiation, and survival. It has previously been shown that targeted disruption of a certain transcription factor often causes a complete or selective defect of hematopoietic cells. For example, AML-1−/−, CBFβ−/−, c-myb−/−, or GATA-2−/− mice have been reported to die at approximately embryonic days 12 to 15, possibly because of the entire absence of definitive hematopoiesis in the fetal liver.1 Furthermore, genes encoding these transcription factors have proved to be the most frequent targets of chromosomal translocations in human acute leukemias—for example, t(8;21)(q22;q22), AML-1-ETO; inv16(p13;q22), CBFβ-MYH11; t(12;21)(p13;q22), TEL-AML1; t(1;14)(p32;q11); and SCL-TCRα.2 These lines of evidence indicated that the appropriately regulated expression and function of these transcription factors are required for normal hematopoiesis.
The GATA family is composed of 6 members and is indispensable for the development and subsequent growth and differentiation of diverse cell types.3 GATA family proteins possess 2 highly conserved zinc finger domains, both of which include the configuration Cys-X2-Cys-X17-Cys-X2-Cys. The carboxyl (C) finger is absolutely required for DNA binding, whereas the amino (N) finger stabilizes the DNA binding and confers full specificity.4,5 The first cloned member of this family, GATA-1, was identified as an erythroid nuclear protein that binds to consensus GATA motifs, (A/T)GATA(A/G), in globin gene promoters, enhancers, and locus control regions. It has been shown that GATA-1 is expressed at high levels in erythroid cells, megakaryocytes, and mast cells and at lower levels in hematopoietic progenitor cells and Sertoli cells of testis.3 In contrast, GATA-2 is ubiquitously expressed, and GATA-3 is exclusively expressed, in T lymphocytes.3 In chimeric mice generated from mutant embryonic stem (ES) cells lacking GATA-1, the mutant cells did not contribute to erythropoiesis.6 In addition, GATA-1–null ES cells were unable to differentiate into mature erythroid cells in vitro.7,8 Thus, GATA-1 is considered to be essential for the terminal differentiation of erythroid progenitor cells. Recently, Shivdasani et al9 reported that lineage-selective GATA-1 knock-out mice exhibited striking thrombocytopenia and severe anemia, which were accompanied by the increased proliferation and impaired cytoplasmic maturation of megakaryocytes.9 Furthermore, Takahashi et al10 demonstrated that heterozygous mutant mice chimeric for the GATA-1 gene displayed marked splenomegaly, anemia, and thrombocytopenia. These results further suggest that GATA-1 plays essential roles not only in erythropoiesis but also in megakaryopoiesis and thrombopoiesis.
PU.1 is a member of the Ets family transcription factors that specifically bind to the GGAA/T motif in the target DNA.11Structural and functional analyses of PU.1 revealed that DNA binding is executed through the C-terminal Ets homology region and that the transactivating domain is located in the N-terminal region.12 In addition, there is a central region, called a PEST domain, rich in proline, glutamic acid, serine, and threonine residues.12 Expression of PU.1 is restrictedly detected in hematopoietic tissues, especially with high levels in monocytic, granulocytic, and B-lymphoid lineages.11 Several presumptive target genes of PU.1 have been identified, some of which are essential for the growth and survival of the cells in these lineages.13 In agreement with these findings, PU.1-targeted mice showed defects in the development of multiple hematopoietic lineages, including B and T lymphocytes, monocytes, and granulocytes.14,15 In addition, PU.1 was shown to have the potential to impose myeloid lineage commitment on multipotent hematopoietic progenitors,16 suggesting that PU.1 is a master regulator of myeloid lineage commitment.
The growth and differentiation of hematopoietic stem/progenitor cells are regulated by a number of growth factors. Among them, thrombopoietin (TPO) is a fundamental regulator of megakaryopoiesis and thrombopoiesis. In previous studies,17-19 others and we have shown that Ras/MAPK (mitogen-activated protein kinase) activation is involved in TPO-induced megakaryocytic differentiation. However, Ras activation is not necessarily coupled with megakaryocytic differentiation in other cell types. For example, Ras activation was shown to promote terminal differentiation toward macrophages in a monocytic cell line, U937.20 In addition, activated Ras was reported to induce cell growth but not to affect the stage of maturation in a murine myeloid cell line, 32D cl3, and a murine pro B cell line, Ba/F3.21 22 Therefore, it remains largely unknown which instinctive cell properties are prerequisite for Ras-mediated megakaryocytic differentiation. Because instinctive characters of hematopoietic cells are primarily controlled by lineage-specific transcription factors such as GATA-1 and PU.1, in the current study we investigated the roles of these transcription factors in Ras-mediated megakaryocytic differentiation. We here found that GATA-1 enabled the myeloid cell line 32D cl3 to undergo megakaryocytic differentiation in response to constitutive Ras activation by H-rasG12V. In addition, though H-rasG12Vinduced megakaryocytic differentiation of an erythroid cell line K562, the introduction of PU.1-and c-myb into H-rasG12V–expressing K562 cells led to granulocytic and macrophage maturation, respectively. In these cells, the transcriptional activity of GATA-1 was suppressed by PU.1 or c-Myb and vice versa. Thus, we here provide unique evidence that GATA-1 activities are required for Ras-mediated megakaryocytic differentiation and that the cell lineage and differentiation may be determined by the combined effects of lineage-specific transcription factors, including GATA-1, PU.1, and c-Myb.
Materials and methods
Reagents and antibodies
Recombinant human (rh) and recombinant murine (rm) IL-3 were provided by Kirin Brewery Company (Tokyo, Japan). AP2 (anti-human GP IIb/IIIa complex) antibody was provided by Dr T. Kunicki (Scripps Research Institute, La Jolla, CA). Anti-CD61 antibody and anti-glycophorin A antibody were purchased from DAKO Japan Company (Tokyo, Japan); anti-CD14, anti-CD33, and anti-CD11b antibodies were purchased from Coulter Immunology (Hialeah, FL); anti-GATA-1, anti-PU.1, anti-Myb, and anti-actin antibodies were purchased from Santa Cruz Biotechnology (California, CA); anti-Ras antibody was purchased from Oncogene Research Product (Cambridge, MA); anti-hemagglutinin (HA) monoclonal antibody was purchased from Boehringer Mannheim (Mannheim, Germany); and anti-Flag antibody was purchased from Eastman Kodak (Rochester, NY).
Cell lines and cultures
32D cl3, a murine IL-3–dependent myeloid cell line, and F-36P, a human IL-3–dependent erythroleukemia cell line, were cultured in RPMI 1640 (Nakarai Tesque, Kyoto, Japan) supplemented with 10% fetal calf serum (FCS; Flow, North Ryde, Australia) in the presence of rmIL-3 (1 ng/mL) and rhIL-3 (5 ng/mL), respectively.
Plasmid constructions and cDNA
Full-length human GATA-1 and murine PU.1 cDNA were subcloned into pcDNA3 (Invitrogen, De Schelp, Netherlands) to generate each expression vector. To construct ΔNZF of GATA-1 and ΔAD of PU.1, cDNA coding amino acids 194 to 250 and 33 to 101 were eliminated by polymerase chain reaction, respectively. This cDNA was subcloned into pcDNA3-HA or pcDNA3-Flag to generate N-terminus epitope-tagged expression vectors. An expression vector of c-myb, pact-c-myb, was kindly provided from Dr S. Ishii (Tsukuba Life Science Center; RIKEN, Ibaraki, Japan). Probes for platelet factor 4 (PF4) and GPIIb were kindly provided from Dr Y. Furukawa (Jichi Medical School, Tochigi, Japan) and Dr Y. Tomiyama (Osaka University, Osaka, Japan), respectively.
Morphologic analysis
The morphology of the cultured cells was analyzed by staining the cytospin preparations with May–Grünwald–Giemsa.
Benzidine staining
Cultured cells were suspended in staining solution, which is a 14:1 mixture of benzidine solution (3% benzidine in acetic acid) and 31% H2O2, for 5 minutes, and subjected to cytospin centrifugation. The black-stained cells were counted as benzidine-positive cells by microscopy.
Flow cytometry
Surface phenotypes of cells were examined by indirect immunofluorescence with FACSort (Beckon Dickinson, Oxnard, CA). DNA content of cultured cells was quantitated by propidium iodide staining.23
Northern blot analysis
The isolation of total cellular RNA and the method used for Northern blot analysis were described previously.23
Immunoprecipitation and immunoblotting
Preparation of cell lysates, immunoprecipitation, gel electrophoresis, and immunoblotting were performed as described previously.23 Immunoreactive proteins were visualized with the enhanced chemiluminescence detection system (Dupont NEN, Boston, MA).
Lac-inducible system and plasmids
To express a target cDNA, we used a LacSwitch II (Stratagene, La Jolla, CA) inducible expression system in which the expression of the target cDNA is induced by isopropyl-β-D-thiogalactopyranoside (IPTG) treatment. In short, F-36P, K562, and 32D cl3 cells were initially transfected with an expression vector of Lac-repressor (Lac-R), pCMV-LacI, by electroporation. After selection with hygromycin (0.5 mg/mL), one clone, in which Lac-R was most intensely expressed, was further transfected with a Lac-inducible expression vector of H-rasG12V, pOPRSVI-H-rasG12V.17After selection with G418 (1.0 mg/mL), the induction levels of H-rasG12V were examined before and after IPTG treatment (0.5 mmol/L) in each clone by Western blot analysis. To further transfect an expression vector of PU.1, c-myb, or GATA-1 into hygromycin- and G418-resistant cells, an expression vector of blasticidin S deaminase pSV2bsr (Kaken Pharma, Tokyo, Japan) was cotransfected with an appropriate expression vector and cultured with 30 μg/mL blastcidin S hydrochloride (Funakoshi, Tokyo, Japan).
