Abstract
Human coagulation factor XI (FXI) is a plasma serine protease composed of 2 identical 80-kd polypeptides connected by a disulfide bond. This dimeric structure is unique among blood coagulation enzymes. The hypothesis was tested that dimeric conformation is required for normal FXI function by generating a monomeric version of FXI (FXI/PKA4) and comparing it to wild-type FXI in assays requiring factor IX activation by activated FXI (FXIa). FXI/PKA4 was made by replacing the FXI A4 domain with the A4 domain from prekallikrein (PK). A dimeric version of FXI/PKA4 (FXI/PKA4-Gly326) was prepared as a control. Activated FXI/PKA4 and FXI/PKA4-Gly326 activate factor IX with kinetic parameters similar to those of FXIa. In kaolin-triggered plasma clotting assays containing purified phospholipid, FXI/PKA4 and FXI/PKA4-Gly326 have coagulant activity similar to FXI. The surface of activated platelets is likely to be a physiologic site for reactions involving FXI/FXIa. In competition binding assays FXI/PKA4, FXI/PKA4-Gly326, and FXI have similar affinities for activated platelets (Ki = 12-16 nM). In clotting assays in which phospholipid is replaced by activated platelets, the dimeric proteins FXI and FXI/PKA4-Gly326 promote coagulation similarly; however, monomeric FXI/PKA4 has greatly reduced activity. Western immunoblot analysis confirmed that activated monomeric FXI/PKA4 activates factor IX poorly in the presence of activated platelets. These findings demonstrate the importance of the dimeric state to FXI activity and suggest a novel model for factor IX activation in which FXIa binds to activated platelets by one chain of the dimer, while binding to factor IX through the other.
Introduction
A paradigm in the field of blood coagulation is that the zymogen of a plasma protease is activated by limited proteolysis on a phospholipid surface, in the presence of a protein cofactor and divalent cations.1-5 In vivo, appropriate phospholipid surfaces are provided by activated platelets and cell membranes of damaged tissues. Formation of surface-bound protease-substrate complexes increases the rate of zymogen activation, concentrates procoagulant reactions to sites of vessel injury, and minimizes spread of thrombogenic proteases beyond wound sites. An apparent exception to this model is the activation of factor IX by activated factor XI (FXIa). In in vitro coagulation systems, such as the activated partial thromboplastin time (aPTT) assay, activation of factor IX by FXIa requires calcium ions.6-8 However, phospholipids known to promote activation of factor X and prothrombin, such as brain cephalin, have little effect on the reaction.9 10 Furthermore, a candidate protein cofactor to promote surface assembly of a FXIa-based factor IX–activating complex has not been identified. These observations suggest, counterintuitively, that factor IX activation by FXIa proceeds to a significant extent in the fluid phase of blood.
Zymogen factor XI (FXI) and FXIa do bind to activated platelets in a process that is saturable and reversible and that requires the protein cofactor high molecular weight kininogen (HK) and zinc ions.11,12 Evidence strongly suggests that the platelet surface is a physiologic environment for reactions involving FXI. When bound to activated platelets, FXI activation by the proteases thrombin, factor XIIa, and FXIa is greatly accelerated.13,14Furthermore, prothrombin may substitute for HK as a cofactor for FXI/FXIa binding to platelets,13,15 providing an explanation for the lack of excessive bleeding in patients congenitally deficient in HK.16 Given these data and the observation that factor IX binds to activated platelets,17 it is likely that the surface of activated platelets is a physiologic environment for activation of factor IX by FXIa.
The FXI polypeptide is composed of an N-terminal noncatalytic heavy chain and a C-terminal trypsin-like catalytic light chain.18,19 The heavy chain consists of 4 homologous subunits called apple domains (designated A1 to A4, from the N-terminus),19,20 a feature FXI shares with the plasma protease prekallikrein (PK).21,22 Mapping studies identified key amino acids in the A2 and A3 domains that are required for normal binding to factor IX.23,24 Amino acids involved in FXI binding to activated platelets have also been localized to the A3 domain.15,25 The putative factor IX and platelet-binding sites partially overlap, raising a question as to the mechanism by which FXIa would bind simultaneously to its substrate and a platelet surface.23,26 A unique feature of FXI structure may offer a solution to this dilemma. The protein is a disulfide bond-linked dimer comprised of 2 of the polypeptides described above.18-20 In this study we describe the preparation and characterization of a monomeric version of FXIa and demonstrate that FXIa must be a dimer to properly promote coagulation in the presence of activated platelets. The findings suggest a novel model for a factor IX activation complex on platelets in which one heavy chain of FXIa binds to the platelet, and the other binds to factor IX.
