Abstract
The role of chemokine–matrix interactions in integrin-dependent T-cell migration was examined to address the critical question of how chemokines provide directional information. The chemokine SDF-1α binds fibronectin (Fn) with a low nanomolar Kd(equilibrium dissociation constant). SDF-1α presented by Fn induced directed migration. Spatial concentration gradients of chemokine were not required to maintain directed migration. Fn-presented chemokine induced the polarization of cells, including the redistribution of the SDF-1α receptor, to the basal surface and leading edge of the cell. A new model for directed migration is proposed in which the co-presentation of an adhesive matrix and chemokine provides the necessary positional information independent of a soluble spatial gradient.
Introduction
The importance of chemokines is apparent in the complexity of the roles played by these molecules and their receptors, members of the G-protein–linked 7-transmembrane receptor family.1 In addition to the critical role of these molecules in leukocyte traffic and development in the immune system,2,3 chemokine receptors are widely expressed and have broader functions than previously thought.1 For example, stromal cell–derived factor 1α (SDF-1α) and its receptor, CXCR4,4 have been exploited by T-cell tropic strains of human immunodeficiency virus (HIV) such that CXCR4 is a coreceptor for viral entry and infection by these strains. SDF-1α and a ligand analog that binds CXCR4 or CCR5 block HIV entry into T cells.4,5 It has been reported that CXCR4 is expressed on endothelial cells.6 Although its role in endothelial cell biology is not yet known, null mutations to CXCR4 or SDF-1α in the mouse are homozygous lethal and show defects in the vascularization of the gastrointestinal tract and profound hematopoietic and neural defects.7-9
Chemokines promote directed cell migration, such as that of leukocytes, into sites of inflammation. The existing paradigm holds that chemokines diffuse away from secreting cells to establish a soluble gradient. Receptive cells some distance from the source then detect the gradient of chemokine through either a spatial or a temporal mechanism10 11 and migrate up the gradient to the source.
This model has some limitations, most notably in a flowing medium such as blood, where establishing such a gradient would seem precluded. It has been reported that chemokines and similar molecules, including tumor necrosis factor α, IL-8, and MIP-1β, can bind to either extracellular matrix (ECM) or cell surfaces by glycosaminoglycans such as heparins or receptors such as Duffy antigen/receptor for chemokines.12-18 This has led to the proposal that chemokines deposited on cells or on ECM may induce migration and, therefore, may allow them to function under flowing conditions.3
Under nonflowing conditions, the nature of diffusion limits the distance from the secreting cell at which effective doses of the chemokine can be established (estimated at 250 μm19). In a simple diffusion model, the concentration of a chemokine in solution will decrease as an inverse-square function of the distance from its source. Accordingly, the gradient will be shallow at a distance from the source and increasingly steep the closer the cell moves toward the source. Thus, the prediction is that the cell will respond to both shallow and steep gradients over a relatively wide range of concentrations. However, in vitro data demonstrate that cell migration is inhibited by relatively small increases in concentrations over those optimal for promoting migration4 20 (also see below). Interactions of chemokines with exposed ECM or cell surface macromolecules could regulate diffusion and, hence, the exposure of migrating cells to chemokine gradients.
In the present work, we examined the ability of SDF-1α to promote the migration of the human T-cell line Jurkat on fibronectin (Fn). We found that SDF-1α bound to Fn specifically and saturably. Moreover, SDF-1α bound by Fn was extremely efficient at promoting migration in the absence of any soluble SDF-1α. Thus, SDF-1α is presented by the matrix in a functional manner that is sufficient to support directed migration. The CXCR4 receptor polarized to the leading edge of migrating cells, especially the basal surface in contact with the matrix and the presented chemokine. Based on these data we propose that chemokine-directed migration does not absolutely require a soluble gradient; rather, cells can receive spatial information directly from matrix-presented chemokine.
Materials and methods
Binding assays
Surface plasmon resonance.
Fibronectin (Gibco/BRL, Rockville, MD) at a concentration of 5 μg/mL was coupled to a Biacore (San Diego, CA) CM-5 chip by primary amines using the EDC/NHS (1-ethyl-3-(3-dimethylaminopropyl) carbodiimide/N-hydroxysuccinimide) chemistry protocol recommended by the manufacturer. The control flow cell had no protein coupled to it. After washes, coupling of 6423 RU (1000 RU = 1 ng/mm2) of Fn was achieved. SDF-1α, at concentrations of 128 nmol/L or 256 nmol/L, was injected in several separate runs, and on-rates and off-rates were estimated using Bia-eval software (Biocore). Between runs, the flow cells were washed for at least 10 minutes with 1 mol/L NaCl to remove any remaining SDF-1α, and they were re-equilibrated with analyte buffer. Analysis was performed to estimate the number of binding-site classes and, at these concentrations of SDF-1α, only 1 class was detected. At higher concentrations of SDF-1α, additional classes of lower affinity sites were revealed.
Direct equilibrium binding.
Sodium iodide I 125–SDF-1α was generated from chemically synthesized SDF-1α by the Iodobead method (Pierce, Rockford, IL) and purified from free 125I on a PD-10 column (Amersham Pharmacia, Piscataway, NJ). Protein concentration was determined by the BCA protein assay method (Pierce), and specific activity of the product was 1.28 × 103 cpm/pmol SDF-1α (measured on a Wallac 1480 automatic γ counter with a counting efficiency for 125I of 80%, yielding a calculated 1.6 × 103dpm/pmol).
For the solid-phase assay, noncharged polystyrene tubes (Pharmingen, San Diego, CA) were coated with 250 μL of a 20 nmol/L solution of Fn in phosphate-buffered saline (PBS) for 4 hours at 37°C. Tubes were washed with PBS and exposed to 100 μL of 125I–SDF-1α solution in PBS for 1 hour at room temperature. Specific binding reached a maximum by 30 minutes, and incubation times of up to 2 hours were used with no increase in specific binding. Tubes were washed 6 times with PBS/0.1% Tween-20 and counted in the γ counter.
EMSA.
Electrophoretic mobility shift assay (EMSA) was performed by mixing the indicated concentrations of 125I–SDF-1α and ECM protein in PBS at room temperature for 1 hour, followed by electrophoresis under native conditions (Tris–glycine buffer; Bio-Rad, Hercules, CA) in a 4% to 20% polyacrylamide gel (Novex, San Diego, CA). The gel was then fixed, dried, and exposed to a storage-fluor phosphor-imaging screen. Bands were visualized and quantified on a Cyclone PhosphorImager (Packard Instruments, Meriden, CT). To generate a standard curve for quantification, a sample treated identically to those described were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), fixed, dried, and exposed to the PhosphorImager plate at the same time as the native gel. A plot of the input SDF-1α concentration versus the PhosphorImager “counts” present in the SDF-1α on this gel resulted in the standard curve.
