It has been proposed that there are at least 2 classes of dendritic cells (DCs), CD8α+ DCs derived from the lymphoid lineage and CD8α− DCs derived from the myeloid lineage. Here, the abilities of lymphoid- and myeloid-restricted progenitors to generate DCs are compared, and their overall contributions to the DC compartment are evaluated. It has previously been shown that primitive myeloid-committed progenitors (common myeloid progenitors [CMPs]) are efficient precursors of both CD8α+ and CD8α− DCs in vivo. Here it is shown that the earliest lymphoid-committed progenitors (common lymphoid progenitors [CLPs]) and CMPs and their progeny granulocyte-macrophage progenitors (GMPs) can give rise to functional DCs in vitro and in vivo. CLPs are more efficient in generating DCs than their T-lineage descendants, the early thymocyte progenitors and pro-T cells, and CMPs are more efficient DC precursors than the descendant GMPs, whereas pro-B cells and megakaryocyte-erythrocyte progenitors are incapable of generating DCs. Thus, DC developmental potential is preserved during T- but not B-lymphoid differentiation from CLP and during granulocyte-macrophage but not megakaryocyte-erythrocyte development from CMP. In vivo reconstitution experiments show that CLPs and CMPs can reconstitute CD8α+ and CD8α− DCs with similar efficiency on a per cell basis. However, CMPs are 10-fold more numerous than CLPs, suggesting that at steady state, CLPs provide only a minority of splenic DCs and approximately half the DCs in thymus, whereas most DCs, including CD8α+ and CD8α− subtypes, are of myeloid origin.
Introduction
Dendritic cells (DCs) are key regulators of the immune system. They are capable of stimulating lymphocytes to generate potent cell-mediated and humoral immune responses to foreign antigens, and can educate T cells to tolerate self-antigens.1-5 This variety of DC functions may result from the heterogeneity of DC subsets. It has been reported that there are at least 2 phenotypically different DC subsets in mice—CD8α+CD11b−and CD8α−CD11b+ DC.6
The CD8α+CD11b− DCs have been considered to be related to the T-lymphoid lineage because early thymocyte progenitors (TP) (CD4loCD44+CD25−c-Kit+) and thymic pro-T cells (CD44+ CD25+c-Kit+) contain precursors capable of differentiating into DCs of this phenotype when transplanted and because most thymic DCs are CD8α+CD11b−.7,8 TP and pro-T cells reportedly give rise to DCs in vitro without the need of granulocyte-macrophage colony-stimulating factor (GM-CSF) or IL-4,9 both of which are important for the in vitro development of myeloid-derived DCs.10-14
One approach to infer cell lineage relationships is to examine cell subsets in mutant mice. Mice deficient in the transcripion factors RelB,15 PU.1,16 and Ikaros17lack only CD8α− DCs supporting, but not directly demonstrating, the model for different developmental pathways of CD8α+ and CD8α− DCs. In contrast, other mutants give results inconsistent with a tight relation of T cells and DCs through a common progenitor in the thymus. Two recent studies show a developmental dissociation of CD8α+ DCs and T cells in c-Kit−γc−18 and Notch1−/−19 mice. Both mutant animals are completely deficient in T cells but show normal percentages of CD8α+DCs in thymus and spleen. Although the complete absence of TP was not formally proven in these mice, TP may not be a required stage for thymic DC development. Furthermore, the TP population is likely heterogeneous, containing precursors for both thymocytes and some myelomonocytic cells.7 20
Thus, a genetic approach to dissect lineage relationships between myeloid and lymphoid progenitors and the 2 phenotypically distinct DC subsets is inconclusive.21 We believe the most direct way to identify lineage relationships is to isolate clonogenic precursors to purity and to test their full developmental capacity. We have recently isolated clonogenic common lymphoid progenitors (CLPs)22 and clonogenic common myeloid progenitors (CMPs) and their downstream granulocyte-macrophage (GMPs) and megakaryocyte-erythrocyte progenitors (MEPs)23 from mouse bone marrow. Differentiation potentials of CLP, CMP, and GMP are restricted to either lymphoid lineages (T, B, natural killer [NK] cells) or myeloid lineages. We have previously demonstrated that CMPs are highly efficient precursors of both CD8α+ and CD8α− DCs.24 Here, we compared the DC developmental potentials of the lymphoid precursors CLPs, TPs, pro-T cells, and pro-B cells, and the myeloid precursors CMPs, GMPs, and MEPs. We demonstrated directly that CLPs can give rise to functional DCs in vivo and in vitro with greater efficiency than downstream TPs and pro-T cells. In addition, CMPs and GMPs give rise to DCs, whereas DC differentiation activity was undetected in pro-B cells and MEP. Thus, there is a DC developmental pathway independent of myeloid differentiation, and this potential is preserved from CLP to pro-T stages but not in B-cell progenitors. We also show that these lymphoid-derived DCs include CD8α+ and CD8α− DCs but are likely to constitute only a minority of DCs in spleen and approximately half the DCs in thymus, whereas CMPs appear to be the major source of CD8α+ and CD8α− DCs throughout the body.