Materials and methods
Preparation and activation of recombinant proteins
Expression constructs for wild-type FXI and for chimeras FXI/PKA4 and FXI/PKA3 (FXI with the A4 or A3 domain, respectively, replaced with the corresponding domain from PK) have been described.27 A dimeric version of FXI/PKA4 (FXI/PKA4-Gly326) was created by changing Cys326 to glycine, using a Chameleon site-directed mutagenesis kit (Stratagene, LaJolla, CA). FXI with Cys321 replaced by alanine (FXI-Ala321) was made in a similar manner. Proteins were expressed in 293 fibroblasts23,27and purified from 500-2000 mL serum-free conditioned media by antibody affinity chromatography.27 Proteins were checked for purity by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and protein concentration was determined by dye binding assay (BioRad, Hercules, CA). Proteins were activated by diluting to 100-200 μg/mL in 25 mM Tris-HCl pH = 7.4, 100 mM NaCl (TBS) containing 5 μg/mL human FXIIa, and incubating at 37°C. Conversion of the 80-kd zymogen to the 45- and 35-kd chains of FXIa was followed by reducing SDS-PAGE.
Plasma proteins
FXI and HK were purified from human plasma by published methods.28,29 All proteins appear as single bands on Gelcode Blue (Pierce, Rockford, IL)–stained gels. Specific activity of human FXI (230 U/mg) was determined by aPTT assay, using FXI-deficient plasma as substrate and pooled normal plasma as standard (1 U FXI activity/mL plasma).30 Human plasmas were from George King Biomed (Overland Park, KS). For competition binding assays, human FXI was labeled with 125I by a modification of the Iodigen method.31 Specific activity of 125I-FXI was 29.4 μCi/μg protein. Human PK, as well as factors IX, IXa, X, and XIIa, was purchased from Enzyme Research Laboratories (South Bend, IN).
Gel filtration chromatography
Protein (10-20 μg) in 100-200 μL of TBS underwent size-fractionation on a Superose-12 gel filtration column (Amersham Pharmacia Biotech, Piscataway, NJ) fitted to a BioLogic FPLC workstation (BioRad, Richmond, CA). The column was equilibrated with 50 mM sodium phosphate pH 7.3 and 150 mM NaCl. Fractions of eluate (500 μL) were collected. Retention times of proteins (determined by OD 280 nm) were compared to a series of protein standards. The identity of the eluted protein was confirmed by performing Western immunoblot analysis on column fractions (data not shown).
Chromogenic substrate assays
Chromogenic substrates S-2366 (L-pyroglutamyl-L-prolyl-L-arginine-P-nitroanaline) and S-2765 (N-α-benzyoxycarbonyl-D-arginyl-glycyl-L-arginine-P-nitroanaline) were from DiaPharma (Westchester, OH).
Cleavage of S-2366 by FXIa.
Activated proteins were diluted to 0.5 μg/mL in TBS with 0.1% bovine serum albumin (TBSA) containing 50-1000 μM S-2366, and change in absorbance at 405 nm was followed on a microtiter plate reader. Michaelis-Menten constants (Km and Vmax) were determined by standard methods, using the average of 2 separate experiments. Values for Vmax were converted to nM pNA/sec, using an extinction coefficient of 9800 optical density units (405 nm)/mole of pNA. Turnover number (kcat) was calculated from the ratio of Vmax to enzyme concentration.
Activation of factor IX by FXIa.