Luciferase assay
At first, the murine minimal JunB promoter (−42 to +136) was subcloned into the luciferase plasmid to construct JunB-MP-Luc. To generate functional and nonfunctional reporter genes for GATA-1, PU.1, and c-Myb, concatamerized double-stranded oligonucleotides were subcloned just upstream of minimal JunB promoter in JunB-MP-Luc, and their sequences were as follows: a wild-type (WT) and a mutated (MT) reporter gene for GATA-1 (3× WT-Mα-Luc, 5′-GGGCAACTGATAAGGATTCC-3′; 3× MT-Mα-Luc, MT, 5′-GGGCAACTGGTCAGGATTCC-3′; recognition sites are underlined); reporter genes for PU.1 (3× WT-MHC-Luc,5′-AAAGAGGAACTTGG-3′; 3× MT-MHC-Luc,5′-AAAGAGCTACTTGG-3′); and reporter genes for c-Myb (4× WT-MBS-Luc,5′-CTCTACACCCTAACTGACACACATTCT-3′; 4× MT-MBS-Luc,5′-CTCTACACCATCACAAACACACATTCT-3′). Luciferase assay was performed with the Dual-Luciferase Reporter System (Promega, Madison, WI) as previously described.24 Briefly, the cultured cells were transfected with an appropriate reporter gene together with pRL-CMV-Rluc, an expression vector of renillaluciferase, by electroporation. In addition, luciferase assays were performed in NIH3T3 cells by the calcium phosphate coprecipitation method.24 Relative firefly luciferase activities were calculated by normalizing transfection efficiency according to therenilla luciferase activities. Experiments were performed in triplicate, and the similar results were obtained from at least 3 independent experiments.
Transient transfection into 293T cells
293T Cells were transfected with expression vectors of GATA-1, PU.1, and c-myb alone or in combination with pcDNA3-GFP (an expression vector of green fluorescence protein) by the calcium phosphate coprecipitation method. After 48 hours, total cellular lysates or nuclear extracts were isolated. Transfection efficiencies were monitored by the intensities of green fluorescence protein by flow cytometric analyses.
Electrophoretic mobility shift assay
Nuclear extracts were prepared from 293T cells transfected with expression vectors of GATA-1, PU.1, and c-myb alone or in combination. Sequences of probes and competitors are as follows: probe for GATA-1 (Mα), 5′-GATCTCCGGCAACTGATAAGGATTCCCTG-3′; mutated GATA-1 competitor, 5′-GATCTCCGGCAACTCAGAAGGATTCCCTG-3′; probe for PU.1 (MHC, major histocompatibility complex), 5′-TCGAAAAGAGGAACTTGGGTA-3′; mutated PU.1 competitor, 5′-TCGAAAAGACGTACTTGGGTA-3′. The binding reaction and electrophoresis were performed as previously described.24In competition assays, nuclear extracts were preincubated with a 200-fold molar excess of unlabeled competitor before the binding reaction. In a supershift assay, nuclear proteins were preincubated with 1 μg anti-GATA-1 antibody or anti-PU.1 antibody for 30 minutes at 4°C, followed by the binding reaction.
Results
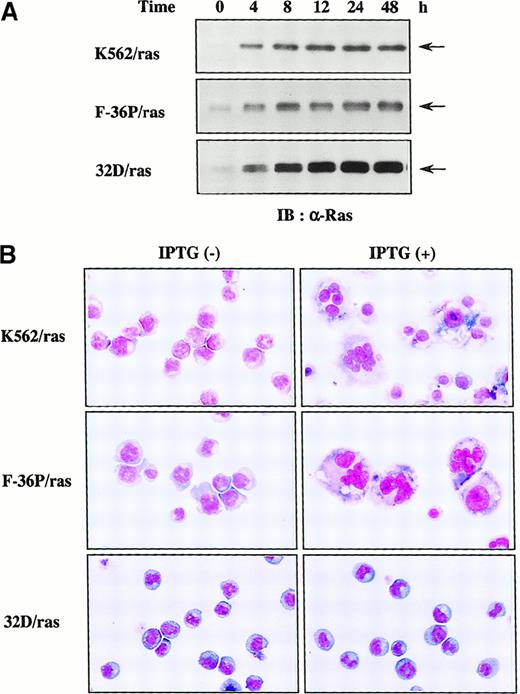
H-rasG12V induces megakaryocytic differentiation of K562 and F-36P but not of 32D cl3
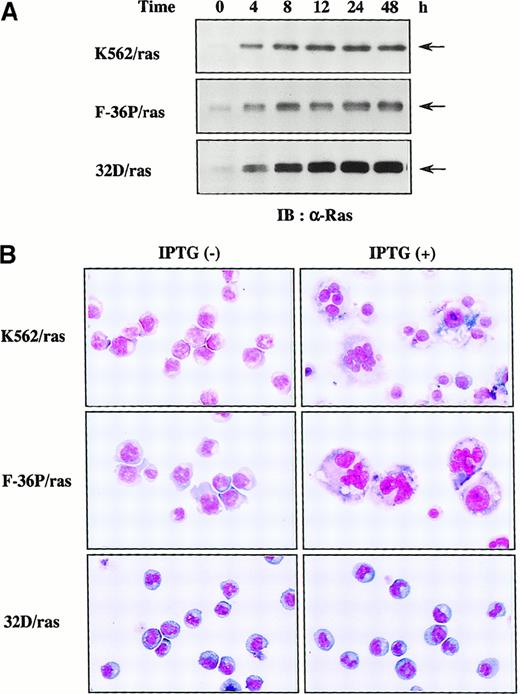
To examine which types of hematopoietic cells are able to undergo megakaryocytic differentiation in response to H-rasG12V, we expressed H-rasG12V in human erythroleukemia cell lines K562 and F-36P and also in a murine myeloid cell line, 32D cl3, by using a Lac-inducible system in which expression of the target protein was induced by IPTG treatment. The clones were designated K562/ras, F-36P/ras, and 32D/ras, respectively. In all clones, IPTG treatment led to the efficient induction of H-rasG12V protein in Western blot analysis with anti-Ras antibody (Figure1A), whereas actin, a loading control, was found to be expressed at similar levels in each lane (data not shown). As shown in Figure 1, panel B, K562/ras, F-36P/ras, and 32D/ras were absolutely composed of small, undifferentiated blast cells before IPTG treatment. After 5-day culture with IPTG, a substantial fraction (60%-70%) of F-36P/ras and K562/ras showed morphologic alterations indicative of megakaryocytic maturation, whereas most of 32D/ras still revealed blastoid features.
Inducible expression of H-rasG12V in K562/ras, F-36P/ras, and 32D/ras.
(A) Western blot analysis on Ras expression during IPTG treatment. Each clone was treated with 0.5 mmol/L IPTG for the time indicated. Total cell lysates were subjected to SDS-PAGE and probed with anti-Ras antibody. (B) Light micrograph of K562/ras, F-36P/ras, and 32D/ras before and after 5-day culture with IPTG. Cytocentrifugation preparation from each culture was stained with May–Grünwald–Giemsa (original magnification, ×100).
Inducible expression of H-rasG12V in K562/ras, F-36P/ras, and 32D/ras.
(A) Western blot analysis on Ras expression during IPTG treatment. Each clone was treated with 0.5 mmol/L IPTG for the time indicated. Total cell lysates were subjected to SDS-PAGE and probed with anti-Ras antibody. (B) Light micrograph of K562/ras, F-36P/ras, and 32D/ras before and after 5-day culture with IPTG. Cytocentrifugation preparation from each culture was stained with May–Grünwald–Giemsa (original magnification, ×100).
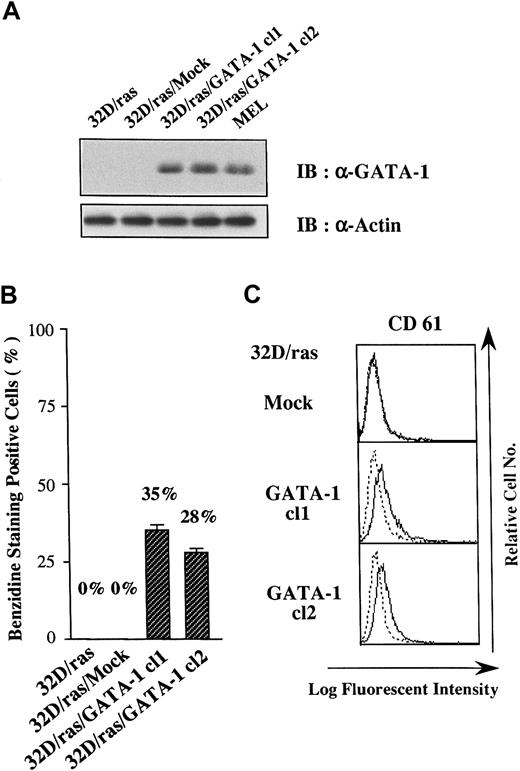
Ectopic overexpression of GATA-1 reprograms 32D/ras into erythrocytic/megakaryocytic lineage
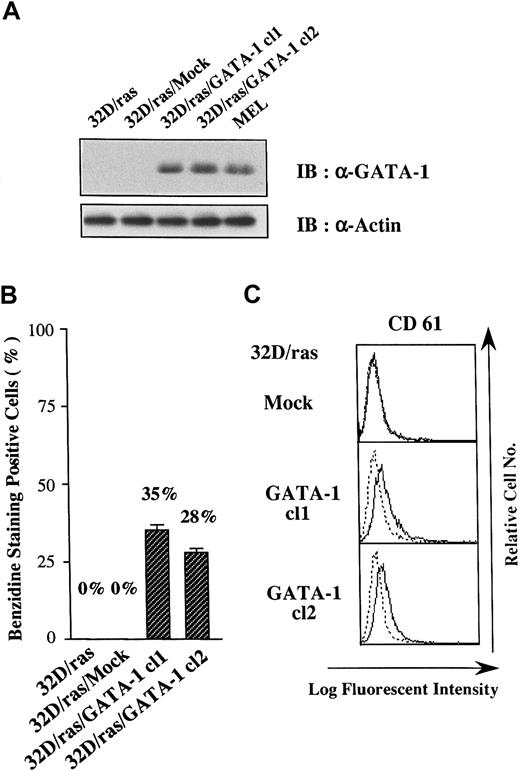
Because GATA-1 has been shown to change the lineage phenotype of hematopoietic cells,25-27 we introduced GATA-1 into 32D/ras. As shown in Figure 2A, GATA-1 protein was hardly detected in a parental clone (32D/ras) and a control clone transfected with an empty vector (32D/ras/Mock), but it was expressed in GATA-1 transfectants 32D/ras/GATA-1 cl1 and cl2 at high levels almost similar to those in a murine erythroleukemia cell line (MEL). We initially explored the expression of β-globin, a marker of mature erythroid cells, in GATA-1 transfectants by benzidine staining. Although 32D/ras and 32D/ras/Mock were essentially negative on benzidine staining, approximately 30% of 32D/ras/GATA-1 became positive. As shown in Figure 2, panel C, a megakaryocytic antigen CD61 (GP IIIa) was scarcely expressed on 32D/ras/Mock, whereas a weak but easily detectable level of CD61 expression was observed on GATA-1 transfectants. These results suggested that the ectopic overexpression of GATA-1 reprogrammed 32D/ras toward the erythrocytic/megakaryocytic lineage.