Materials and methods
Animals
Antibodies, cell staining, and sorting
The following antibodies were used: M1/70 (anti–Mac-1, anti-CD11b), 8C5 (anti–Gr-1), 6B2 (anti-B220), KT-31 (anti-CD3), GK1.5 (anti-CD4), 53-6.7 (anti-CD8α), anti-TER119, A7R34 (anti–IL-7Rα, CD127), 2B8 (anti–c-Kit, CD117), 3C11 (anti–c-Kit, CD117), E13–161-7 (anti–Sca-1), 19XE5 (anti–Thy-1.1), A20.1.7 (anti–CD45.1), AL1-4A2 (anti–CD45.2), NLDC145 (anti-DEC205) (Serotec, Oxford, United Kingdom), and goat anti–rat IgG (PE or Cy5-PE conjugated) (Caltag, Burlingame, CA). Antibodies purchased from Pharmingen were R6-60.2 (anti-IgM), 2.4G2 (anti-FcγRII/III), RAM34 (anti-CD34), NK1.1 (anti-NK cell marker), S7 (anti-CD43), 7D4 (anti-IL2Rα, CD25), 1D3 (anti-CD19), AF6-120.1 (anti-IAb), HL3 (anti-CD11c), 16-10A1 (anti-CD80), GL1(anti-CD86), and 3/23 (anti-CD40) (San Diego, CA). For visualization of biotinylated antibodies, streptavidin-conjugated phycoerythrin (PE), Cy5-PE, and Texas red (Gibco, Rockville, MD) were used.
CLPs, TPs, pro-T cells, pro-B cells, CMPs, GMPs, and MEPs were sorted as reported elsewhere.8,22,23 25 Thorough methodologies for staining and sorting CLPs, CMPs, GMPs, and MEPs can be found athttp://www.metazoa.com UPL 2030 and UPL 2001. In brief, for CLP and the myeloid progenitors, lineage-positive (Lin+) (CD3, CD4, CD5, CD8, B220, IgM, CD19, Mac-1, Gr-1, Ter119) cells were partially removed by immunomagnetic depletion, and CLPs were sorted as lineage-negative (Lin−) IL-7R+Thy-1−Sca-1lo c-Kitlo cells. Myeloid progenitors were sorted as Lin−Sca-1−c-Kit+CD34+FcγRlo(CMP), Lin−Sca-1−c-Kit+CD34+FcγRhi (GMP), and Lin−Sca-1−c-Kit+CD34−FcγRlo(MEP) cells. TP (CD3−CD8−CD4loc-Kit+CD25−) and pro-T cells (CD3−CD8−CD4− c-Kit+CD25+) were sorted after immunomagnetic c-Kit enrichment (biotinylated anti–c-Kit) (3C11 streptavidin microbeads [Miltenyi Biotec, Bergisch-Gladbach, Germany]). Pro-B cells (CD43+B220+IgM−NK1.1−or CD19+CD43+IgM−) were sorted after partial immunomagnetic removal of Gr.1+ and Mac-1+ cells. For all experiments, dead cells were excluded by propidium iodide staining, and appropriate isotype-matched, irrelevant control monoclonal antibodies were used to determine the level of background staining.