Activation of factor IX by activated proteases was evaluated as described.23 Briefly, enzyme (0.2 μg/mL) was incubated at 37°C for 1 minute with factor IX (0.05-1.0 μM) in TBSA containing 5 mM CaCl2. Activation was stopped by adding EDTA to 25 mM. The reaction was diluted 1:100 in TBSA, and 10 μL was added to 50 μL of a mixture of factor VIII (8 U/mL; Recombinate Baxter/Hyland, Glendale, CA), CaCl2 (10 mM), and rabbit brain cephalin, and to 30 μL factor X (450 nM). Incubations were at 37° C for 2.5 minutes, and then EDTA was added as above. Fifty microliters of each reaction was mixed with 50 μL 1.0 mM S-2765, and change in absorbance at 405 nm was followed on the microtiter plate reader. Results were compared to a control curve constructed with purified factor IXa. Michaelis-Menten constants were determined, using averages from 3 separate experiments. Rabbit brain cephalin was made from rabbit brain acetone extract (Sigma, St Louis, MO) by the method of Bell and Alton.32
Platelet-binding experiments
Gel-filtered human platelets were prepared from fresh blood as previously described.33 Thrombin receptor agonist SFLLRN-amide was prepared at the Protein Chemistry Facility of the University of Pennsylvania.25 Binding experiments were performed by a modification of published methods.12Briefly, platelets (108/mL) were activated by 5 μM SFFLRN-amide for 5 minutes at 37°C and then supplemented with ZnCl2 (25 μM), CaCl2 (2 mM), HK (50 nM), and 125I-labeled plasma-derived FXI (22 nM) either in the presence or absence of recombinant proteins. Incubation was continued for 30 minutes at 37°C. Aliquots (100 μL) were layered on Dow Corning methyl silicon oil (3 parts 550 density oil:2 parts 200 density oil) and platelets were separated from unbound protein by centrifugation in a microfuge. 125I-FXI bound to the platelet pellet was measured with a Wallace 1470 Wizard gamma counter. The concentration of cold protein that displaced 50% of bound125I-FXI (IC50) was determined by plotting125I-FXI bound to platelets against the concentration of competing ligand. Ki was calculated, using the equation IC50 = (1 + [S]/Kd) Ki, where S is the concentration of 125I-FXI (22 nM) and Kd is the binding constant for FXI determined by direct binding experiments (10 nM).
Activity of recombinant proteins in plasma clotting assays
Coagulant activities for zymogen FXI/PKA4 and FXI/PKA4-Gly326 were determined by aPTT assay. HEPES-Tyrode buffer pH 7.4 (100 μL) containing 1 nM FXI/PKA4 or FXI/PKA4-Gly326 was mixed with 50 μL FXI-deficient plasma. To this mixture was added 50 μL kaolin (5 mg/mL) in HEPES-Tyrode buffer pH 7.4 containing either phospholipid (inosithin 0.04%; Accurate Chemicals, Westbury, NY) or activated platelets (108/mL). Incubation was for 5 minutes at 37°C, followed by addition of 50 μL 50 mM CaCl2. Time to fibrin clot formation was determined on a fibrometer. All proteins were tested in triplicate and were compared to a standard curve prepared with wild-type FXI. One nanomolar wild-type FXI was assigned an activity of 1.00 (100%). FXIa and activated chimeric enzymes were tested in a similar manner, except that phospholipid or platelet suspensions did not contain kaolin, and incubation at 37°C was for 60 seconds prior to addition of CaCl2. A standard curve was prepared with wild-type FXIa (1 nM FXIa was assigned an activity of 1.00 or 100%).
Western immunoblot analysis of factor IX activation by FXIa
Human factor IX (150 nM) was incubated at 37°C with 1 nM wild-type FXIa or activated FXI/PKA4 in TBSA that contained 2 mM CaCl2. At various time points, 10 μL samples were removed into 5 μL SDS-sample buffer (500 mM Tris-HCl pH 6.8, 40% glycerol, 10% SDS). A second set of experiments was carried out under similar conditions, except that reactions included HK (50 nM), ZnCl2 (25 μM), and activated platelets (0.5 × 108/mL). Samples were size-fractionated on 12% polyacrylamide gels, followed by transfer to nitrocellulose membranes. Blots were developed with a goat antihuman factor IX polyclonal immunoglobulin G (Affinity Biologicals, Hamilton, Ontario, Canada), using an enhanced chemiluminescence Western blotting detection kit (Amersham Pharmacia Biotech).