Effects of GATA-1 transgene on lineage phenotype of 32D/ras.
(A) Expression of GATA-1 in 32D/ras, 32D/ras/Mock, 32D/ras/GATA-1, and MEL. Total cell lysates were subjected to SDS-PAGE and probed with anti-GATA-1 and anti-actin antibodies. (B) Benzidine staining of the cultured cells. Cells were suspended in benzidine staining solution and subjected to cytospin centrifugation. Black-stained cells were counted as benzidine-positive cells by microscopy. Results are shown as the mean ± SD of triplicate experiments. (C) Flow cytometric analyses on CD61 expression. Expression of CD61 was examined by staining with anti-CD61 antibody () and control antibody of the same isotype (····).
Effects of GATA-1 transgene on lineage phenotype of 32D/ras.
(A) Expression of GATA-1 in 32D/ras, 32D/ras/Mock, 32D/ras/GATA-1, and MEL. Total cell lysates were subjected to SDS-PAGE and probed with anti-GATA-1 and anti-actin antibodies. (B) Benzidine staining of the cultured cells. Cells were suspended in benzidine staining solution and subjected to cytospin centrifugation. Black-stained cells were counted as benzidine-positive cells by microscopy. Results are shown as the mean ± SD of triplicate experiments. (C) Flow cytometric analyses on CD61 expression. Expression of CD61 was examined by staining with anti-CD61 antibody () and control antibody of the same isotype (····).
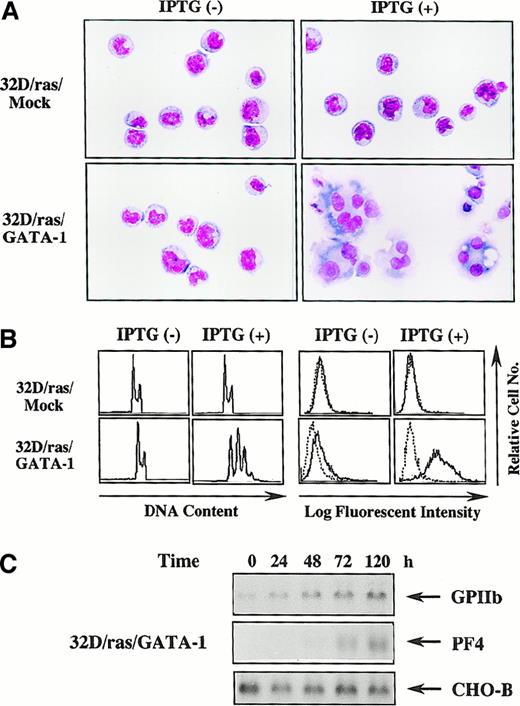
GATA-1–transfected 32D/ras undergoes megakaryocytic differentiation in response to H-rasG12V
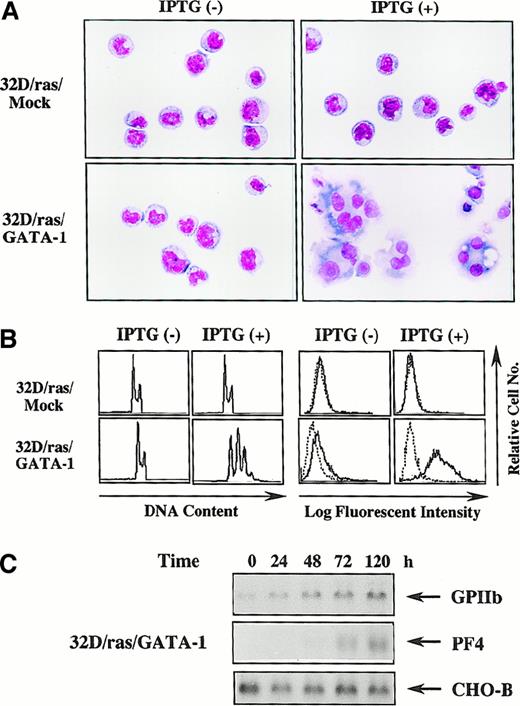
Next, we investigated whether biologic responses of 32D cl3 to H-rasG12V were modulated by GATA-1. Like parental 32D/ras (Figure 1B), an apparent morphologic change was not induced by IPTG in 32D/ras/Mock (Figure 3A, upper panel), whereas 32D/ras/GATA-1 underwent morphologic changes indicating megakaryocytic maturation after a 5-day IPTG treatment (Figure 3A, lower panel). DNA content analysis showed that IPTG treatment resulted in polyploid formation (2N, 32%; 4N, 38%; 8N, 24%; 16N, 6%) in 32D/ras/GATA-1 but not in 32D/ras/Mock (Figure 3B, left panel). In addition, CD61 expression was augmented after IPTG treatment in 32D/ras/GATA-1, whereas its expression was hardly detected in 32D/ras/Mock before and after IPTG treatment (Figure 3B, right panel). In addition, the expression of GPIIb mRNA was up-regulated and that of PF4 mRNA was induced after IPTG treatment in 32D/ras/GATA-1 (Figure3C), yet these expressions were scarcely detected in 32D/ras/Mock before and after IPTG treatment (data not shown). These results suggested that ectopically introduced GATA-1 enabled 32D/ras to differentiate into mature megakaryocytes in response to H-rasG12V.
GATA-1–transfected 32D/ras undergoes megakaryocytic differentiation in response to H-rasG12V.
(A) Light micrograph of 32D/ras/Mock and 32D/ras/GATA-1 before and after 5-day culture with IPTG. Cells were cultured with or without IPTG for 5 days and subjected to analysis. Cytocentrifugation preparation was stained with May–Grünwald–Giemsa (original magnification, ×100). (B) Flow cytometric analyses on DNA content and surface expression of CD61. DNA content of cultured cells was quantitated by propidium iodide staining and analyzed on FACSort. Surface expression of CD61 was analyzed by staining with anti-CD61 monoclonal antibody () and control antibody of the same isotype (····). (C) Changes in expression of GPIIb and PF4 mRNA in 32D/ras/GATA-1 during IPTG treatment.
GATA-1–transfected 32D/ras undergoes megakaryocytic differentiation in response to H-rasG12V.
(A) Light micrograph of 32D/ras/Mock and 32D/ras/GATA-1 before and after 5-day culture with IPTG. Cells were cultured with or without IPTG for 5 days and subjected to analysis. Cytocentrifugation preparation was stained with May–Grünwald–Giemsa (original magnification, ×100). (B) Flow cytometric analyses on DNA content and surface expression of CD61. DNA content of cultured cells was quantitated by propidium iodide staining and analyzed on FACSort. Surface expression of CD61 was analyzed by staining with anti-CD61 monoclonal antibody () and control antibody of the same isotype (····). (C) Changes in expression of GPIIb and PF4 mRNA in 32D/ras/GATA-1 during IPTG treatment.