Cells were sorted and analyzed using a highly modified dual-laser FACS (488-nm argon laser, 599-nm dye laser) (FACS Vantage; Becton Dickinson Immunocytometry Systems, Mountain View, CA). All cultured and transplanted progenitors and all cells for polymerase chain reaction (PCR) analysis were purified by sorting and resorting to obtain precise numbers of cells essentially pure for the indicated surface marker phenotype. For single-cell and limiting-dilution assays, the re-sort was performed using an automatic cell deposition unit system (Becton Dickinson). The specific number of cells sorted in each well was confirmed by visual inspection under an inverted microscope. For analysis of cultured or in vivo reconstituted cells, antibody staining was used as indicated in “Results.”
In vitro differentiation assay
CLPs, TPs, pro-T cells, pro-B cells, and the myeloid progenitors were cultured in Iscoves modified Dulbecco medium (Gibco) supplemented with 10% fetal calf serum (FCS), 10−4 M 2-mercaptoethanol, sodium pyruvate, and antibiotics. Murine IL-1β (10 ng/mL), IL-3 (20 ng/mL), IL-4 (10 ng/mL), IL-7 (10 ng/mL), steel factor (SLF) (10 ng/mL), tumor necrosis factor α (TNF-α) (10 ng/mL), Flt3-L (40 ng/mL), GM-CSF (10 ng/mL), and M-CSF (10 ng/mL) were used as indicated (all cytokines from R&D Systems, Minneapolis, MN). Cells were cultured in 60-well Terasaki trays (up to 103cells/well) or in 96-well plates (up to 104 cells/well). Cluster formation was evaluated using inverted phase-contrast microscopy. To use cells in further assays, DC clusters were disrupted by adding 0.1 vol of 0.1 M EDTA and by repeated pipetting. All cultures were incubated at 37°C in a humidified chamber under 7% CO2. Lineage-reduced bone marrow–derived DCs were generated in culture as described11 using GM-CSF (10 ng/mL) with the addition of IL-4 (10 ng/mL).
Mixed leukocyte reaction
Graded numbers (103-104) of irradiated (25 Gy) DCs either harvested from cultures or sorted from reconstituted animals (all C57Bl/Ka) were plated in U-bottom 96-well plates together with 2 × 105 splenocytes or 1 × 105 lymph node cells of BALB/c mice in a final volume of 200 μL. Culture media consisted of RPMI 1640 and 10% FCS. No cytokines were added. Cells were cultured for 4 days and pulsed with 1 μCi3H-thymidine per well during the last 16 hours of culture.3H-thymidine incorporation was measured on a β-plate counter (Wallac, Gaithersburg, MD).
RT-PCR analysis
Total RNA was extracted from 103 double-sorted CLPs taken from bone marrow, 103 double-sorted major histocompatibility complex (MHC) class II+CD11c+CD8α+ and MHC class II+CD11c+CD8α− cells from spleen, 103 double-sorted MHC class II+CD11c+CD8α+ from thymus, and 103 double-sorted MHC class II+CD11c+ DC from day 4 of CLP culture. These RNA samples were reverse-transcribed to cDNA as described previously.23,26 cDNA was analyzed for the presence of MHC class II activator (CIITA), Epstein-Barr virus–induced molecule-1 ligand chemokine (ELC), RelB, PU.1, CD8α, and CD3ε. PCR primer sequences and annealing temperatures used were as follows: CIITA-F, CCA GAA CTG GTT GTA GAG CC; CIITA-R, CAG CTT GCT AGG CTC CAA TT (tm, 65°C), resulting in a 500-bp PCR product27; ELC-F, GGT GCT AAT GAT GCG GAA GAC; ELC-R, AGA CAC AGG GCT CCT TCT GGT (tm, 65°C), resulting in a 250-bp PCR product28; RelB-F, GAT CCA CAT GGA ATC GAG AG; RelB-R, AGA TGT CCG ATT CAG GAT GA (tm, 60°C), resulting a 575-bp PCR product29; PU.1-F, TGG AAG GGT TTT CCC TCA CC; PU.1-R, TGC TGT CCT TCA TGT CGC CG (tm, 60°C), resulting in a 615-bp PCR product30; CD8α-F, CAC GAA TAA TAA GTA CGT TCT CAC C; CD8α-R, ATG TAA ATA TCA CAG GCG AAG TCC A (tm, 60°C), resulting in a 268-bp PCR product; CD3ε-F, GAA AGA ATC AGG CTG CTC AGA; and CD3ε-R, TGG AGA TGG TGA TGA CCA TCC GA (tm, 60°C), resulting in a 539-bp PCR product. TheHPRT gene was also amplified as a control. PCR amplification consisted of an initial denaturation step at 94°C for 3 minutes, followed by 32 cycles at 94°C for 30 seconds, annealing for 30 seconds, and extension at 72°C for 90 seconds in each cycle. PCR products were electrophoresed on an ethidium bromide–stained 2.0% agarose gel. PCR amplification was repeated at least twice for at least 2 separately prepared cDNA samples for each experiment.