Results
Recombinant proteins
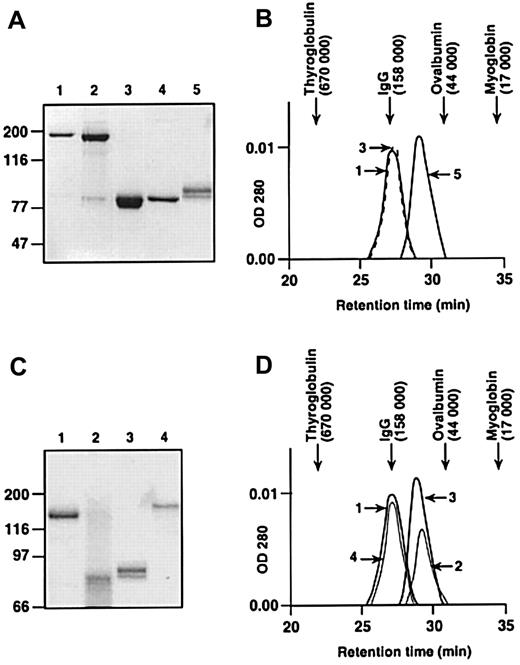
Human FXI is a 160-kd disulfide bond-linked dimer comprised of 2 identical 80-kd polypeptides (Figure 1A, lanes 1 and 4).18,20 The A4 domain mediates dimer formation, with Cys321 in A4 forming the inter-chain disulfide bond.34 FXI in which Cys321 is replaced by alanine (FXI-Ala321, Figure 1A, lane 3) is an 80-kd protein on nonreducing SDS-PAGE; however, gel filtration experiments performed under conditions of physiologic salt concentration and pH demonstrate that it is the same size as plasma factor XI (Figure 1B). This suggests that the protein is a noncovalently associated dimer. These data confirm earlier work, demonstrating that an inter-chain disulfide bond is not required for dimer formation.34 In contrast, PK (Figure1A, lane 5), which is structurally homologous to FXI,20,22has a higher retention time on gel filtration (molecular mass ∼90 kd; Figure 1), demonstrating it is a monomer.34
SDS-PAGE and size-exclusion chromatography of plasma-derived and recombinant proteins.
(A) SDS-polyacrylamide gel of proteins stained with Gelcode Blue. All lanes contain unreduced protein except for lane 4. Lane 1, human plasma FXI; lane 2, human recombinant FXI; lane 3, recombinant FXI-Ala321; lane 4, human plasma FXI (reduced); and lane 5, human plasma PK. (B) Retention times of proteins shown in panel A size-fractionated on a Superose-12 gel filtration column. The numbers next to the protein peaks correspond to the numbers used in panel A. The peak for human plasma FXI (lane 1) is drawn as a solid line and FXI-Ala321 (lane 3) as a dashed line. (C) Nonreducing SDS-polyacrylamide gel of chimeric proteins stained with Gelcode Blue. Lane 1, human plasma FXI; lane 2, FXI/PKA4; lane 3, human plasma PK; and lane 4, FXI/PKA4-Gly326. (D) Retention times of proteins shown in panel C size-fractionated on a Superose-12 gel filtration column. The numbers next to the protein peaks correspond to numbers in panel C. For panels A and C, positions of molecular mass standards in kilodaltons are shown at the left of the figures. For panels B and D, retention times of molecular mass standards are shown at the top of the figures.
SDS-PAGE and size-exclusion chromatography of plasma-derived and recombinant proteins.
(A) SDS-polyacrylamide gel of proteins stained with Gelcode Blue. All lanes contain unreduced protein except for lane 4. Lane 1, human plasma FXI; lane 2, human recombinant FXI; lane 3, recombinant FXI-Ala321; lane 4, human plasma FXI (reduced); and lane 5, human plasma PK. (B) Retention times of proteins shown in panel A size-fractionated on a Superose-12 gel filtration column. The numbers next to the protein peaks correspond to the numbers used in panel A. The peak for human plasma FXI (lane 1) is drawn as a solid line and FXI-Ala321 (lane 3) as a dashed line. (C) Nonreducing SDS-polyacrylamide gel of chimeric proteins stained with Gelcode Blue. Lane 1, human plasma FXI; lane 2, FXI/PKA4; lane 3, human plasma PK; and lane 4, FXI/PKA4-Gly326. (D) Retention times of proteins shown in panel C size-fractionated on a Superose-12 gel filtration column. The numbers next to the protein peaks correspond to numbers in panel C. For panels A and C, positions of molecular mass standards in kilodaltons are shown at the left of the figures. For panels B and D, retention times of molecular mass standards are shown at the top of the figures.
To prepare monomeric FXI, the A4 domain was replaced with PKA4. The resulting protein, FXI/PKA4, as expected, is an 80-kd protein on nonreducing SDS-PAGE (Figure 1C, lane 2). In gel filtration experiments, FXI/PKA4 has a similar retention time to PK, indicating it is a monomer (Figure 1D). FXI/PKA4 is expressed poorly by 293 fibroblasts (< 100 ng/mL conditioned media).27 This is consistent with data showing that mutations interfering with FXI intracellular dimerization result in poor protein expression.35 36 Dimeric FXI/PKA4 (FXI/PKA4-Gly326) was made by replacing Cys326 in FXI/PKA4 with glycine. Cys326 in PKA4 is normally paired with Cys321 to form an intra-chain disulfide bond. Its removal leaves Cys321 free to form an inter-chain disulfide-link with Cys321 on another polypeptide. FXI/PKA4-Gly326 is expressed by 293 cells at similar levels to wild-type FXI (data not shown), and the expressed protein is entirely dimeric (Figure 1C, lane 4, and 1D). Parenthetically, this suggests that elements in FXI distinct from the A4 domain are involved in promoting dimer formation.