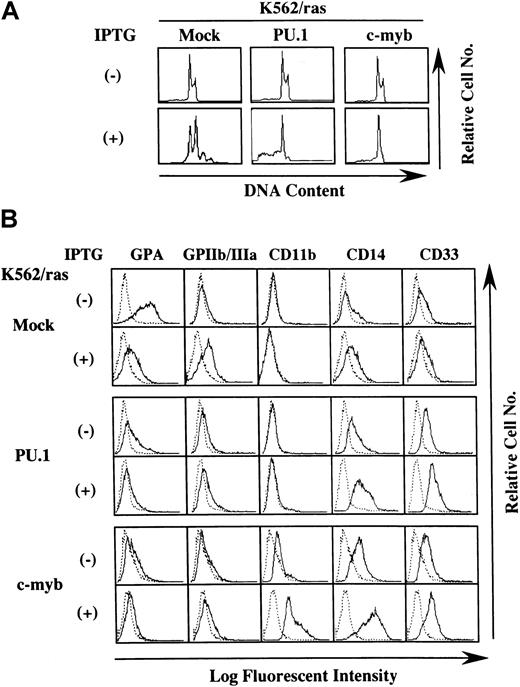
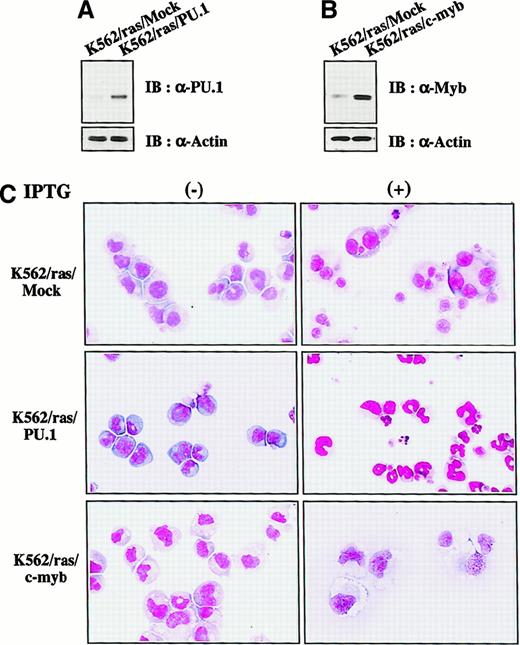
Granulocyte and macrophage differentiation of PU.1- and c-myb–transfected K562 after induction of H-rasG12V
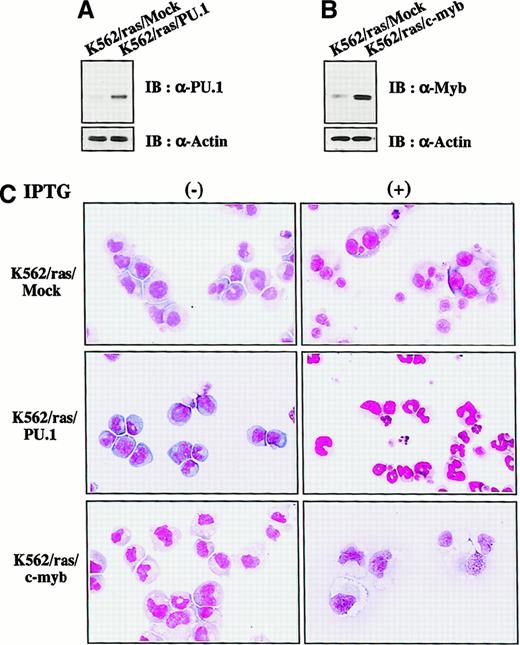
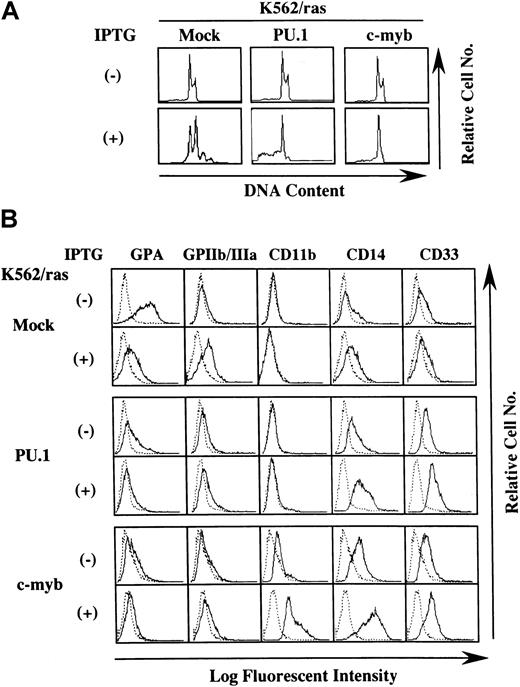
We next investigated whether H-rasG12V–mediated megakaryocytic differentiation was influenced by other transcription factors. PU.1 and c-myb were introduced into K562/ras, and the clones were designated K562/ras/PU.1 and K562/ras/c-myb, respectively. As shown in Figure 4A, PU.1 protein was efficiently expressed in K562/ras/PU.1, but endogenous PU.1 was detected only faintly in a control clone K562/ras/Mock transfected with an empty expression vector. Although c-Myb was expressed in K562/ras/Mock endogenously, the more increased level of c-Myb was expressed in K562/ras/c-myb (Figure 4B). Without IPTG treatment, K562/ras/PU.1, K562/ras/c-myb, and K562/ras/Mock were primarily composed of blastoid cells, and no significant difference was observed between the clones (Figure 4C, left panel). After a 5-day IPTG treatment, megakaryocytic maturation was observed in K562/ras/Mock just as parental K562/ras (Figure 4C, right panel). In contrast, a significant proportion of K562/ras/PU.1 revealed morphologic changes suggesting granulocytic maturation. Furthermore, a noticeable proportion of K562/ras/PU.1 underwent apoptosis after IPTG treatment, which was characterized by shrunken cell size, nuclear fragmentation, or both. Moreover, K562/ras/c-myb revealed macrophage-like features characterized by enlarged cell size and vacuoles in the cytoplasm after IPTG treatment. In agreement with morphologic changes, DNA content analysis showed that 5-day culture with IPTG induced polyploid formation up to 16N in K562/ras/Mock (Figure5A). In K562/ras/PU.1, the proportion of cells in S or G2/M phase was reduced from 40% to 7% by IPTG treatment. In addition, IPTG treatment yielded apoptotic cells in approximately 45% of cultured cells, which was detected as a subdiploid fraction, and H-rasG12V induced growth suppression in K562/ras/c-myb (proportion of cells in S or G2/M phase: IPTG −42% vs IPTG +5%; Figure 5A). This severe growth inhibition in K562/ras/PU.1 and K562/ras/c-myb after IPTG treatment was coupled with terminal differentiation of these clones.
Enforced expression of PU.1 and c-Myb in K562/ras.
(A, B) Western blot analyses on PU.1 and c-Myb expression. Expression levels of PU.1 and c-Myb were examined in each clone by Western blot analyses on total cellular lysates. (C) Light micrograph of K562/ras/Mock, K562/ras/PU.1, and K562/ras/c-myb before and after 5-day culture with IPTG. Cells were cultured with or without IPTG for 5 days. Cytocentrifugation preparation was stained with May–Grünwald–Giemsa (original magnification, ×100).
Enforced expression of PU.1 and c-Myb in K562/ras.
(A, B) Western blot analyses on PU.1 and c-Myb expression. Expression levels of PU.1 and c-Myb were examined in each clone by Western blot analyses on total cellular lysates. (C) Light micrograph of K562/ras/Mock, K562/ras/PU.1, and K562/ras/c-myb before and after 5-day culture with IPTG. Cells were cultured with or without IPTG for 5 days. Cytocentrifugation preparation was stained with May–Grünwald–Giemsa (original magnification, ×100).
Effects of PU.1 and c-Myb on the lineage phenotype of K562.
(A) DNA content analysis before and after 5-day IPTG treatment. DNA content of the cultured cells was examined by propidium iodide staining and analyzed with flow cytometry. (B) Flow cytometric analysis on the surface expression of GPA, GPIIb/IIIa, CD11b, CD14, and CD33 before and after 5-day IPTG treatment. Surface phenotypes of the cultured cells were examined with the indirect immunofluorescence method by staining with the indicated antibody () and control antibody of the same isotype (····).
Effects of PU.1 and c-Myb on the lineage phenotype of K562.
(A) DNA content analysis before and after 5-day IPTG treatment. DNA content of the cultured cells was examined by propidium iodide staining and analyzed with flow cytometry. (B) Flow cytometric analysis on the surface expression of GPA, GPIIb/IIIa, CD11b, CD14, and CD33 before and after 5-day IPTG treatment. Surface phenotypes of the cultured cells were examined with the indirect immunofluorescence method by staining with the indicated antibody () and control antibody of the same isotype (····).
Next, we examined the surface expression of lineage-specific antigens such as an erythroid marker glycophorin A (GPA), a megakaryocytic marker GPIIb/IIIa, and the myeloid/macrophage markers CD11b, CD14, and CD33 (Figure 5B). Without IPTG treatment, GPA was expressed intensely on K562/ras/Mock, whereas its expression decreased distinctly on K562/ras/PU.1 and K562/ras/c-myb. In contrast, expression levels of CD14 and CD33 were up-regulated slightly in K562/ras/PU.1 and K562/ras/c-myb. In addition, CD11b expression emerged on K562/ras/c-myb. Consistent with the findings on morphologic and DNA content analyses, K562/ras/Mock showed phenotypic changes indicative of megakaryocytic maturation in response to H-rasG12V—that is, the up-regulation of GPIIb/IIIa and the down-regulation of GPA—whereas expression levels of CD11b, CD14, and CD33 were not affected. By contrast, H-rasG12V augmented CD14 and CD33 expression and reduced GPA expression in K562/ras/PU.1. In K562/ras/c-myb, IPTG treatment enhanced the expression of CD11b, CD14, and CD33. Together with the findings on morphologic analyses, these results suggested that the overexpression of PU.1 and c-myb changed the phenotype of K562 to myeloid/monocytic lineage and allowed K562 to differentiate into granulocytes and macrophages in response to H-rasG12V, respectively.
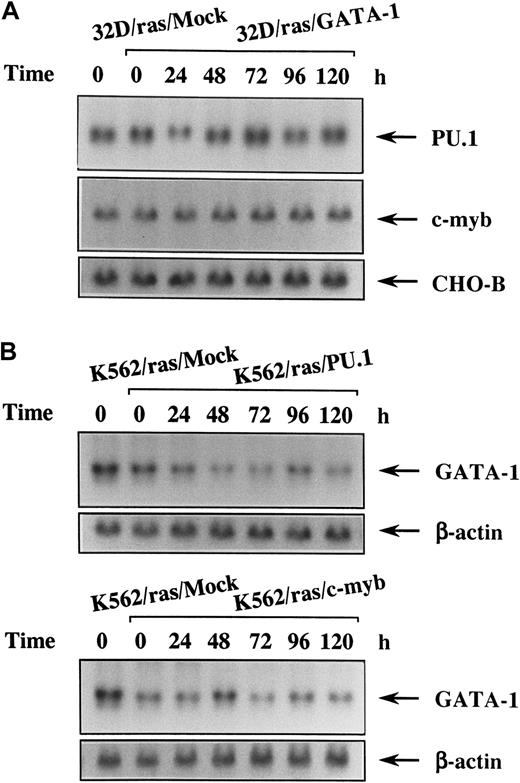
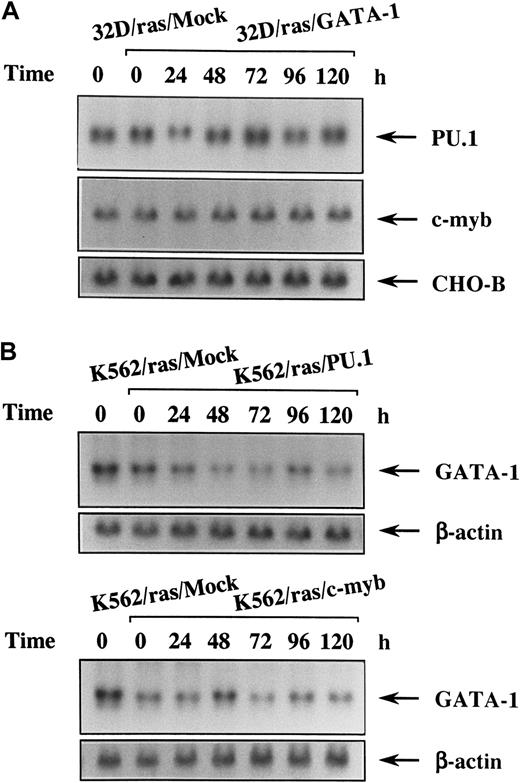
Sustained expression of PU.1 and c-myb in 32D/ras/GATA-1 and that of GATA-1 in K562/ras/PU.1 and K562/ras/c-myb during IPTG treatment
Because 32D cl3 is a myeloid cell line and expresses PU.1 and c-myb originally, we investigated changes of these expressions in 32D/ras/GATA-1 during H-rasG12V–induced megakaryocytic differentiation by Northern blot analysis. As shown in Figure6, panel A, expression levels of PU.1 and c-myb did not reveal an apparent difference between 32D/ras/Mock and 32D/ras/GATA-1 before IPTG treatment (Figure 6A, left 2 lanes). These expressions did not show significant change during H-rasG12V–induced megakaryocytic differentiation of 32D/ras/GATA-1 for up to 120 hours. We next examined the expression of endogenous GATA-1 during H-rasG12V–induced granulocytic differentiation of K562/ras/PU.1 and of macrophage differentiation of K562/ras/c-myb. Before IPTG treatment, GATA-1 expression was slightly reduced in K562/ras/PU.1 and K562/ras/c-myb compared with that in K562/ras/Mock (Figure 6B, left 2 lanes). After IPTG treatment, GATA-1 expression was down-regulated modestly in K562/ras/PU.1 and K562/ras/c-myb (Figure 6B). However, GATA-1 expression was still detectable even after 120 hours of IPTG treatment, at which time granulocytic or macrophage differentiation was already obvious with morphologic and surface phenotypic analyses (Figures 4C, 5B). Coincident with the findings on Northern blot analysis, each protein was expressed at comparable levels on Western blot analysis (data not shown).