In vivo reconstitution assays
For reconstitution assays, purified progenitors were injected into the retro-orbital venous sinus of 8- to 12-week-old, lethally irradiated (9.5 Gy) congenic mice, which differed only at the CD45 allele, together with 2 × 105 host CD45-type whole bone marrow cells. The presence of donor-derived DCs was evaluated at different time points. DCs were isolated as described elsewere.7,31 Briefly, spleens and thymi were cut in small fragments and digested under repeated agitation for 30 minutes. at 37°C in RPMI 1640 supplemented with 10% FCS, collagenase (1 mg/mL) (Collagenase D; Boehringer Mannheim, Mannheim, Germany), and DNAse (50 μg/mL) (DNAse I from bovine pancreas grade II; Boehringer Mannheim). After organ digestion, EDTA was added at a concentration of 10 mM to disrupt DC complexes, and it was maintained in the medium throughout the subsequent procedures. Debris was removed by filtration, and red cells were osmotically lysed. Initially, DCs were enriched by gradient centrifugation using Nycodenz medium (Nycomed AS, Oslo, Norway) followed by immunomagnetic depletion of the remaining T cells, B cells, and neutrophils as described.7 31 However, using this method, we had low DC recovery rates. Therefore, we used CD11c+ selection for DC enrichment in all subsequent experiments, which resulted in high recovery rates. Single-cell suspensions were incubated for 10 minutes with mouse immunoglobulin to block Fc receptors. This was followed by 25-minute incubation with CD11c (N418) MicroBeads (Miltenyi Biotec) and positive selection over a magnetic column (MiniMACS and MidiMACS; Miltenyi Biotec) according to the manufacturer's instructions. Finally, cells were fluorescence-labeled and analyzed or sorted as indicated in “Results.” To evaluate DC percentages of total cells, staining and analysis were performed after the digestion of organs without further DC enrichment.
Results
CLPs differentiate into DCs in vitro
CLPs were sorted and cultured at 100 to 1000 cells per well in Terasaki 60-well plates containing a variety of recombinant cytokines (Table 1). Cells with cytoplasmic extensions characteristic of DC morphology were observed starting on day 1.5 of culture and formed expanding clusters ranging from 5 to more than 50 cells. Cluster size appeared to reflect DC density rather than proliferation on a single-cell basis; clusters with more than 20 cells were rarely observed in cultures started with fewer than 500 CLPs. The maximum number of cells was reached on day 4 to 5 of culture; thereafter, most cells died with approximately 10% of cells remaining on day 12 of culture. On day 4, the number of clusters containing 10 or more DCs in cultures grown from 103 CLPs was used to determine the effect of cytokines on DC differentiation and proliferation (Table 1). The highest cluster number and expansion (approximately 5-fold) of the initially plated cells was achieved with the combination of IL-1β, IL-3, IL-4, IL-7, SLF, Flt3-L, and TNF-α (Table 1, row 1). With this cytokine combination, approximately 90% of the cells showed typical DC morphology (Figure1A-B and Figure 2). The removal of IL-7 from the cocktail caused a massive drop in cluster formation. The addition of myeloid-directed cytokines, such as GM-CSF and M-CSF, did not stimulate dendritic cluster formation from CLPs. Nor did DCs develop when IL-7 was added to GM-CSF and IL-4 (Table 1, row 12). No adherent macrophages developed in any cytokine combination, in accordance with our previous findings.22 Thus, the cytokine cocktail with the highest cluster formation capacity on day 4 (Table 1, row 1) was used for all in vitro experiments.