FXIa activity in purified protein assays
Activated proteases were studied in 2 purified protein systems. In the first system, the capacities of the proteases to cleave the chromogenic substrate S-2366 were tested. Kinetic parameters for S-2366 cleavage are similar for all proteins tested (Table1), indicating that the catalytic domains are intact. Kinetic parameters for activation of factor IX by recombinant proteases were determined by a 3-stage assay.23 27 The results demonstrate that activated FXI/PKA4 and FXI/PKA4-Gly326 activate factor IX similarly to FXIa (Table 1). This indicates that both chimeric molecules bind normally to, and have normal catalytic activity toward, factor IX when activation takes place in solution.
FXI binding to platelets
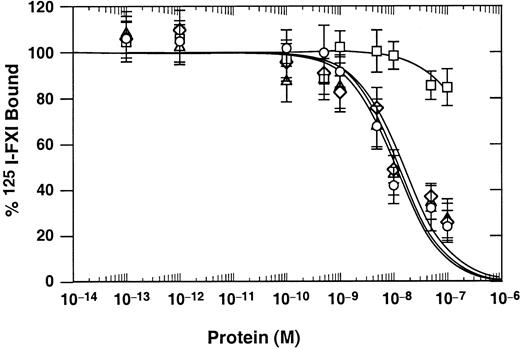
FXI/PKA4 and FXI/PKA4-Gly326 were tested for their capacity to compete with 125I-labeled FXI for binding to activated platelets (Figure 2). Results were compared to those for plasma-derived FXI (positive control) and to FXI/PKA3 (negative control). FXI/PKA3, a chimera consisting of FXI with the A3 domain replaced by the PKA3 domain,27 binds poorly to platelets because PKA3 lacks critical amino acids required for platelet binding.15,25 The Ki for FXI/PKA4 (14 nM) and FXI/PKA4-Gly326 (16 nM) are similar to the value for plasma FXI (12 nM), indicating the 3 proteins bind to platelets with similar avidity. In contrast, the Ki for FXI/PKA3 is more than 500 nM, a result similar to reported values for PK binding to platelets.12
Binding of FXI and chimeric proteins to activated platelets.
FXI and FXI/PK chimeras were used in competition binding studies as competitive ligands for 22 nM 125I-FXI binding to activated platelets in the presence of 50 nM HK and 25 μM ZnCl2. Plasma-derived FXI (○), FXI/PKA3 (■), FXI/PKA4 (▵), and FXI/PKA4-Gly326 (⋄). Data are mean values (± SEM) of total binding results from 3 separate experiments, each performed in triplicate.
Binding of FXI and chimeric proteins to activated platelets.
FXI and FXI/PK chimeras were used in competition binding studies as competitive ligands for 22 nM 125I-FXI binding to activated platelets in the presence of 50 nM HK and 25 μM ZnCl2. Plasma-derived FXI (○), FXI/PKA3 (■), FXI/PKA4 (▵), and FXI/PKA4-Gly326 (⋄). Data are mean values (± SEM) of total binding results from 3 separate experiments, each performed in triplicate.
Activity of FXI and FXIa in plasma clotting assays
In contact activation-initiated clotting assays, such as the aPTT, a negatively charged substance is used to initiate coagulation, and phospholipid is required for several enzymatic steps.3 Activated platelets may serve as a source of phospholipid. Clot formation in this type of assay depends on factor IX activation by FXIa. In aPTT assays using purified phospholipid (inosithin), FXI/PKA4 and FXI/PKA4-Gly326 correct the defect in FXI-deficient plasma similarly to wild-type FXI (Table2, column 1). In contrast, when the phospholipids are replaced by activated platelets, only the dimeric protein FXI/PKA4-Gly326 shows significant activity (Table 2, column 2). In the presence of activated platelets, the activity of monomeric FXI/PKA4 is below the lower limit of detection of the assay.