Changes in expression of the lineage-specific transcription factors during megakaryocyte, granulocyte, or macrophage differentiation in each transfectant.
(A) Expression of PU.1 and c-myb mRNA in 32D/ras/GATA-1 during IPTG treatment. 32D/ras/GATA-1 cells were treated with IPTG for the indicated times and subjected to Northern blot analysis. (B) Expression of GATA-1 mRNA in K562/ras/PU.1 and K562/ras/c-myb during IPTG treatment. K562/ras/PU.1 and K562/ras/c-myb were treated with IPTG, and the expression of GATA-1 mRNA was examined by Northern blot analysis.
Changes in expression of the lineage-specific transcription factors during megakaryocyte, granulocyte, or macrophage differentiation in each transfectant.
(A) Expression of PU.1 and c-myb mRNA in 32D/ras/GATA-1 during IPTG treatment. 32D/ras/GATA-1 cells were treated with IPTG for the indicated times and subjected to Northern blot analysis. (B) Expression of GATA-1 mRNA in K562/ras/PU.1 and K562/ras/c-myb during IPTG treatment. K562/ras/PU.1 and K562/ras/c-myb were treated with IPTG, and the expression of GATA-1 mRNA was examined by Northern blot analysis.
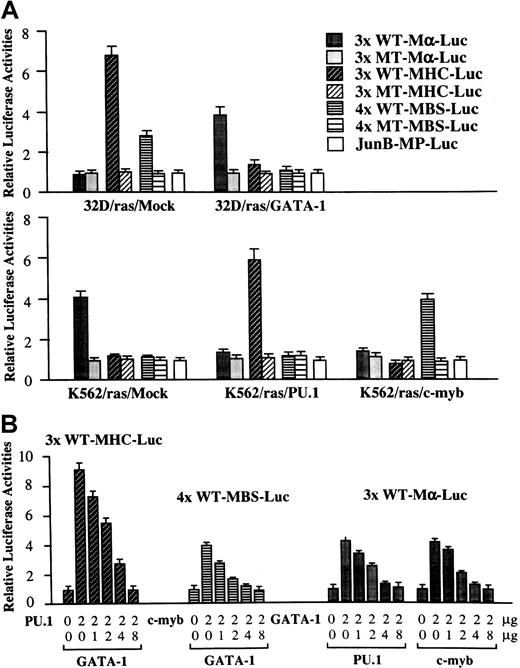
GATA-1 inhibits PU.1 and c-myb activities reciprocally
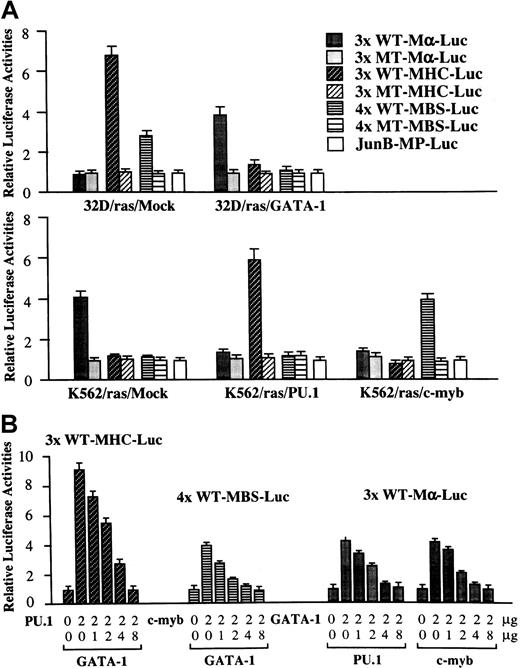
Next, we performed luciferase assays with functional and nonfunctional reporter genes for GATA-1 (3× WT-Mα-Luc and 3× MT-Mα-Luc), PU.1 (3× WT-MHC-Luc and 3× MT-MHC-Luc), and c-Myb (4× WT-MBS-Luc and 4× MT-MBS-Luc) in 32D/ras/Mock, 32D/ras/GATA-1, K562/ras/Mock, K562/ras/PU.1, and K562/ras/c-myb (Figure7A). In 32D/ras/Mock, reporter genes for PU.1 and c-Myb were activated 6.8-fold and 2.8-fold, respectively, with reference to the activities of the mutant reporter genes, whereas those for GATA-1 were scarcely activated. In contrast, PU.1 and c-Myb activities were inhibited drastically in 32D/ras/GATA-1 instead of GATA-1 activities (relative luciferase activities: PU.1, 1.4-fold; c-Myb, 1.1-fold; GATA-1, 3.8-fold). In K562/ras/Mock, endogenous GATA-1 activated 3× WT-Mα-Luc by 4.1-fold, yet PU.1 and c-Myb activities were hardly detectable (PU.1, 1.1-fold; c-Myb, 1.1-fold). However, GATA-1 activities were reduced to 1.2-fold in both K562/ras/PU.1 and K562/ras/c-myb, but ectopically introduced PU.1 and c-Myb stimulated 3× WT-MHC-Luc by 5.8-fold and 4× WT-MBS-Luc by 4.0-fold, respectively. These results raised the possibility that GATA-1 may antagonize PU.1 and c-Myb and, conversely, that PU.1 and c-Myb may antagonize GATA-1. To examine this possibility, we performed luciferase assays in NIH3T3 cells. As shown in Figure 7, panel B, both PU.1-induced 3× WT-MHC-Luc activities and c-Myb–induced 4× WT-MBS-Luc activities were repressed by cotransfected GATA-1 in a dose-dependent manner. GATA-1–stimulated 3× WT-Mα-Luc activities were reduced by cotransfected PU.1 or c-myb, depending on their doses. In these experiments, neither expression vector affected the luciferase activities of mutant reporter genes for GATA-1, PU.1, or c-Myb (data not shown). Furthermore, negative cross-talk was observed among other members of GATA and Ets families, because PU.1-, Ets-1, or Ets-2–induced luciferase activities were dose-dependently inhibited by GATA-1, GATA-2, or GATA-3 in NIH3T3 cells (data not shown).
Functional interaction, GATA-1–PU.1, and GATA-1–c-Myb.
(A) GATA-1, PU.1, and c-Myb activities in each transfectant. Twenty micrograms of functional and nonfunctional reporter gene for GATA-1 (3× WT Mα-Luc and 3× MT Mα-Luc), PU.1 (3× WT-MHC-Luc and 3× MT-MHC-Luc), c-Myb (4× WT-MBS-Luc and 4× MT-MBS-Luc), or a backbone reporter gene (JunB-MP-Luc) was cotransfected with 3 μg pRL-CMV-Rluc by electroporation. After 36 hours, the cells were subjected to the measurement of the firefly and the renilla luciferase activities. The relative firefly luciferase activities were calculated by normalizing transfection efficiency according to therenilla luciferase activities. Results are shown as the mean ± SD of triplicate experiments. (B) Reciprocal inhibition, GATA-1–PU.1, and GATA-1–c-Myb in NIH3T3 cells. NIH3T3 cells were cotransfected with various amounts of expression vectors, 1.0 μg of an appropriate reporter gene, and 10 ng of pRL-CMV-Rluc by the calcium phosphate coprecipitation method. After 36 hours of culture, cells were subjected to luciferase assay.
Functional interaction, GATA-1–PU.1, and GATA-1–c-Myb.
(A) GATA-1, PU.1, and c-Myb activities in each transfectant. Twenty micrograms of functional and nonfunctional reporter gene for GATA-1 (3× WT Mα-Luc and 3× MT Mα-Luc), PU.1 (3× WT-MHC-Luc and 3× MT-MHC-Luc), c-Myb (4× WT-MBS-Luc and 4× MT-MBS-Luc), or a backbone reporter gene (JunB-MP-Luc) was cotransfected with 3 μg pRL-CMV-Rluc by electroporation. After 36 hours, the cells were subjected to the measurement of the firefly and the renilla luciferase activities. The relative firefly luciferase activities were calculated by normalizing transfection efficiency according to therenilla luciferase activities. Results are shown as the mean ± SD of triplicate experiments. (B) Reciprocal inhibition, GATA-1–PU.1, and GATA-1–c-Myb in NIH3T3 cells. NIH3T3 cells were cotransfected with various amounts of expression vectors, 1.0 μg of an appropriate reporter gene, and 10 ng of pRL-CMV-Rluc by the calcium phosphate coprecipitation method. After 36 hours of culture, cells were subjected to luciferase assay.