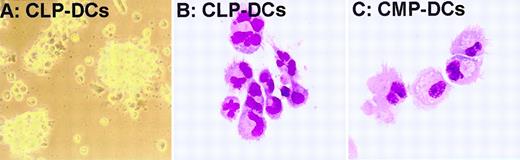
Morphology of in vitro–generated DCs from CLPs and CMPs.
(A) Typical clusters of DCs developing from 1 × 103 CLPs cultured with IL-1β, IL-3, IL-4, IL-7, SLF, TNF-α, and Flt3-L. Clusters were photographed directly in the Terasaki tray on an inverted microscope (objective × 40) on day 4 of culture. (B, C) Cells developing from 100 CLPs in the above cocktail on day 4 or from 5000 CMPs in IL-3, IL-4, SLF, TNF-α, Flt-3L, and GM-CSF on day 6 were dissociated with EDTA, spun onto slides, and stained with Giemsa (objective × 60 oil). Almost all CLP-derived cells (B) and a substantial fraction of CMP-derived cells (C) displayed typical DC morphologies varying in size and nuclear shape.
Morphology of in vitro–generated DCs from CLPs and CMPs.
(A) Typical clusters of DCs developing from 1 × 103 CLPs cultured with IL-1β, IL-3, IL-4, IL-7, SLF, TNF-α, and Flt3-L. Clusters were photographed directly in the Terasaki tray on an inverted microscope (objective × 40) on day 4 of culture. (B, C) Cells developing from 100 CLPs in the above cocktail on day 4 or from 5000 CMPs in IL-3, IL-4, SLF, TNF-α, Flt-3L, and GM-CSF on day 6 were dissociated with EDTA, spun onto slides, and stained with Giemsa (objective × 60 oil). Almost all CLP-derived cells (B) and a substantial fraction of CMP-derived cells (C) displayed typical DC morphologies varying in size and nuclear shape.
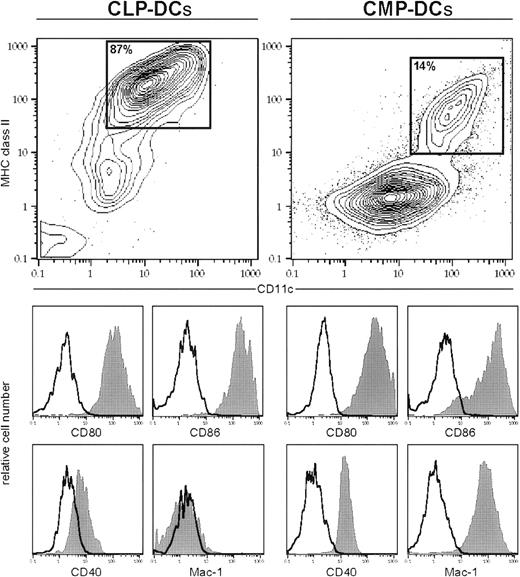
Phenotypic characterization of in vitro CLP- and CMP-derived DCs.
Cells were cultured as described in “Material and methods.” All cells were stained with anti–MHC class II and anti-CD11c (contour plot) plus 1 or 2 additional markers (histograms) by day 4 (CLP cultures) or day 6 (CMP cultures), respectively. Filled histograms show specific staining, and open histograms show isotype-matched controls of gated population in the contour plot (MHC class II+CD11c+). CLPs and CMPs do not express MHC class II (not shown).
Phenotypic characterization of in vitro CLP- and CMP-derived DCs.