These data indicate that activated FXI/PKA4 bound to platelets does not properly activate factor IX. Alternatively, zymogen FXI/PKA4 may not be activated well in this system. To distinguish between these possibilities, a modified clotting assay was performed in which coagulation is initiated by FXIa or activated chimera rather than by kaolin. Poor activation of FXI/PKA4 is not an issue in this case as the protease is added in the active form. Activated FXI/PKA4 and activated FXI/PKA4-Gly326 demonstrate significant activity in the presence of phospholipid (Table 2, column 3). However, consistent with the results obtained using zymogens in the aPTT assay, activated FXI/PKA4 has little activity in the presence of activated platelets compared to its dimeric counterpart FXI/PKA4-Gly326 (Table 2, column 4).
Factor IX activation by FXIa in the presence of platelets
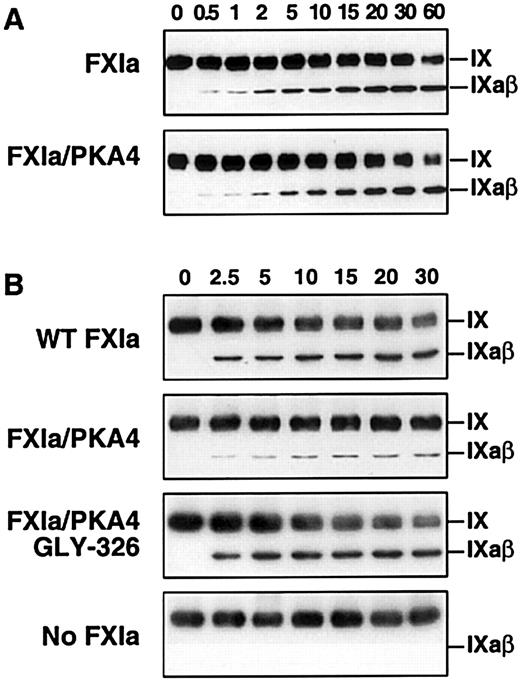
During activation of factor IX (molecular mass 55 kd), an approximately 11-kd activation peptide is released to generate the active enzyme, factor IXaβ (45 kd).6-8 Therefore, the activation of factor IX by FXIa can be directly observed by Western immunoblot assay under nonreducing conditions. Wild-type FXIa and activated FXI/PKA4 activate factor IX to factor IXaβ similarly in the absence of platelets (Figure 3A). In contrast, and consistent with the results of the clotting assays, activation by activated FXI/PKA4 is significantly reduced compared to wild-type FXIa and activated XI/PKA4-Gly326 when activated platelets are included in the reaction (Figure 3B). It is not clear if the relatively small amount of factor IXaβ generated by FXI/PKA4 represents enzyme activity on the platelet surface, or activation in solution phase by FXI/PKA4 that has not bound to the platelet.
Western immunoblot analysis of factor IX activation by FXIa in the presence of platelets.
(A) Factor IX (150 nM) was incubated with 1 nM wild-type FXIa or activated FXI/PKA4 in TBSA containing 2 mM CaCl2. At the indicated time points, samples were removed into sample buffer and processed as described in “Materials and methods.” (B) Reactions were run in the same manner as in panel A with the following additions: HK 50 nM, ZnCl2 (25 μM), and activated platelets (0.5 × 108/mL). The positions of zymogen (FIX) and activated (FIXaβ) factor IX are shown to the right of each blot. Time in minutes is shown across the top of each panel.
Western immunoblot analysis of factor IX activation by FXIa in the presence of platelets.
(A) Factor IX (150 nM) was incubated with 1 nM wild-type FXIa or activated FXI/PKA4 in TBSA containing 2 mM CaCl2. At the indicated time points, samples were removed into sample buffer and processed as described in “Materials and methods.” (B) Reactions were run in the same manner as in panel A with the following additions: HK 50 nM, ZnCl2 (25 μM), and activated platelets (0.5 × 108/mL). The positions of zymogen (FIX) and activated (FIXaβ) factor IX are shown to the right of each blot. Time in minutes is shown across the top of each panel.