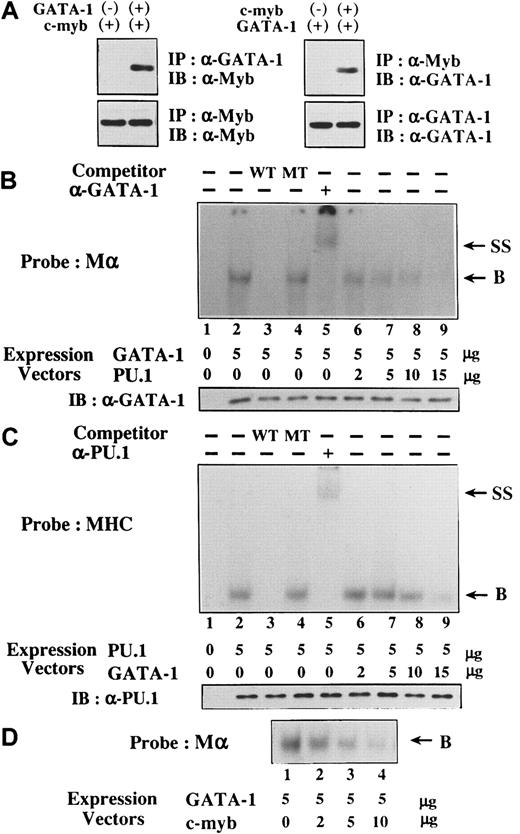
Mechanisms of reciprocal inhibition of GATA-1–PU.1 and GATA-1–c-Myb
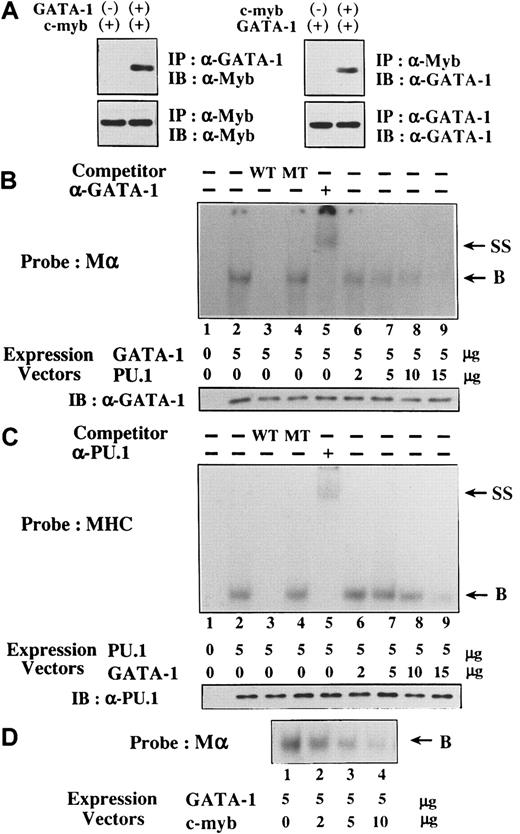
We examined the mechanisms of the reciprocal inhibition of GATA-1–PU.1 and GATA-1–c-Myb. At first, 293T cells were transfected with expression vectors of HA-tagged GATA-1, Flag-tagged PU.1, or both, and the formation of GATA-1-PU.1 complex was examined by a coimmunoprecipitation method. In agreement with recent findings,28-30 GATA-1 and PU.1 were found to associate through the 2 zinc finger domains of GATA-1 and the Ets domain of PU.1 (data not shown). Similarly, we found that c-Myb was coimmunoprecipitated with GATA-1 and vice versa by the coimmunoprecipitation method (Figure 8A).
Mechanisms of reciprocal inhibition, GATA-1–PU.1, and GATA-1–c-Myb.
(A) GATA-1 and c-Myb form the complex in vivo. 293T cells were transfected with GATA-1 or PU.1, and total cellular lysates were prepared. Formation of the GATA-1–c-Myb complex was examined by coimmunoprecipitation method. (B-D) The influence of the interaction GATA-1–PU.1 and GATA-1–c-Myb on the respective DNA-binding activities. Nuclear extracts were isolated from 293T cells transfected with various amounts of GATA-1, PU.1, or c-myb expression vectors as indicated. DNA-binding activities of GATA-1 and PU.1 were examined with the probe of Mα and MHC, respectively. In competition assays, nuclear extracts were preincubated with a 200-fold molar excess of unlabeled competitor oligonucleotides before the binding reaction. In a supershift assay, the nuclear proteins were preincubated with 1 μg anti-GATA-1 antibody or anti-PU.1 antibody for 30 minutes at 4°C and subjected to the binding reaction (B, DNA-binding complex; SS, supershifted DNA-binding complex). Expression levels of GATA-1 and PU.1 were also examined by Western blot analysis in each transfectant.
Mechanisms of reciprocal inhibition, GATA-1–PU.1, and GATA-1–c-Myb.
(A) GATA-1 and c-Myb form the complex in vivo. 293T cells were transfected with GATA-1 or PU.1, and total cellular lysates were prepared. Formation of the GATA-1–c-Myb complex was examined by coimmunoprecipitation method. (B-D) The influence of the interaction GATA-1–PU.1 and GATA-1–c-Myb on the respective DNA-binding activities. Nuclear extracts were isolated from 293T cells transfected with various amounts of GATA-1, PU.1, or c-myb expression vectors as indicated. DNA-binding activities of GATA-1 and PU.1 were examined with the probe of Mα and MHC, respectively. In competition assays, nuclear extracts were preincubated with a 200-fold molar excess of unlabeled competitor oligonucleotides before the binding reaction. In a supershift assay, the nuclear proteins were preincubated with 1 μg anti-GATA-1 antibody or anti-PU.1 antibody for 30 minutes at 4°C and subjected to the binding reaction (B, DNA-binding complex; SS, supershifted DNA-binding complex). Expression levels of GATA-1 and PU.1 were also examined by Western blot analysis in each transfectant.
Next, we examined whether the interaction between GATA-1 and PU.1 affects the respective DNA-binding activities. Nuclear extracts were isolated from 293T cells transfected with GATA-1 and PU.1 expression vectors. Although the nuclear extract prepared from mock (an empty expression vector)-transfected 293T cells did not show any binding activity to the probe for GATA-1 (Mα) (Figure 8B, lane 1), the nuclear extract from GATA-1–transfected 293T cells formed the DNA-binding complex (lane 2). This complex was effectively erased by an excessive amount of WT Mα competitor (lane 3) but not by MT Mα competitor (lane 4), suggesting that the complex was formed in a sequence-specific manner. In addition, this complex was super-shifted by anti–GATA-1 antibody (lane 5), indicating that this complex was formed from GATA-1. When an expression vector of PU.1 was cotransfected with that of GATA-1, the amounts of GATA-1 binding to the probe were significantly reduced, depending on the doses of PU.1 expression vector (lanes 6 to 9), though GATA-1 was expressed equally in each nuclear extract (Figure 8B, lower panel). Similarly, the sequence-specific DNA binding of PU.1 to the MHC probe was detected in the nuclear extracts from PU.1-transfected 293T cells with the competition and supershift assays (Figure 8C, lanes 2 to 5). DNA binding of PU.1 was partially inhibited by the cotransfected GATA-1 expression vector in a dose-dependent manner (Figure 8C, lanes 6 to 9), despite the same levels of PU.1 expression in each transfectant (Figure 8C, lower panel). However, GATA-1 was less effective in inhibiting DNA-binding activities of PU.1 because a significant amount of PU.1 could still bind to the MHC probe. Therefore, it was speculated that, besides inhibiting DNA-binding activities of PU.1, GATA-1 might suppress PU.1 activities through additional mechanisms. Furthermore, as a result of the interaction, c-Myb inhibited DNA-binding activities of GATA-1 in a dose-dependent manner (Figure 8D).
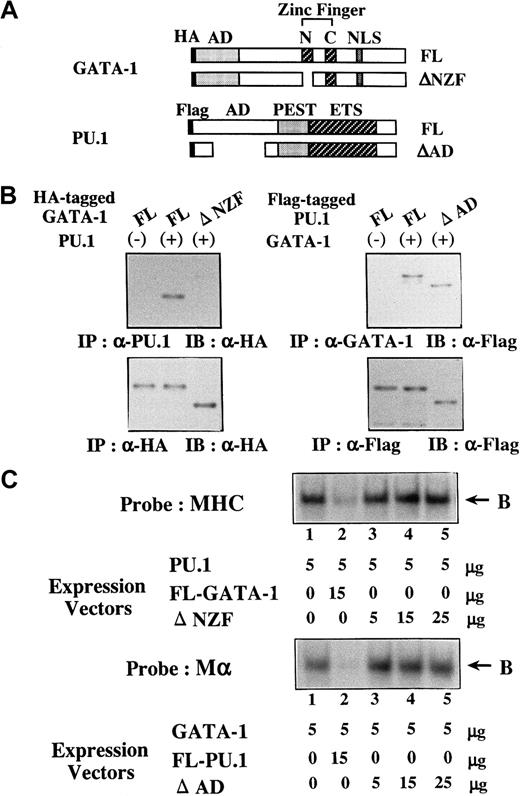
Characterization of ΔNZF-GATA-1 and ΔAD-PU.1
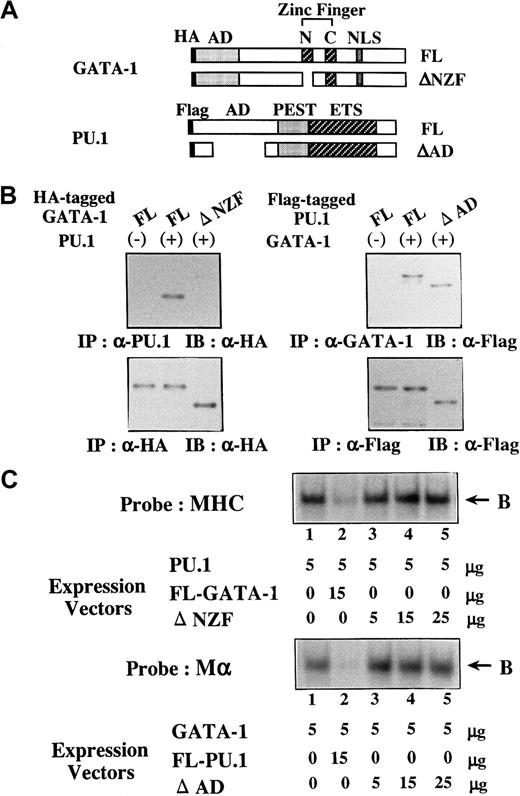
To further assess the biologic significance of antagonizing effects between GATA-1 and PU.1, we generated several mutants of PU.1 and GATA-1. Of these mutants, ΔNZF-GATA-1, which lacks N-zinc finger (NZF) of GATA-1 (Figure 9A), had little suppressive effect on PU.1 activities, whereas its transcriptional activity on 3× WT-Mα-Luc was almost similar to that of full-length (FL)-GATA-1 (data not shown). In addition, ΔAD-PU.1, which lacks a part of the activating domain (AD) of PU.1 (Figure 9A), hardly affected GATA-1 activities but showed transcriptional activity on 3× WT-MHC-Luc approximately 50% that of FL-PU.1 (data not shown). To characterize these mutants, the formation of the GATA-1–PU.1 complex was examined by coimmunoprecipitation. We transfected FL-PU.1 together with HA-tagged FL-GATA-1 or HA-tagged ΔNZF-GATA-1 into 293T cells. Although FL-GATA-1 and ΔNZF-GATA-1 were expressed at similar levels on Western blot analysis of the anti-HA–immunoprecipitated proteins with anti-HA antibody (Figure 9B, left, lower panel), FL-GATA-1 but not ΔNZF-GATA-1 was coimmunoprecipitated with PU.1 (Figure 9B, left, upper panel). We transfected FL-GATA-1 in combination with Flag-tagged FL-PU.1 or Flag-tagged ΔAD-PU.1 into 293T cells and found that both FL-PU.1 and ΔAD-PU.1 were coimmunoprecipitated with GATA-1 with similar efficiency (Figure 9B, right, upper panel). Next, we prepared nuclear extracts from 293T cells transfected with these expression vectors. Although FL-GATA-1 significantly inhibited the DNA binding of PU.1, ΔNZF-GATA-1 could scarcely affect the DNA binding (Figure 9C, upper panel). In contrast to FL-PU.1, ΔAD-PU.1 scarcely inhibited the DNA binding of GATA-1 (Figure 9C, lower panel), though ΔAD-PU.1 could bind to GATA-1 as efficiently as FL-PU.1 (Figure 9B, right upper panel). These results suggested that NZF of GATA-1 is necessary for its binding to PU.1 and its inhibition of the DNA-binding activities of PU.1 and that the AD of PU.1 is required for the inhibition of the DNA-binding activities of GATA-1 but not for the interaction between GATA-1 and PU.1.