Transwell migration assays
The Transwell (Corning Costar, Cambridge, MA) assay was performed essentially as described previously.21 To coat only one side of the membrane, dry inserts were placed in 24-well plate wells, each of which contained a 75-μL drop of 20 nmol/L Fn. This procedure allowed not only the bottom of the membrane to be coated but the insides of pores as well because of the capillary action. However, the top of the membrane is not wet in this process. After a 1-hour incubation at 37°C, the inserts were coated similarly with the indicated concentrations of SDF-1α for 1 hour at 37°C and were washed in PBS; finally, the entire insert was coated top and bottom with Fn. The top and bottom chambers were filled with AIM-V serum-free medium (Gibco/BRL), and cells were inoculated into the top chamber and allowed to migrate for 4 hours or as indicated. Migrated cells were detected by counting cells in the bottom chamber with a Coulter counter model Z-1. Alternatively, inserts were coated with Fn, and soluble SDF-1α was added at the indicated concentrations to the bottom chamber before the addition of cells.
Adhesion assay
We used the adhesion assay of Calof and Lander22 as modified previously by us.21 Briefly, a silicone gasket in the format of a 96-well manifold is used to form 20-μL microwells on a plastic plate. The plastic in the wells is coated with the appropriate matrix and SDF-1α concentration as described for the migration assays, followed by blocking with 1% heat-denatured bovine serum albumin. 35S-methionine–labeled cells are deposited in the microwells and centrifuged briefly to bring them in contact with the plate. After adhesion at 37°C for 1 hour, the top of the gasket is sealed with a second plate, inverted, and centrifuged for 5 minutes at the indicated G-force to remove nonattached cells. The top plate is removed while immersed in PBS to allow nonadhered cells to float away; the remaining cells are fixed with 10% formalin for 15 minutes, air dried, exposed to a phosphor-imaging screen, and quantitated on a Molecular Dynamics PhosphorImager with Imagequant software (Amersham Pharmacia). Results are presented as percentage of input cells that remain adhered. For each point, the average ±1 SD is presented (n = 6).
Stripe-migration assay and confocal microscopy
We modified the assay of Calof et al23 to use the silicone gaskets to deposit a stripe of SDF-1α–coated Fn on a coverslip surrounded by uniform Fn. Cells were then deposited in a drop either at one end of the stripe of SDF-1α or at a point several millimeters away from the stripe of SDF-1α. Adhesion was allowed to proceed at 37°C for 1 hour. Coverslips were immersed in AIM-V medium and incubated for 2 additional hours at 37°C. In some experiments, the second incubation was performed in a flow chamber with a flow rate of 3 μL/(min · mm2) orthogonal to the direction of migration. Cells then were fixed and stained for photography under light microscopy using the Leukostat staining kit (Fisher Diagnostics, Tustin, CA) or fixed in the CS buffer of Conrad et al24 for antibody staining and confocal microscopy. For confocal microscopy, coverslips were blocked first in PBS/50 mmol/L glycine and then in PBS/5% donkey serum for 1 hour at room temperature. Cells were stained with a primary monoclonal mouse antihuman CXCR4 antibody (clone 12G5; Pharmingen) for 1 hour at room temperature (2.5 μg/500 μg) and, after washing, were detected with a donkey antimouse LSRC-labeled secondary antiserum (Jackson Laboratories, West Grove, PA). Controls were made with isotype and species-matched antibodies. After mounting in an antifade medium (Immuno Fluor; ICN, Costa Mesa, CA) the cells were analyzed on a Zeiss Axiovert microscope (Carl Zeiss, Thornwood, NY) with a scanning laser confocal attachment (MRC 1024; Bio-Rad) at 400× magnification.
Quantification of polarization
The image presented in Figure 4 was imported into Imagequant software (Molecular Dynamics) as a TIFF file. A 4-sector, pie-shaped grid was placed over each cell and oriented so that the number of pixels in the north, east, west, and south quadrants could be quantified. These data were collected for every cell in the image that could be unambiguously separated from neighboring cells (n = 55 for cells on Fn alone, and n = 54 for cells on Fn-presented chemokine). The pixel value for each direction was divided by that of the opposite direction either one-by-one (eg, north/south) or two-by-two [eg, (north+west)/(south+east)]—the latter to ensure that cells polarized between 2 primary compass directions were not missed. The largest ratio for each cell was taken as the polarization index. A cell was considered polarized if the polarization index was 3.0 or greater. A polarization index of 3.0 as the cutoff for polarity was chosen because the nonpolarized cells defined an obvious group in which the mean value for the polarization index shown for each cell was 1.8 ± 0.4 SD. Thus, the cutoff for polarization was 3 SD above the mean polarization index for the nonpolarized group and allowed us a greater than 99% confidence level that they represented different distributions.
Determination of soluble SDF-1α concentration
For enzyme-linked detection of soluble SDF-1α, we used a plate-binding assay and SDF-1α synthesized with a single biotin molecule added to the side-chain amine of K68. Known input concentrations of SDF-1α-biotin were placed in the wells of Costar EIA (enhanced immunoabsorbance) plates that were precoated with fibronectin or other matrices and incubated for 1 hour at 37°C. After incubation, the plates were washed 3 times with PBS and the bound chemokine molecules detected with streptavidin-conjugated horseradish peroxidase (Gibco BRL) and ABTS peroxidase substrate chromogen (Zymed Laboratory, South San Francisco, CA), according to the supplier's instructions. The absorbance was read at 405 nm. We generated a standard curve from these values and determined that our detection limit was 1 nmol/L. Simultaneously, the medium from the bottom of Transwell plates to which various concentrations of SDF-1α-biotin had been added was subjected to the same procedure, and OD 405 from these samples was compared to the standard curve.
In a separate assay, 125I–SDF-1α as described above was incubated in triplicate at various concentrations in the bottom of a Transwell chamber previously coated with Fn as described. After 1 hour, aliquots were removed from the bottom or top chamber and counted in the γ counter. Actual concentrations were determined by specific activity and by direct comparison to standard concentrations counted at the same time. The amount of free 125I in the preparation was determined by subjecting aliquots to SDS-PAGE and following that by fixing, drying, and quantifying gels with the Cyclone PhosphorImager. Approximately 4% of the total 125I was present as free salt.
Results
SDF-1α binds to Fn
SDF-1α, made by complete chemical synthesis,25binds specifically and saturably to Fn with a low nanomole per literKd (equilibrium dissociation constant) as determined both by surface plasmon resonance and by 2 equilibrium binding assays. Using surface plasmon resonance, we determined on and off rates for several concentrations of SDF-1α. These values were between 2.5 and 2.6 × 105 (mol/L)−1sec−1 for the on rate and between 4.5 and 6.5 × 10−3 sec−1 for the off rate (SD < 10%, all determinations). From these kinetic parameters, we calculated a thermodynamic Kd of approximately 20 nmol/L.