Discussion
The formation of protease-substrate complexes on activated platelets and damaged tissue is crucial for normal hemostasis.1-5 In complexes involving vitamin K–dependent proteases (prothrombin and factors VII, IX, and X), both protease and substrate bind to phospholipid in an interaction involving the N-terminal “Gla-domain” of each protein.2,3Gla-domains contain 10 to 12 glutamic acid residues that undergo post-translational modification by addition of a carboxyl group to the γ-carbon.37 This modification is necessary for calcium-dependent protein binding to phospholipid. FXI is the only serine protease required for normal coagulation that lacks a Gla-domain.19,20 This may explain why phospholipid has little effect on factor IX activation by FXIa. Although FXI may not interact with purified phospholipid, it is clear that FXI and FXIa bind to activated platelets.11,12 Amino acids in the C-terminus of the A3 domain are required for platelet binding.15,25Curiously, this region may overlap with the factor IX binding site. As data suggest that the platelet surface is a physiologic site for FXI activation and activity, this finding raises a question as to the manner in which FXIa would bind simultaneously to its substrate and a platelet surface.23 26 A possible answer may be found in the dimeric structure of FXI.
Human FXI is a disulfide-bond linked homodimer,19,20 a unique feature among coagulation proteases. Meijers et al34 demonstrated that the A4 domain is involved in dimer formation,34 with Cys321 involved in the inter-chain disulfide bond.20,34 To test the significance of the dimeric state to FXI function, a FXI monomer is required for comparison. Taking advantage of the homology between FXI and PK (a monomeric protein), we generated the monomeric chimera FXI/PKA4. As in FXI, there is a cysteine at position 321 in PKA4; however, it is involved in an intra-chain bond with Cys326, a residue unique to PK. FXI/PKA4 is expressed poorly in 293 cells,27 consistent with work showing that dimerization is necessary for proper protein secretion.35,36 Indeed, Meijers et al34postulated that FXI may be dimeric to facilitate intracellular processing and secretion. In our experiments with FXI/PK chimeras, arrangements placing the A3 domain of FXI in a monomeric protein result in poor expression (D.G., unpublished observation, November 1999). This suggests that FXIA3, in contrast to PKA3, prefers to be a component of a dimer. This is supported by the normal expression of FXI/PKA4-Gly326 in 293 cell culture. Despite lacking FXIA4, a free cysteine at Cys321 facilitates dimer formation (possibly driven by the A3 domain) and improves the poor expression seen with FXI/PKA4.
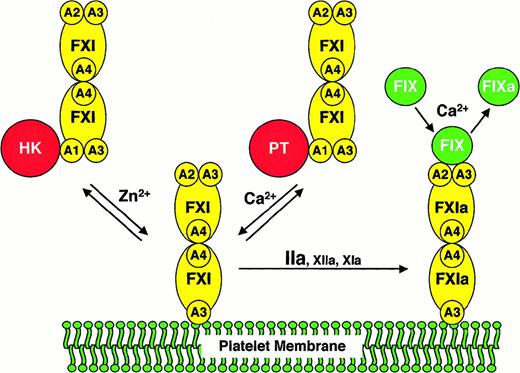
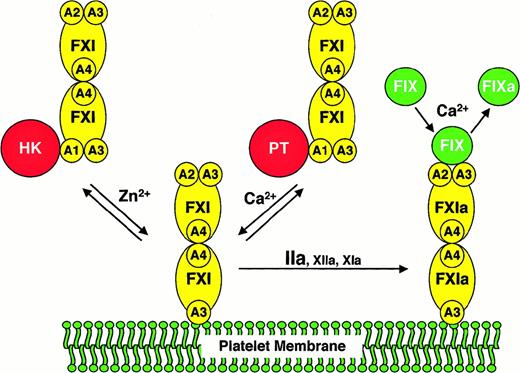
We compared FXI/PKA4 to wild-type FXI and FXI/PKA4-Gly326 in a series of assays requiring factor IX activation by FXIa. In the absence of activated platelets (either in purified protein or plasma clotting assays) the 3 molecules performed similarly. In contrast, when activated platelets are used as a lipid source, monomeric FXI/PKA4 demonstrates a defect in factor IX activation. Several scenarios must be considered as explanations for this observation. FXI/PKA4 may be activated poorly on the platelet surface in an aPTT assay. However, FXI/PKA4 has poor activity even when added to the assay in the activated state. Similarly, it is difficult to invoke abnormalities in FXI/PKA4 platelet binding because FXI/PKA4 and wild-type FXI have similar affinities for platelets. A hypothesis that fits all experimental data well is that monomeric FXIa is unable to interact simultaneously with activated platelets and factor IX. The findings are consistent with a scenario in which FXIa binds to a platelet by one polypeptide of the dimer, while interacting with factor IX through the other. A model based on this hypothesis is shown in Figure4.
A model for the activation of FXI and factor IX on activated platelets.