Characterization of ΔNZF-GATA-1 and ΔAD-PU.1.
(A) Structure of ΔNZF-GATA-1 and ΔAD-PU.1. (B) Coimmunoprecipitation analyses on the binding activities of ΔNZF-GATA-1 to PU.1 and ΔAD-PU.1 to GATA-1. 293T cells were transfected with HA-tagged FL-GATA-1, HA-tagged ΔNZF-GATA-1, Flag-tagged FL-PU.1, and Flag-tagged ΔAD-PU.1 in various combinations as indicated. Formation of the GATA-1–PU.1 complex was examined by coimmunoprecipitation. (C) Effects of ΔNZF-GATA-1 and ΔAD-PU.1 on the DNA-binding activities of PU.1 and GATA-1, respectively. 293T cells were transfected with ΔNZF-GATA-1, FL-GATA-1, ΔAD-PU.1, and FL-PU.1 in various combinations at the doses indicated. Effects of ΔNZF-GATA-1 (upper panel) and ΔAD-PU.1 (lower panel) on the DNA-binding activities of PU.1 and GATA-1 were examined with the probe of MHC and Mα, respectively. B, DNA-binding complex.
Characterization of ΔNZF-GATA-1 and ΔAD-PU.1.
(A) Structure of ΔNZF-GATA-1 and ΔAD-PU.1. (B) Coimmunoprecipitation analyses on the binding activities of ΔNZF-GATA-1 to PU.1 and ΔAD-PU.1 to GATA-1. 293T cells were transfected with HA-tagged FL-GATA-1, HA-tagged ΔNZF-GATA-1, Flag-tagged FL-PU.1, and Flag-tagged ΔAD-PU.1 in various combinations as indicated. Formation of the GATA-1–PU.1 complex was examined by coimmunoprecipitation. (C) Effects of ΔNZF-GATA-1 and ΔAD-PU.1 on the DNA-binding activities of PU.1 and GATA-1, respectively. 293T cells were transfected with ΔNZF-GATA-1, FL-GATA-1, ΔAD-PU.1, and FL-PU.1 in various combinations at the doses indicated. Effects of ΔNZF-GATA-1 (upper panel) and ΔAD-PU.1 (lower panel) on the DNA-binding activities of PU.1 and GATA-1 were examined with the probe of MHC and Mα, respectively. B, DNA-binding complex.
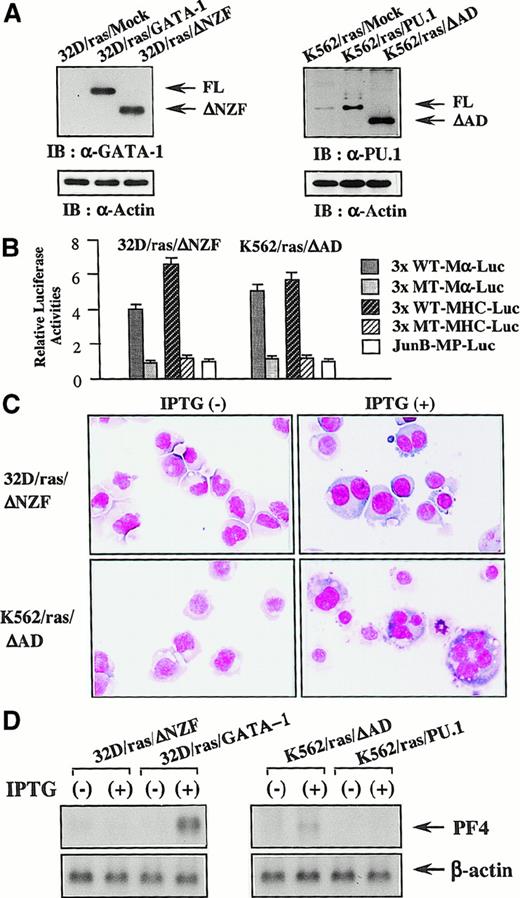
ΔNZF-GATA-1 and ΔAD-PU.1 were less effective in reprogramming the lineage of 32D cl3 and K562
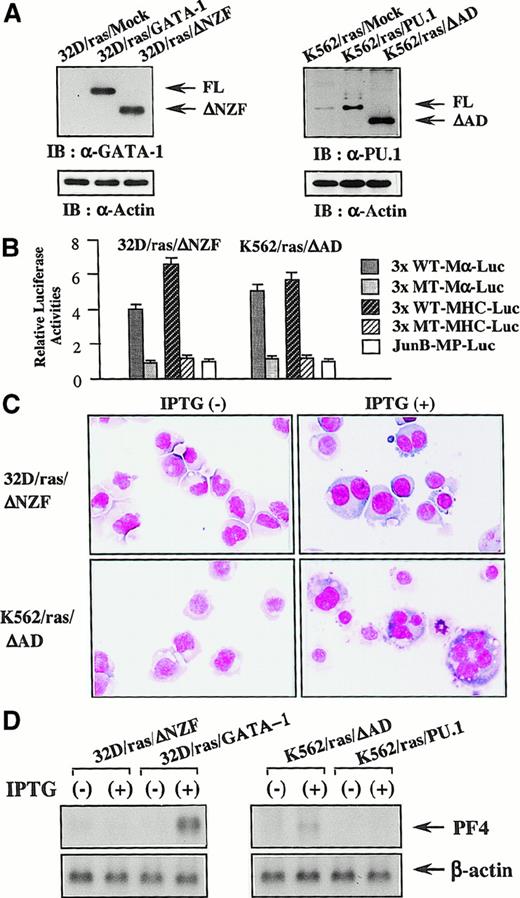
Next, we introduced ΔNZF-GATA-1 into 32D/ras and ΔAD-PU.1 into K562/ras; these clones were named 32D/ras/ΔNZF and K562/ras/ΔAD, respectively. Western blot analysis of 32D/ras/ΔNZF and K562/ras/ΔAD showed that ΔNZF-GATA-1 and ΔAD-PU.1 were expressed in each clone at similar or increased levels compared with full-length products (Figure 10A). GATA-1 and PU.1 activities were detected in 32D/ras/ΔNZF (GATA-1 activities, 4.1-fold; PU.1 activities, 6.3-fold, with reference to the respective nonfunctional reporter gene; Figure 10B), whereas PU.1 activities were severely reduced in 32D/ras/GATA-1 (GATA-1 activities, 3.8 fold; PU.1 activities, 1.4 fold; Figure 7A). PU.1 and GATA-1 activities were detected in K562/ras/ΔAD (GATA-1 activities, 5.2-fold; PU.1 activities, 5.6 fold; Figure 10B), whereas GATA-1 activities were intensely inhibited in K562/ras/PU.1 (GATA-1 activities, 1.2-fold; PU.1 activities, 5.8 fold; Figure 7A). Although the induced expression of H-rasG12V for 5 days led to the development of fully matured megakaryocytes from 32D/ras/GATA-1 (Figure 3A), it was less effective in promoting maturation of 32D/ras/ΔNZF (Figure 10C, upper panel). In addition, IPTG treatment failed to induce PF4 mRNA in 32D/ras/ΔNZF, whereas it was effectively induced in 32D/ras/GATA-1 (Figure 10D, left panel). CD61 induction and polyploid formation were perturbed distinctly in 32D/ras/ΔNZF (data not shown). In K562/ras/ΔAD morphology, 5-day treatment with IPTG still induced megakaryocytic maturation (Figure 10C, lower panel), whereas K562/ras/PU-1 underwent granulocytic maturation (Figure 4C). In accordance with the morphologic findings, PF4 mRNA was induced by IPTG treatment in K562/ras/ΔAD but not in K562/ras/PU-1 (Figure 10D, right panel). Together, these results suggested that ΔNZF-GATA-1 and ΔAD-PU.1, which respectively had little inhibitory activity on PU.1 and GATA-1, were less effective in reprogramming the lineage phenotype in spite of the retained transcriptional activities. Similar findings on luciferase assays and responses to IPTG were also observed in other stable transformants with ΔNZF-GATA-1 and ΔAD-PU.1 (data not shown).
Reprogramming activities of ΔNZF-GATA-1 and ΔAD-PU.1.