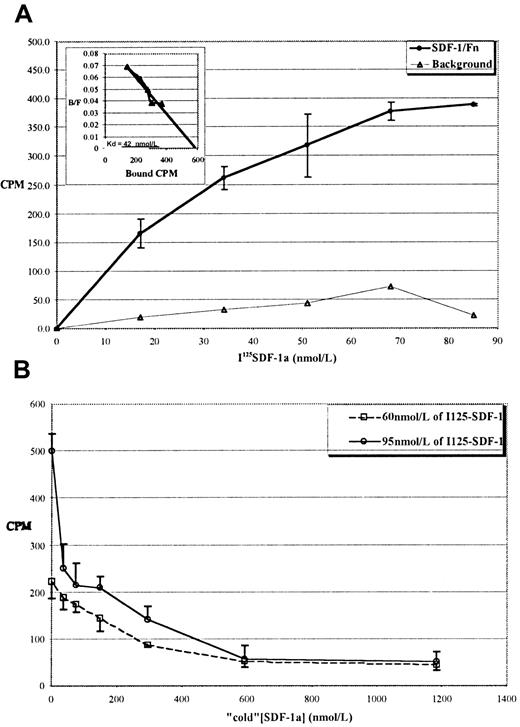
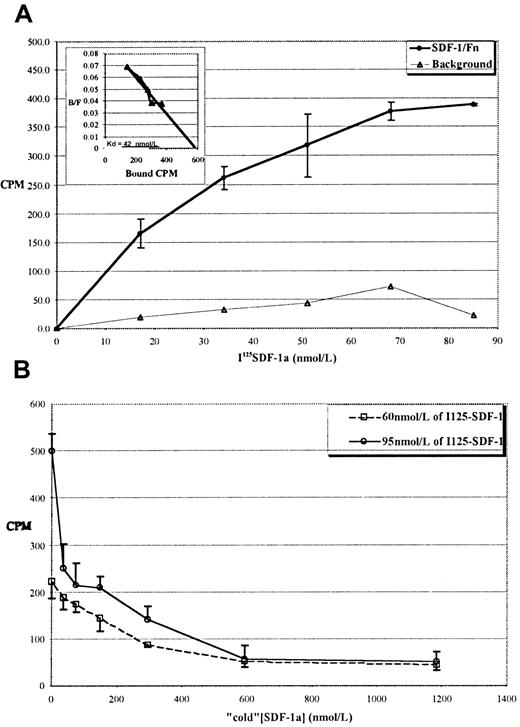
We next performed equilibrium-binding studies with125I-labeled SDF-1α as an independent measure of theKd. Figure 1A shows a typical binding curve for the interaction of 125I SDF-1α with immobilized Fn. Inspection of the graph shows half-maximal binding to be at approximately 20 nmol/L, and Scatchard analysis of the data (inset) revealed a Kd of approximately 40 nmol/L. This binding was inhibitable by excess “cold” SDF-1α, as shown in Figure 1B.
Specific binding of 125I SDF-1α to immobilized Fn shows a 40 nmol/L Kd.
(A) Typical equilibrium-binding isotherm for 125I–SDF-1α to Fn-coated tubes. ○: 125I–SDF-1α binding to Fn. ▵: 125I–SDF-1α binding to noncoated tubes. Values reported are the mean of 4 independent assays for each concentration ±1 SD. Inset: Scatchard transform of the data in the main panel, showing a calculated Kd of approximately 40 nmol/L. (B) Binding is inhibited by “cold” SDF-1α. Two concentrations of 125I–SDF-1α (60 or 95 nmol/L) were incubated in Fn-coated tubes with increasing concentrations of cold SDF-1α. Values reported for both panels are the mean of 4 independent assays for each concentration ±1 SD.
Specific binding of 125I SDF-1α to immobilized Fn shows a 40 nmol/L Kd.
(A) Typical equilibrium-binding isotherm for 125I–SDF-1α to Fn-coated tubes. ○: 125I–SDF-1α binding to Fn. ▵: 125I–SDF-1α binding to noncoated tubes. Values reported are the mean of 4 independent assays for each concentration ±1 SD. Inset: Scatchard transform of the data in the main panel, showing a calculated Kd of approximately 40 nmol/L. (B) Binding is inhibited by “cold” SDF-1α. Two concentrations of 125I–SDF-1α (60 or 95 nmol/L) were incubated in Fn-coated tubes with increasing concentrations of cold SDF-1α. Values reported for both panels are the mean of 4 independent assays for each concentration ±1 SD.
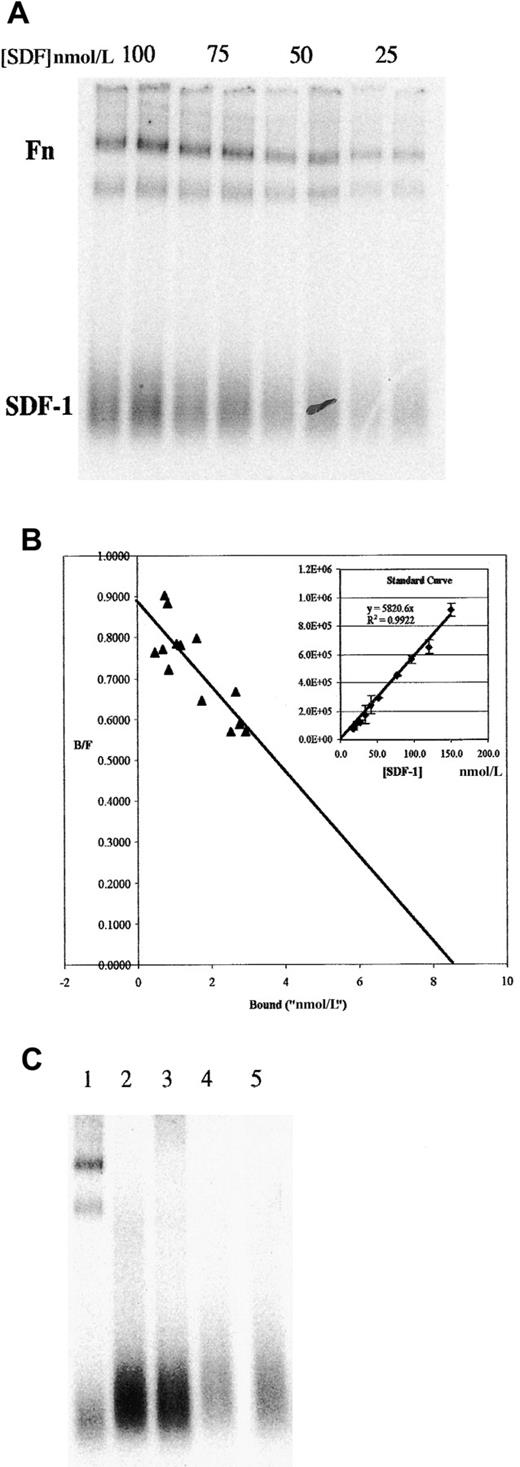
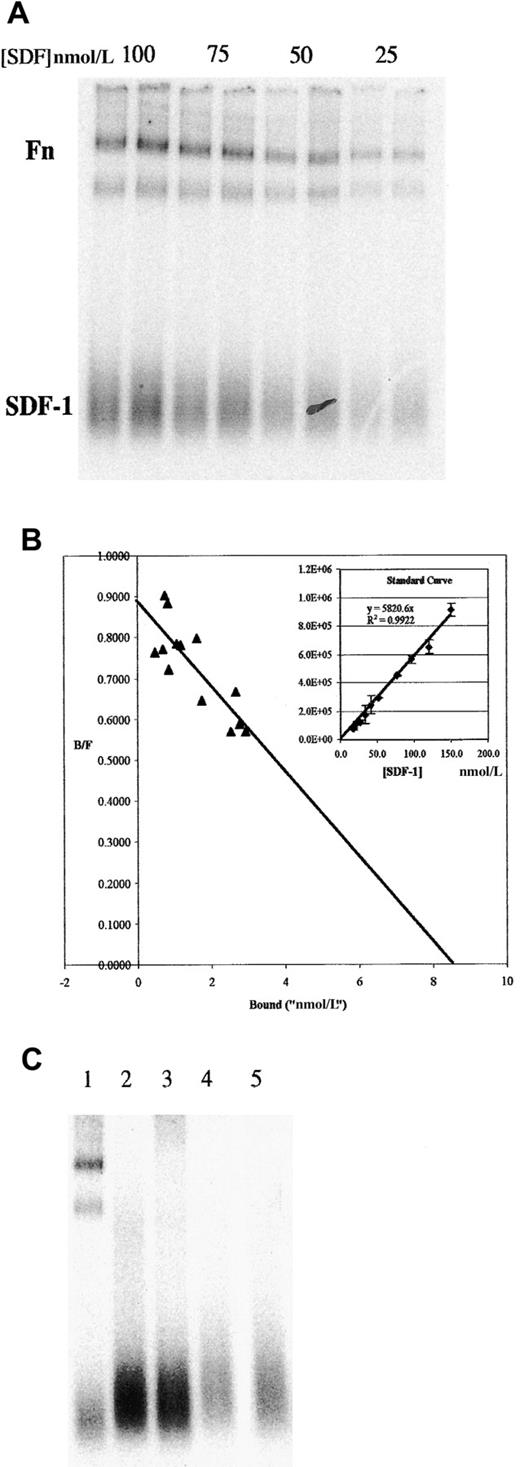
We also examined the interactions of SDF-1α with soluble Fn by using an adaptation of the EMSA. 125I–SDF-1α and Fn were incubated in solution and then electrophoresed in a nondenaturing polyacrylamide gel. These were then visualized by phosphorimaging. A typical experiment is shown in Figure 2A. Under native conditions, free SDF-1α migrates as a broad band at the bottom of the gel (under SDS-PAGE conditions, a single band at the correct molecular weight is detected). 125I–SDF-1α can also be seen associated with 2 bands at the top of the gel, corresponding to the major bands in this preparation of Fn (identity of these bands as Fn was confirmed by Western blot analysis; data not shown). In these experiments, duplicate lanes represented independent binding assays. Note that molecular weight markers were omitted from this figure because molecular weight does not strictly correlate with mobility in a native gel. However, the lower of the 2 Fn bands comigrated with myosin, which was approximately the same size as monomer Fn.