The FXI or FXIa molecule is a dimer composed of 2 identical 80-kd polypeptides, each containing 4 apple domains (designated A1 through A4), and one trypsin-like catalytic domain. For the sake of clarity, schematic representations of FXI are shown only with apple domains involved in the relevant interaction. FXI binds to the surface of an activated platelet through one of its A3 domains in a reaction that requires either HK and Zn2+, or prothrombin (PT) and Ca++. HK or PT (shown interacting with the A1 domain of FXI in the solution phase of the diagram) appears to be required for FXI to be in the proper conformation for binding to the platelet but does not appear to be a necessary component of a platelet-binding site for FXI. It is not known if FXI interacts with either HK or PT on the platelet surface. Furthermore, it is not clear if HK and PT interact with one or both polypeptides of the FXI dimer. FXI bound to the platelet is activated to FXIa by thrombin, factor XIIa, or factor XIa.13,14 Factor IX binds to FXIa through the heavy chain not involved in binding to the platelet. Available data suggest that the factor IX binding site on FXIa involves components of the A2 and A3 domains.23,24,27 FXIa then converts factor IX (FIX) to factor IXa (FIXa) in a reaction requiring calcium ions.6Abbreviations: FXI, factor XI; FXIa, factor XIa; FIX, factor IX; FIXa, factor IXaβ; A1 through A4, factor XI apple domains 1 through 4, respectively; HK, high molecular weight kininogen; PT, prothrombin.
A model for the activation of FXI and factor IX on activated platelets.
The FXI or FXIa molecule is a dimer composed of 2 identical 80-kd polypeptides, each containing 4 apple domains (designated A1 through A4), and one trypsin-like catalytic domain. For the sake of clarity, schematic representations of FXI are shown only with apple domains involved in the relevant interaction. FXI binds to the surface of an activated platelet through one of its A3 domains in a reaction that requires either HK and Zn2+, or prothrombin (PT) and Ca++. HK or PT (shown interacting with the A1 domain of FXI in the solution phase of the diagram) appears to be required for FXI to be in the proper conformation for binding to the platelet but does not appear to be a necessary component of a platelet-binding site for FXI. It is not known if FXI interacts with either HK or PT on the platelet surface. Furthermore, it is not clear if HK and PT interact with one or both polypeptides of the FXI dimer. FXI bound to the platelet is activated to FXIa by thrombin, factor XIIa, or factor XIa.13,14 Factor IX binds to FXIa through the heavy chain not involved in binding to the platelet. Available data suggest that the factor IX binding site on FXIa involves components of the A2 and A3 domains.23,24,27 FXIa then converts factor IX (FIX) to factor IXa (FIXa) in a reaction requiring calcium ions.6Abbreviations: FXI, factor XI; FXIa, factor XIa; FIX, factor IX; FIXa, factor IXaβ; A1 through A4, factor XI apple domains 1 through 4, respectively; HK, high molecular weight kininogen; PT, prothrombin.
Several assumptions were made in preparing the model that require further testing and may, therefore, not be accurate. For example, it is likely that factor IX is bound to the platelet through its Gla-domain during activation, rather than being in solution as shown in Figure 4. In addition, it is not clear if one or both FXIa catalytic domains interact with the substrate. A study by Wolberg et al38suggested that the 2 proteolytic cleavages made in factor IX during activation by FXIa may require both FXIa catalytic domains. Finally, for most reactions involving vitamin K–dependent coagulation proteases, phospholipid or activated platelets are both suitable surfaces. In contrast, our results strongly indicate that FXIa behaves differently in the presence of activated platelets compared to purified phospholipid. This suggests that FXIa may be interacting with a platelet membrane protein rather than the phospholipid component of the platelet membrane as shown in Figure 4. In this regard, preliminary data indicating that FXI binds to glycocalycin, the extramembrane portion of glycoprotein Ib, are of interest (P.N.W., unpublished observations, June 1999). Additional work will be required to validate this model; however, it offers a reasonable explanation as to why FXI, alone among coagulation proteases, is dimeric.
The authors are grateful to Dr George J. Broze Jr for his thoughtful reading of the manuscript and to Jean McClure for graphics work.
Supported by grants HL58837 and HL02917 (D.G.) and by grants HL46213, HL56153, and HL56914 (P.N.W.) from the National Heart, Lung, and Blood Institute. D.G. is an Established Investigator of the American Heart Association.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David Gailani, Division of Hematology/Oncology, Vanderbilt University, 538 MRB II, 2220 Pierce Ave, Nashville, TN 37232-6305; e-mail:dave.gailani@mcmail.vanderbilt.edu.