Cells were cultured as described in “Material and methods.” All cells were stained with anti–MHC class II and anti-CD11c (contour plot) plus 1 or 2 additional markers (histograms) by day 4 (CLP cultures) or day 6 (CMP cultures), respectively. Filled histograms show specific staining, and open histograms show isotype-matched controls of gated population in the contour plot (MHC class II+CD11c+). CLPs and CMPs do not express MHC class II (not shown).
Although CLPs expressed no detectable MHC class II or CD11c (not shown), there was a gradual up-regulation of MHC class II and CD11c during the culture period. MHC class II+ CD11c+cells were positive for DC-related antigens, including CD40, CD80, CD86 (Figure 2). CLP-derived DCs expressed low levels of DEC205 and were negative for other lineage markers, such as Mac-1, Gr-1, B220, and CD3 (Figure 2 and not shown). It has been reported that CD8α is expressed on DCs in vivo but not on DCs developed in vitro from TP.9 Consistent with these data, the DCs derived from cultured CLPs did not express CD8α (not shown).
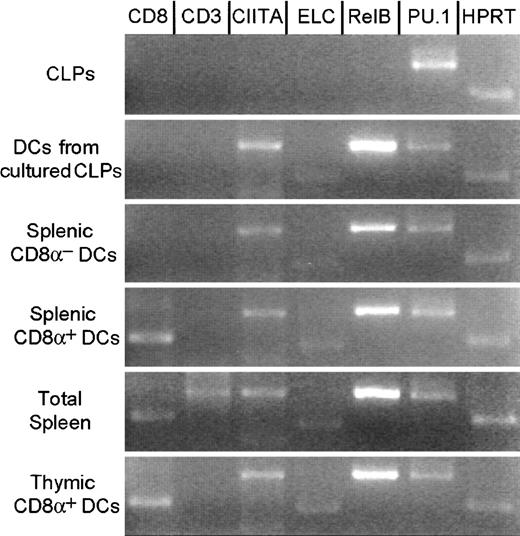
DCs in T-cell areas of lymph nodes express ELC28 and the MHC class II activator (CIITA) gene. RelB andPU.1 are reported to be critical genes in the development of CD8α−, but not CD8α+, DCs.15 16 We therefore tested the expression of ELC,CIITA, RelB, and PU.1 in CLPs and CLP-derived DCs (MHC class II+ CD11c+) by RT-PCR 4 days after the initiation of cultures. CLP-derived DCs, but not freshly isolated CLPs, expressed ELC and CIITA, showing that DC commitment occured during culture. CLP-derived DCs expressed RelB and PU.1, as do all CD8α+ and CD8α− DC populations (Figure 3). Expression profiles of these genes did not fit with their proposed role in DC subset specification (lymphoid vs myeloid, CD8α+ vs CD8α−) but were consistent with some role in the development or function of all types of DCs.
Gene expression in CLPs and DCs.
RT-PCR analysis of CLPs, CLP-derived DCs in culture, CD8α+ and CD8α− splenic DCs, total splenic cells, and CD8α+ thymic DCs sorted from healthy animals. Although CLPs do not express CIITA, ELC, andRelB, CLP-derived DCs in culture, splenic and thymic DCs express these genes. No DC subsets expressed CD3. Uncultured splenic and thymic DCs expressed CD8α+ mRNA. PCR conditions were as described in “Materials and methods.”
Gene expression in CLPs and DCs.
RT-PCR analysis of CLPs, CLP-derived DCs in culture, CD8α+ and CD8α− splenic DCs, total splenic cells, and CD8α+ thymic DCs sorted from healthy animals. Although CLPs do not express CIITA, ELC, andRelB, CLP-derived DCs in culture, splenic and thymic DCs express these genes. No DC subsets expressed CD3. Uncultured splenic and thymic DCs expressed CD8α+ mRNA. PCR conditions were as described in “Materials and methods.”