Binding of 125I–SDF-1α to Fn in solution as determined by EMSA shows a Kd of approximately 10 nmol/L.
(A) Indicated concentrations of 125I–SDF-1α were incubated in solution with 200 nmol/L Fn, then run on a native 4% to 20% PAGE gel. Duplicate lanes represent independent binding assays performed simultaneously. Position of free SDF-1α and Fn are indicated (both high-molecular-weight bands are positive in Fn Western blot; the lower of the 2 comigrates with myosin, approximately 250 kd). (B) Similar gels were used to measure the amount of125I–SDF-1α in the bound and free fractions, as described in the text, and were plotted in a Scatchard analysis. Inset: typical standard curve used to determine the bound and free fractions. (C) Binding to other ECM proteins was examined by EMSA. Lane 1, Fn; lane 2, collagen IV; lane 3, laminin 1; lane 4, laminin 2; lane 5, no ECM protein (125I–SDF-1α alone).
Binding of 125I–SDF-1α to Fn in solution as determined by EMSA shows a Kd of approximately 10 nmol/L.
(A) Indicated concentrations of 125I–SDF-1α were incubated in solution with 200 nmol/L Fn, then run on a native 4% to 20% PAGE gel. Duplicate lanes represent independent binding assays performed simultaneously. Position of free SDF-1α and Fn are indicated (both high-molecular-weight bands are positive in Fn Western blot; the lower of the 2 comigrates with myosin, approximately 250 kd). (B) Similar gels were used to measure the amount of125I–SDF-1α in the bound and free fractions, as described in the text, and were plotted in a Scatchard analysis. Inset: typical standard curve used to determine the bound and free fractions. (C) Binding to other ECM proteins was examined by EMSA. Lane 1, Fn; lane 2, collagen IV; lane 3, laminin 1; lane 4, laminin 2; lane 5, no ECM protein (125I–SDF-1α alone).
The EMSA procedure allows direct measurement of the amount of bound and free SDF-1α within each lane. For quantification, an SDS-PAGE gel of identically treated samples was exposed to the imaging plate at the same time as the native gel. The input concentration of SDF-1α in the SDS-PAGE gel was then plotted against the counts detected in the SDF-1α band to generate a standard curve (Figure 2B inset shows a typical standard curve). Scatchard analysis was performed on data obtained from gels similar to those shown in Figure 2, panel A, but including many more points over multiple gels. When the standard curve generated as described is used to estimate the bound and free fraction and the results are plotted in Scatchard analysis, aKd of approximately 10 nmol/L is obtained (Figure 2B).
To determine the specificity of the interaction, we screened other ECM proteins in the EMSA assay. Figure 2, panel C shows that SDF-1α does not bind appreciably to collagen IV, laminin 1, or laminin 2. Thus, although this is not an exhaustive survey, the data show that under these conditions the binding of SDF-1α to Fn is not shared by several other ECM proteins typically associated with basement membranes. In other experiments, vitronectin and osteopontin were found to be negative for binding (data not shown).