(A) Expression of ΔNZF-GATA-1 in 32D/ras/ΔNZF and ΔAD-PU.1 in K562/ras/ΔAD. Total cell lysates obtained from each clone were subjected to SDS-PAGE, and the blot was probed with anti-GATA-1 antibody (left panel) or anti-PU.1 antibody (right panel). Expression levels of actin were examined as a loading control. (B) GATA-1 and PU.1 activities in 32D/ras/ΔNZF and K562/ras/ΔAD. Twenty micrograms of each reporter gene was cotransfected with 3 μg pRL-CMV-Rluc by electroporation. After 36 hours, the cells were subjected to the luciferase assays. (C) Light micrograph of 32D/ras/ΔNZF and K562/ras/ΔAD before and after 5-day culture with IPTG. Cells of each clone were cultured with or without IPTG for 5 days. Cytocentrifugation preparation from each culture was stained with May–Grünwald–Giemsa (original magnification, ×100). (D) Changes of PF4 mRNA expression before and after 5-day IPTG treatment. Total cellular RNA was extracted from 32D/ras/ΔNZF, 32D/ras/GATA-1, K562/ras/ΔAD, and K562/ras/PU.1 before and after IPTG treatment, and PF4 mRNA expression was examined by Northern blot analysis.
Reprogramming activities of ΔNZF-GATA-1 and ΔAD-PU.1.
(A) Expression of ΔNZF-GATA-1 in 32D/ras/ΔNZF and ΔAD-PU.1 in K562/ras/ΔAD. Total cell lysates obtained from each clone were subjected to SDS-PAGE, and the blot was probed with anti-GATA-1 antibody (left panel) or anti-PU.1 antibody (right panel). Expression levels of actin were examined as a loading control. (B) GATA-1 and PU.1 activities in 32D/ras/ΔNZF and K562/ras/ΔAD. Twenty micrograms of each reporter gene was cotransfected with 3 μg pRL-CMV-Rluc by electroporation. After 36 hours, the cells were subjected to the luciferase assays. (C) Light micrograph of 32D/ras/ΔNZF and K562/ras/ΔAD before and after 5-day culture with IPTG. Cells of each clone were cultured with or without IPTG for 5 days. Cytocentrifugation preparation from each culture was stained with May–Grünwald–Giemsa (original magnification, ×100). (D) Changes of PF4 mRNA expression before and after 5-day IPTG treatment. Total cellular RNA was extracted from 32D/ras/ΔNZF, 32D/ras/GATA-1, K562/ras/ΔAD, and K562/ras/PU.1 before and after IPTG treatment, and PF4 mRNA expression was examined by Northern blot analysis.
Discussion
During the development of hematopoietic cells from pluripotent stem cells to terminally differentiated cells, lineage-specific transcription factors play crucial roles in lineage determination and subsequent maturation through the transcriptional regulation of lineage-specific genes. In a previous study,17 we found that TPO-induced Ras activation was necessary and sufficient for promoting megakaryocytic differentiation of a human erythroleukemia cell line. However, Ras activation is not specific for megakaryocytic differentiation; it is also observed in other cell types, and it evokes various effects in the cells.20-22 Therefore, it was speculated that there might be a key molecule that is prerequisite for Ras-mediated megakaryocytic differentiation. In previous studies, it has been shown that GATA-1 is unique in its ability to influence the lineage phenotype of hematopoietic cells. For example, enforced expression of GATA-1 reprogrammed transformed chicken myeloblasts into erythroblasts, thromboblasts, or eosinophils.25 Moreover, the overexpression of GATA-1 in the early myeloid 416B and M1 cells provoked megakaryocytic differentiation accompanied by a marked decrease in myeloid surface phenotype.26 27 These findings led us to speculate that GATA-1 might confer the characteristics that facilitate the cells to undergo megakaryocytic differentiation in response to H-rasG12V. In agreement with this hypothesis, ectopically introduced GATA-1 reprogrammed myeloid 32D cl3 into an erythroid/megakaryocytic lineage and caused 32D cl3 to undergo megakaryocytic differentiation in response to H-rasG12V. These results suggested that GATA-1 is one of the key molecules necessary for megakaryocytic differentiation, whereas the direct target gene(s) of GATA-1 remains unknown. In addition, because our preliminary experiments showed that H-rasG12V enhanced GATA-1 activities approximately 2.5-fold, it was suggested that H-rasG12V-evoked megakaryocytic differentiation might result from these augmented GATA-1 activities or from the cooperation of GATA-1 and H-rasG12V-induced unidentified product(s).
PU.1 is known as a pivotal transcription factor involved in the commitment and terminal differentiation of myeloid cells.11,31 It has been reported that constitutive expression of PU.1 inhibits terminal erythroid differentiation of the murine cell line MEL.32-34 The enforced expression of c-Myb, in addition to PU.1, was shown to block the erythroid differentiation of MEL cells.35 36 In this study, we examined whether these transcription factors could affect H-rasG12V–induced megakaryocytic differentiation of K562. As a result, the overexpression of PU.1 and c-Myb changed the phenotype of K562 to myeloid/monocyte lineage and caused K562 to differentiate into granulocytes and macrophages, respectively, in response to H-rasG12V.
For this mechanism, the transcription activity of GATA-1 was antagonized by either PU.1 or c-Myb, and transcriptional activities of PU.1 or c-Myb were also suppressed by GATA-1. Although the expression patterns of lineage-specific transcription factors are known to be regulated reciprocally during the differentiation programs,37 it is also reported that conflicting lineage-specific transcription factors, such as GATA-1 and PU.1, coexist in multipotent hematopoietic progenitor cells.38Therefore, it was suggested that relative levels of these transcription factors may have a great influence on progenitor cells to commit and differentiate toward myeloid or erythroid/megakaryocyte lineage.
Our data also showed that, compared with wild types, ΔNZF-GATA-1 and ΔAD-PU.1 had less activity in switching the lineage phenotypes of 32D cl3 and K562. The deletion mutants, ΔNZF-GATA-1 and ΔAD-PU.1, still retained individual transcriptional activities but showed little or no inhibitory effect on PU.1 and GATA-1, respectively. In a previous study, Visvader et al39 also generated the murine N-zinc finger-defective GATA-1, corresponding to our human ΔNZF-GATA-1, and demonstrated that their mutant GATA-1 was less effective in changing the lineage phenotype and imposing megakaryocytic differentiation of a myeloid cell line 416B, whereas the mutant revealed the similar level of transcriptional activities to full-length GATA-1. Together with their findings, our results raised the possibility that GATA-1 and PU.1 may elicit their full biologic effects not only through the transcriptional activities but also through the antagonizing activities on the conflicting transcription factor.
Regarding the mechanisms of the reciprocal inhibition, GATA-1 and PU.1 were reported to associate in vivo,28-30 and we found similar results in the current study. In this association, the respective DNA-binding domains (zinc finger domains of GATA-1 and Ets domain of PU.1) were to be used as a surface of protein–protein interaction. In previous studies,40-50 the zinc finger domains of GATA-1 and the Ets domain of PU.1 were reported to interact with various types of nuclear proteins: FOG, CREB-binding protein, Sp1, EKLF, U-shaped, and the estrogen receptor for GATA-1; C/EBPα, C/EBPδ, AML1, c-fos, c-jun, and Pip for PU.1. In most cases, GATA-1 and PU.1 activities are reported to be positively or negatively regulated through these interactions, though limited information is available for the biologic significance of the GATA-1–PU.1 complex in this protein assembly. Our data revealed that, as a result of the interaction, DNA-binding activities of GATA-1 and PU.1 were inhibited reciprocally by electrophoretic mobility shift assay. However, GATA-1 was less effective than PU.1 in inhibiting DNA-binding activities of PU.1. More mechanisms might be involved in the suppression of PU.1 activities by GATA-1. In addition, ΔAD-PU.1, which was capable of binding to GATA-1, did not reveal any opposing effects on GATA-1 activities. Although it was able to associate with PU.1 (data not shown), del C80-GATA-1, a deletion mutant of the C-terminal 80 amino acid, was less effective in antagonizing PU.1 activities. Therefore, an as yet unidentified mechanism may be involved in the reciprocal inhibition of transcriptional and DNA-binding activities between GATA-1 and PU.1.
It has been shown that several megakaryocyte-specific genes, such as c-mpl, GPIbα, GPIIb, and GPIX, contain several neighboring Ets-binding sites and GATA-binding motifs in their promoters.51-55 In addition, transient reporter assays demonstrated that these promoters are actually transactivated by Ets and GATA family proteins individually and cooperatively, suggesting that both Ets family proteins and GATA family proteins may contribute to megakaryocyte differentiation.51-55 In contrast to the previous transient assays, however, the expression level of GPIIb was not affected in PU.1-transfected K562 even before the IPTG treatment (granulocytic differentiation). Moreover, besides GATA-1 and PU.1, reciprocal inhibition was also observed between other members of GATA and Ets family proteins. Therefore, special regard should be paid to the reciprocal inhibition of GATA family and Ets family proteins for the assessment of promoter activities, especially by using megakaryocytes.
In agreement with our result indicating the mutual negative regulation between GATA-1 and PU.1, Zhang et al28 have recently shown that both GATA-1 and GATA-2 interact with PU.1 through the C-terminal zinc finger, thereby resulting in reciprocal inhibition. Furthermore, they have shown that GATA-1 inhibits the binding of PU.1 to c-jun, a critical coactivator of PU.1. Rekhtman et al29 and Nerlov et al30 have recently demonstrated that GATA-1 and PU.1 directly interact through both zinc fingers of the GATA-1 and Ets domains of PU.1, which led to functional antagonism of these factors.29,30 In addition, Rekhtman et al29 have shown that PU.1 inhibits erythroid differentiation of a murine erythroid cell line, MEL, by repressing GATA-1 activities. These results, including ours, suggested that direct interaction of these lineage-specific transcription factors profoundly affects the activities of other lineage-specific transcription factors and influences the lineage commitment and subsequent differentiation of hematopoietic cells. Further studies concerning the interaction between GATA-1 and PU.1 using our system should provide more useful information on normal and abnormal hematopoiesis.
Acknowledgment
We thank Dr S. Ishii for providing us with an expression vector of c-myb.
Supported in part by grants from the Japanese Ministry of Education, Science, Sports and Culture, the Japanese Ministry of Health and Welfare, Senri Life Science Foundation, Uehara Memorial Foundation, Naito Foundation, and the Japan Medical Association.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Itaru Matsumura, Department of Hematology and Oncology, Osaka University Medical School, 2-2, Yamada-oka, Suita, Osaka 565, Japan; e-mail: matsumura@bldon.med.osaka-u.ac.jp.