SDF-1α presented by Fn stimulates directed migration
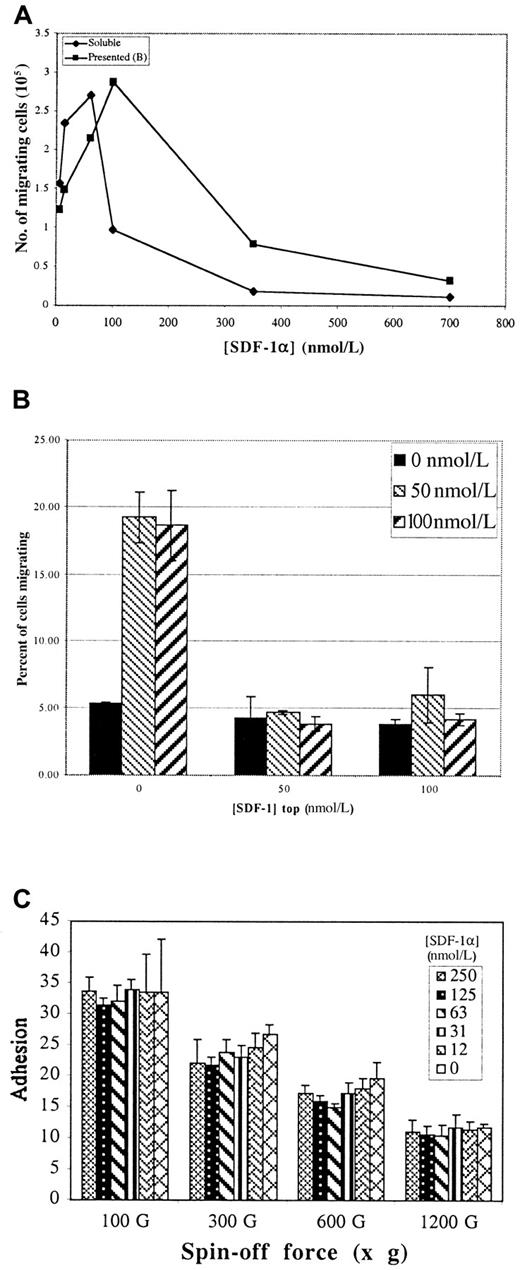
The observation that SDF-1α binds to Fn prompted us to examine the ability of so-called matrix-presented SDF-1α to promote T-cell migration in a modified Boyden chamber assay (Transwell).21 Figure 3, panel A compares the dose–response for SDF-1α–mediated migration of Jurkat T cells on Fn either when SDF-1α is matrix presented on the bottom of the insert or added in solution to the bottom chamber (ie, the conventional assay). In both cases, the membranes were first coated on both sides with 20 nmol/L Fn. In the conventional assay, migration showed the expected bell-shaped dependence on the concentration of SDF-1α.
Effect of SDF-1α on migration and adhesion of Jurkat T cells.
(A) Transwell assays for cell migration in response to either matrix-presented SDF-1α or SDF-1α added in solution to the bottom chamber (as per the standard assay). Both presented and “soluble” show similar dose responses for stimulation and inhibition of migration. (B) Tops and bottoms of Transwell membranes were first coated with Fn, then with the indicated concentrations of SDF-1α. 106 cells were loaded in the top of each well and allowed to migrate 14 hours, and migrated cells were counted. Values represent mean of 3 trials ±1 SD. (C) Percentage of cells adhering to Fn bound with the indicated concentration of SDF-1α over a range of “spin-off” forces. Presented SDF-1α has no effect on either the strength or the efficiency of Jurkat T-cell adhesion. Values represent mean of 6 trials ±1 SD.
Effect of SDF-1α on migration and adhesion of Jurkat T cells.
(A) Transwell assays for cell migration in response to either matrix-presented SDF-1α or SDF-1α added in solution to the bottom chamber (as per the standard assay). Both presented and “soluble” show similar dose responses for stimulation and inhibition of migration. (B) Tops and bottoms of Transwell membranes were first coated with Fn, then with the indicated concentrations of SDF-1α. 106 cells were loaded in the top of each well and allowed to migrate 14 hours, and migrated cells were counted. Values represent mean of 3 trials ±1 SD. (C) Percentage of cells adhering to Fn bound with the indicated concentration of SDF-1α over a range of “spin-off” forces. Presented SDF-1α has no effect on either the strength or the efficiency of Jurkat T-cell adhesion. Values represent mean of 6 trials ±1 SD.
Migration on matrix-presented SDF-1α showed essentially the same dose-dependent response (Figure 3A). Inhibition of migration by relatively small increases of soluble chemokine over optimal concentration has been reported.4 20 The reason for this inhibition is unknown, though the down-regulation of receptor expression, the desensitization of receptor signaling, or both have been suggested. Similar results were obtained when freshly prepared human thymocytes, peripheral blood leukocytes, or CD34 progenitors were used, indicating that these phenomena are not restricted to T-cell lines. Migration of Jurkat T cells was dependent on both α4β1 and α5β1, as demonstrated by inhibition with specific blocking antibodies (data not shown).
Presented SDF-1α stimulates chemotaxis
Directed migration is a complex behavior requiring the regulation of several steps, including adhesion, motility, and spatial orientation. Changes to any of these parameters can result in increased migration, as measured in the standard Transwell assay. For example, the induction of random cell motility, or chemokinesis, could result in more of the input cells reaching the bottom chamber through a random-walk process. Similarly, changes in cell adhesion can either decrease migration by retaining cells in the upper chamber or increase migration by allowing more cell contact with the matrix. By definition, chemotaxis must involve the induction of spatial orientation resulting in directed migration. We performed several experiments to determine which of these parameters was involved in the migration we observed.
Chemokinesis of Jurkat T cells is not enhanced by presented chemokine.
To determine whether chemokinesis is important in the enhanced migration, Transwell assays were performed with SDF-1α presented at various concentrations on each side of the Transwell membrane. If chemokinesis were the mechanism, cells in this assay would show enhanced motility in the presence of SDF-1α, regardless of whether there was a difference between the concentrations presented at the top and the bottom, resulting in more cells reaching the bottom chamber by random walk. However, no increase in migration above that occurring without added SDF-1α was detected in this assay when SDF-1α was present at the top and the bottom of the membrane. Migration was detected only when the chemokine was at the bottom alone (Figure 3B). This result suggests that chemokinesis is not the primary mechanism for the enhanced migration.
SDF-1α has no effect on Jurkat T cell adhesion.
Two components of adhesion, efficiency and strength, can be defined by the resistance of adhered cells to a range of centrifugal forces. Efficiency is given as the percentage cell adhesion at low g-force, and strength is detected as resistance to higher g-forces. Figure 3, panel C shows that neither of these parameters is affected by the presented SDF-1α. Moreover, pretreatment with soluble SDF-1α at concentrations up to 1 μmol/L neither inhibited nor promoted adhesion to Fn (data not shown). Because Jurkat cells are comparable to activated T cells, these results are consistent with a previous report demonstrating that the adhesion of activated lymphocytes is not affected by RANTES and MIP-1β.26 In contrast, the adhesion of resting lymphocytes may be enhanced by chemokines. Alternatively, SDF-1α has been reported to inhibit cytokine-induced adhesion of CD34+ stem cells.27 Thus, though chemokines may have complex effects on cell adhesion that are likely to be important in vivo, they cannot account for either the stimulatory or the inhibitory effects of SDF-1α on Jurkat migration in vitro.
CXCR4 localizes to the leading edge of cells migrating on presented chemokine.
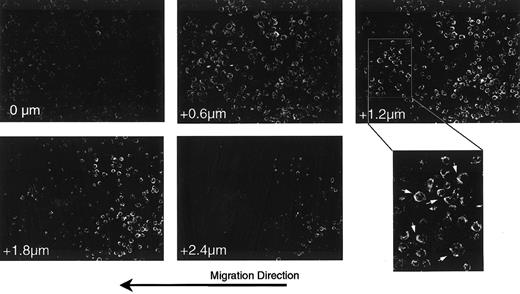
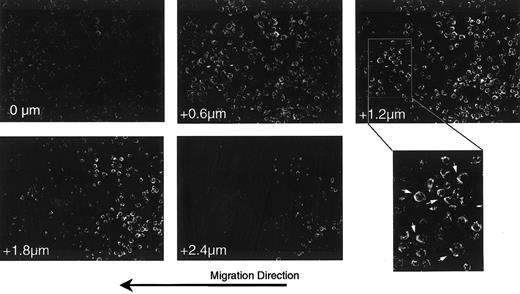
Previous reports28 29 suggest that the localization of chemokine receptors to one edge of the cells defines a leading edge for migration. We examined the cellular localization of CXCR4 on cells migrating on Fn-presented SDF-1α. For this, a uniform stripe of SDF-1α was deposited on a coverslip coated with Fn. Cells were then deposited at the edge of the stripe, allowed to interact with the substrate for 2 hours, then fixed and stained with anti-CXCR4. Figure4 shows a series of Z-section confocal images in a field of cells located at the edge between Fn alone and the SDF-1α–coated Fn. Consistent with Figure 3, panel C, there was no difference in the apparent cell density (a measure of adhesion efficiency) between Fn alone (right half of images) and SDF-1α–coated Fn (left half of images). However, the enlarged region of the +1.2 μm section clearly demonstrates that many of the cells showed polarization of CXCR4 to the leading edge with respect to the direction of migration (arrows).
Polarization of the CXCR-4 receptor in response to presented SDF-1α.
Cells plated at the edge of a stripe of SDF-1α on a uniform field of Fn were fixed and stained for the expression of the SDF-1α receptor, CXCR4.24 Shown are selected sections of a confocal Z-section series through a field of cells at the interface between the SDF-1α–coated Fn stripe (left half) and the Fn alone (right half). Numbers in the lower left of each frame indicate the relative position of the optical section, starting from the basal surface of the cells. The final panel shows an enlargement of a region from the +1.2 μm section in which cells polarized in the direction of overall migration are indicated (arrows). Note that the cells in this field on Fn alone are rounder and thicker than those on SDF-1α–coated Fn, as evidenced by the fact that the former remain in the image at the 1.8- and 2.4-μm sections, whereas most of the latter are no longer detectable. Note also that at the basal surface (0 μm) there is relatively higher staining for CXCR4 on cells on the SDF-1α stripe than on Fn alone.
Polarization of the CXCR-4 receptor in response to presented SDF-1α.
Cells plated at the edge of a stripe of SDF-1α on a uniform field of Fn were fixed and stained for the expression of the SDF-1α receptor, CXCR4.24 Shown are selected sections of a confocal Z-section series through a field of cells at the interface between the SDF-1α–coated Fn stripe (left half) and the Fn alone (right half). Numbers in the lower left of each frame indicate the relative position of the optical section, starting from the basal surface of the cells. The final panel shows an enlargement of a region from the +1.2 μm section in which cells polarized in the direction of overall migration are indicated (arrows). Note that the cells in this field on Fn alone are rounder and thicker than those on SDF-1α–coated Fn, as evidenced by the fact that the former remain in the image at the 1.8- and 2.4-μm sections, whereas most of the latter are no longer detectable. Note also that at the basal surface (0 μm) there is relatively higher staining for CXCR4 on cells on the SDF-1α stripe than on Fn alone.
To quantify the polarization in these images, all cells were overlaid with a pie-shaped grid defining the 4 compass directions, and the pixel density in each quadrant was measured with an image-processing software (Imagequant; Molecular Dynamics). The ratio of each direction over its opposite was calculated, and the largest ratio for each cell is taken as the polarization index. A polarization index of 3.0 or more was taken to indicate polarity in that direction. The results, presented in Table 1, verify the general impression that most cells on the SDF-1α–coated side are polarized and oriented in a direction (west) along the stripe of presented SDF-1α. In contrast, most cells on the Fn-alone side show no polarity.
Additionally, cells on SDF-1α–coated Fn are more spread, as evidenced by a larger projected area and by the fact that they are thinner in the Z-direction. This can be seen readily in the +2.4 μm section, where cells on Fn alone are still visible but those on SDF-1α–coated Fn are not. In comparing Z-section series from several fields, we found that cells on Fn alone were approximately 30% to 50% thicker than those on SDF-1α–coated Fn. This change in morphology, evident in all the cells on the SDF-1α–coated Fn, is consistent with a migratory phenotype.
In contrast to the appearance of the +2.4 μm section, the 0-μm section, defining the basal surface, shows more prominent staining for CXCR4 in the cells on SDF-1α–coated Fn. Thus, our data suggest the recruitment of the SDF-1α receptor to the leading edge and the basal surface of cells migrating in response to matrix-presented chemokine. This is consistent with cells expressing a polarized and migratory phenotype on presented SDF-1α.
Directed migration can occur in the absence of a concentration gradient
The previous experiment demonstrated that after 2 hours of contact with matrix-presented chemokine, many of the cells were oriented away from the edge of the chemokine stripe, indicating directional migration. We next asked whether cells would continue to migrate in the absence of a gradient to reinforce the correct direction. To test this, we again used a uniform stripe of SDF-1α deposited on a coverslip coated with Fn. Cells were plated in a drop at one end of the stripe such that a small subset was in contact with the Fn-presented chemokine at this edge. As shown in Figure 5, panel B, these cells selectively migrated to the stripe during the subsequent 2-hour incubation. Virtually no other cells were found on the coverslip apart from where they were deposited. Cells plated on Fn but without contact with the SDF-1α stripe showed no migration(Figure 5A). The directional migration of cells on the uniform stripe of SDF-1α suggested that a gradient of chemokine is not required to maintain direction, once cells have correctly polarized. It appears from Figure3B and Figure 4, however, that at least an edge between the presented chemokine and no chemokine is required for correct polarization. In future experiments it will be interesting to determine whether a gradient will enhance the efficiency of directed migration by increasing the speed or processivity of the cells.
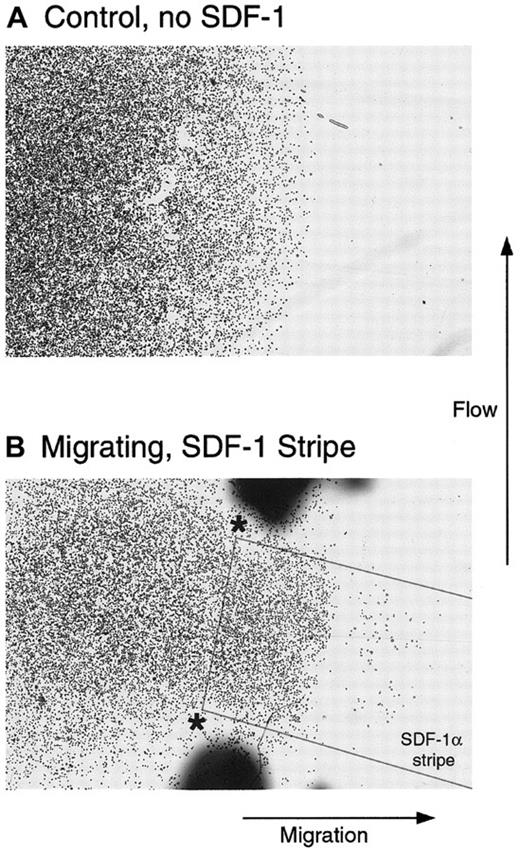
Directed migration of T cells in the absence of a gradient and under flowing conditions.
Cells were plated on an Fn-coated coverslip in contact with Fn alone (A) or overlapping a stripe of SDF-1α coated on the Fn as outlined in the Figure (B). The coverslip was placed in a flow chamber, as described, so that the flow was orthogonal to the direction of migration and was incubated for 2 additional hours. Although the cells in A remained within the boundaries of the original spot, a subset of cells in B clearly demonstrated directed migration to the stripe of SDF-1α.
Directed migration of T cells in the absence of a gradient and under flowing conditions.
Cells were plated on an Fn-coated coverslip in contact with Fn alone (A) or overlapping a stripe of SDF-1α coated on the Fn as outlined in the Figure (B). The coverslip was placed in a flow chamber, as described, so that the flow was orthogonal to the direction of migration and was incubated for 2 additional hours. Although the cells in A remained within the boundaries of the original spot, a subset of cells in B clearly demonstrated directed migration to the stripe of SDF-1α.
One additional possibility for the migration observed with uniformly presented SDF-1α is that the matrix acts as a sink for the chemokine and then slowly releases it into the microenvironment, where it functions at relatively high local concentrations as a soluble gradient. To address this possibility the stripe-migration assays in Figure 5 were performed under flowing conditions (3 μL min−1mm−2) applied orthogonal to the direction of migration. At this flow rate, any molecule of SDF-1α released from the stripe would travel the diameter of a cell in 0.2 seconds or less. In this time frame, no significant diffusion could occur. This would prevent the local accumulation of a soluble SDF-1α gradient and did not appreciably alter the resultant migration compared with similar experiments without flow.
Directed migration occurs in the absence of detectable soluble chemokine
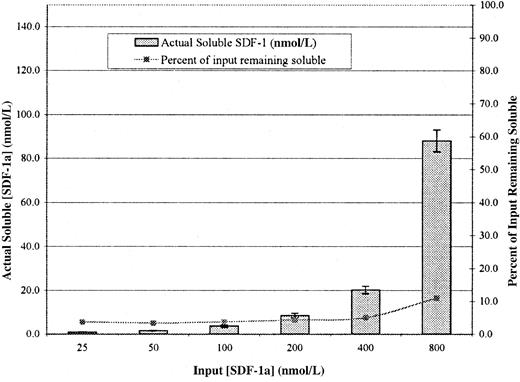
Transwell migration assays are commonly performed by the addition of soluble chemokine to the bottom chamber to create what is thought to be a soluble gradient. Based on our data, we considered the possibility that there is no soluble SDF-1α in a standard Transwell migration assay. To test this, varying concentrations of biotinylated SDF-1α were added to the bottom chamber of Transwells containing Fn-coated membranes. After 1 hour at 37°C, medium was removed from the bottom chamber and the SDF-1α concentration was determined with a plate-binding assay. The concentration of chemokine found in the bottom chamber defined the maximal concentration of soluble gradient existing across the membrane. Simultaneously, we assayed for SDF-1α bound to Fn on the membrane using the same enzymatic detection method. We found that at input concentrations of SDF-1α stimulating directed migration (ie, 10 to 100 nmol/L; Figure 3A), the actual soluble concentration in the bottom compartment was below the level of detection (1.0 nmol/L) for our assay (Table 2). Note that the reported Kd of SDF-1α for CXCR4 on T cells is approximately 1.5 nmol/L.30 Thus, under optimal conditions for migration, the concentration of soluble SDF-1α in the lower chamber is less than 1.0 nmol/L, a value below the reportedKd for receptor binding. In fact, the first nominal concentration at which remaining soluble SDF-1α could be detected was 200 nmol/L, which inhibits migration (Figure 3A).
As an additional test of this surprising result, we performed an analogous experiment using 125I–SDF-1α. As shown in Figure 6, at concentrations known to stimulate migration, the amount of detectable SDF-1α is extremely low. In fact, at input concentrations up to 200 nmol/L, it remained at a constant level of 4% of input (right-hand axis). Although this assay has some advantages over the enzymatic assay shown in Table 2, one disadvantage is that free 125I would be counted as soluble125I–SDF-1α. We determined by SDS-PAGE that the amount of free 125I in our labeled 125I-SDF-1α preparation was approximately 4% and that the actual amount of soluble SDF-1α in the bottom chamber at input concentrations that support migration were again 1 nmol/L or less. In this assay, only at input concentrations of 400 and 800 nmol/L (inhibitory concentrations) does the actual concentration rise above the 4% value.
Actual concentration of soluble SDF-1α in the bottom chamber of a standard Transwell assay.
Transwells were coated with Fn at 5 μg/mL and washed, and the indicated concentrations of 125I–SDF-1α were added to the bottom. After 1 hour, aliquots from the bottom chambers were counted in a γ counter and were compared to the known specific activity to determine the actual soluble SDF-1α concentration. The right-hand Y-axis represents the percentage input remaining soluble. Values represent mean of 3 trials ±1 SD.
Actual concentration of soluble SDF-1α in the bottom chamber of a standard Transwell assay.
Transwells were coated with Fn at 5 μg/mL and washed, and the indicated concentrations of 125I–SDF-1α were added to the bottom. After 1 hour, aliquots from the bottom chambers were counted in a γ counter and were compared to the known specific activity to determine the actual soluble SDF-1α concentration. The right-hand Y-axis represents the percentage input remaining soluble. Values represent mean of 3 trials ±1 SD.
Discussion
Our data indicate that SDF-1α binds to Fn specifically and saturably with high affinity. This “presented” chemokine induces directed migration, primarily by inducing spatial re-orientation of cells. Cell migration in response to presented SDF-1α can occur under flowing conditions, which offers a potential explanation for the ability of chemokines to function in the bloodstream or in other nonstatic conditions. Essentially all the effects previously attributed to soluble chemokine in Transwell assays can be recapitulated by matrix-presented SDF-1α. Moreover, the binding of SDF-1α to Fn within the concentration range of chemokine optimal for migration is so complete that no detectable soluble chemokine is present even in the standard Transwell assay. In light of our data, it is reasonable to consider the possible role of matrix-presented chemokine in previously reported data on directed cell migration.
For example, previously, Nieto et al28 examined the T-cell surface expression of the chemokine receptors CCR2 and CCR5 in response to the chemokines RANTES, IL-8, and MCP-1. They found that, consistent with our results, overall surface expression was reduced and polarization to the basal surfaces and leading edges of the cells had occurred. Although these experiments were designed to examine the effect of a soluble chemokine gradient, the receptor was found concentrated on the basal surface. A possible explanation for their data is that these chemokines were not soluble but were presented by the matrix.
A recent report31 concluded that T cells migrate away from high concentrations of SDF-1α. As shown in Figure 3, panel B, we tested various concentrations of SDF-1α at the top and bottom of Transwell membranes. At approximately 100 nmol/L on the top and no SDF-1α on the bottom, Poznansky et al31 reported optimal migration away from the chemokine. Although there are important differences in the way the assays are performed, it is worth noting that we did not observe the migration away from SDF-1α.
The adhesive interaction of cells with ECM has inherent polarity (basal/apical). Based on our data, we propose the hypothesis that the interaction of cells with matrix-presented chemokine is sufficient to induce polarization of T cells. However, the direction imparted to the cells is a random one. Chemotaxis could occur by a simple directed random-walk model, whereby cells would continue to migrate in the direction in which they initially polarize as long as they continue to receive a chemokine–matrix signal. If that signal is lost or reduced, as would happen if the cells were migrating away from the source, cells would reorient and begin migrating again. Thus, in our Transwell and our stripe-migration assays, a defined edge of the SDF-1α was required to orient the cells correctly. Alternatively, though uniform concentrations appear able to maintain the direction of a polarized cell, the initiation of processive migration may require a gradient or an edge. This model is similar to chemotaxis by bacteria32and is essentially a modification of the temporal sampling model.11 In support of this, Albrecht and Petty33 reported recently that neutrophils respond to a sudden reduction in chemokine levels by reorienting.
This simple model addresses many of the problems inherent in soluble diffusion gradient models. It can work under flowing conditions. In addition, as noted by Francis and Palsson,19 interaction of a diffusing solute with a binding substrate allows meaningful concentrations of the solute to be reached at greater distances from the source. It also can make the concentration more nearly uniform. Thus, a cell need not be able to respond to chemokine over such a wide range of concentrations in both steep and shallow gradients. Finally, in the case of several chemokine receptors that polarize to the leading edge of a migrating cell, such as CXCR4, this concentration of receptors would make detection of a spatial gradient difficult. (Imagine a cell polarized incorrectly, with twice as much chemokine at its trailing edge as at its leading edge. Such a 100% gradient should be easy to detect; however, with 3 times as many receptors on its leading edge, the cell might still perceive more chemokine at its leading edge.)
Several biologic implications and predictions can be based on our model. For example, co-signaling by integrins and chemokine receptors could be necessary to determine cell polarity and directional migration. It is a well-established paradigm that T-cell activation requires a series of co-stimulatory signals, including those from integrins.34-36 Thus, the inhibitory effects of soluble chemokine on T-cell migration may be analogous to the induction of anergy in which activation of the T-cell antigen receptor in the absence of appropriate co-stimulatory signals results in T-cell unresponsiveness. Integrins and chemokine receptors can interact with a large number of potential ligands; how cells prioritize and interpret signals from these interactions is unclear. Signaling complexes comprising multiple receptor–ligand interactions may contribute to the specificity of the overall interaction. For example, correct responses to chemokines might require the expression of a functional integrin for the target matrix, the appropriate chemokine receptor, and the presence of a chemokine capable of binding that matrix.
It has been reported recently that 7-transmembrane receptors, such as CXCR4, signal by 2 distinct pathways. One is dependent on heterotrimeric G proteins and does not require dimerization of the receptor, and the other is dependent on nonreceptor tyrosine kinases and does require receptor dimerization.37 38 It is apparent that interaction with the chemokine presented on ECM could have effects on the ability of the receptor to dimerize and, therefore, on the signals received by the cells. In that regard, it is worth noting that all our quantitative binding assays show evidence of additional binding, with a higher Kd, at concentrations of SDF-1α greater than 120 nmol/L.
Finally, recent reports39,40 indicate a role for matrix and matrix receptors in pathogen/host–cell interactions such that pathogens may be presented by matrix in the process of invading the host. It was recently reported41 that HIV gp120 and gp160 bind Fn and that presentation of retroviral vectors by Fn enhances gene transfer.42-45 Moreover, it is well established that in vitro, SDF-1α competes with T-trophic strains of HIV for binding to CXCR4 expressed by T lymphocytes. In vivo T lymphocytes are likely to encounter HIV within immune compartments that contain a complex organization of extracellular matrix. In this context, our model would predict that this competition may be mediated in vivo by the presentation in matrix of both HIV and SDF-1α.
Supported by National Institutes of Health (NIH) grants AI42384 (D.R.S.) and GM53489 (A.J.P.), Bethesda, MD, and a grant from the Katherine Huber-Steiner Foundation (P.H.), Bern, Switzerland.
A.J.P. and L.J.W.vL. contributed equally to this report.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Daniel R. Salomon, Department of Molecular and Experimental Medicine, The Scripps Research Institute, 10550 N. Torrey Pines Rd, La Jolla, CA.