CLPs are more potent in generating DCs in vitro than early thymocyte progenitors on a single-cell basis
To clarify the efficiency of DC development from CLPs, we determined the ability of single CLPs and TPs to differentiate to DCs. CLPs and TPs were cultured in numbers of 1, 2, and 5 cells per well. Results are shown in Table 2. On day 1, some CLPs and thymocyte progenitors began to increase in size and to show cytoplasmic extensions. Proliferative activity varied widely between wells. On day 4, cells in each well were counted, and DC read-out was evaluated by morphology in culture and by Giemsa staining of cytospin preparations. Eighty-three percent of single CLPs develop into cells with DC morphology. In contrast, 47% of single TPs developed into DCs. From single-sorted CLPs, the average number of DCs per well showing cells with DC morphology was 3.8. From the single-sorted TPs, the average number of DCs per well showing cells with DC morphology was 2.7. When we cultured 2 or 5 cells of each population, both DC plating efficiency and average number of DCs were always higher in CLPs than in TPs (Table 2).
CMPs and GMPs differentiate into DCs in vitro
CMPs and GMPs were sorted and cultured at 2000 to 5000 cells per well in 96-well plates in 100 μL media containing various combinations of the cytokines IL-1, IL-3, IL-4, IL-7, SLF, Flt-3L, TNF-α, and GM-CSF. DC read-out was evaluated by FACS analysis on days 4, 6, 8, and 10. Fresh media were added on days 4 and 8 in ongoing cultures. The most efficient DC read-out from CMPs and GMPs was achieved with a cytokine combination consisting of IL-3, IL-4, SLF, Flt-3L, TNF-α, and GM-CSF on days 6 to 8 of culture (Figure 1C). With this combination, CMPs and GMPs expanded approximately 35 times and 20 times from the input cell numbers and gave rise to approximately 15% and 10% MHC class II+ CD11c+ DCs, respectively. GM-CSF or SLF deletion from the cultures caused the foremost drop in DC generation, proving those the most important of the evaluated cytokines for in vitro DC generation from myeloid-committed progenitors.
Although CMP and GMP, like CLPs, express no detectable MHC class II or CD11c (not shown), those markers were up-regulated during the culture period. The MHC class II+CD11c+ cells expressed Mac-1 and DC-related antigens, including CD40, CD80, and CD86 (Figure2).
In vivo DC differentiation potential of CLPs, TPs, pro-T cells, CMPs, and GMPs
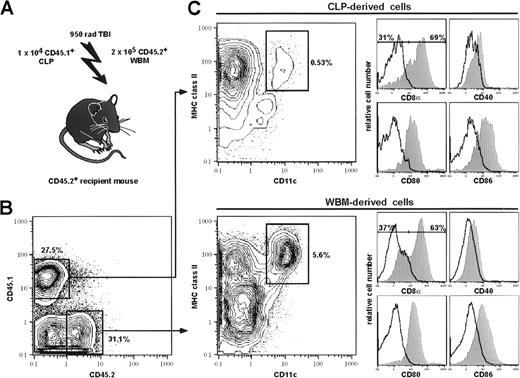
We previously reported that CLPs have potent reconstitution activity for T and B cells; after intravenous injection into sublethally irradiated RAG-2–deficient mice, 1 × 103CLPs give rise to approximately 1.5 × 107B220+ splenic B cells and 1 × 107CD3+ splenic T cells by 2 weeks and 5 weeks, respectively.22 To estimate the contribution of CLPs to the DC compartment in vivo, we transplanted graded numbers (1 × 103, 5 × 103, and 10 × 103) of CLPs to lethally irradiated host BM protected CD45 congenic recipient mice (see “Materials and methods”). Myelo-erythroid progeny were undetected as previously reported.22 Although reasonable DC progeny above background was not detectable with 1 × 103transplanted CLPs, a significant fraction of cells with DC phenotype (MHC class II+CD11c+CD40+CD80+CD86+ CD8α+/−) was seen in spleen, thymus and lymph nodes 14 to 21 days after transplantation of 5 × 103 or more CLPs (Figure4, Table3). Most likely this reflected the fact that CLPs have a large burst size for B and T cells but, like TPs and pro-T cells, a small expansion potential for DC. Therefore, DC progeny became clearly evident only with high progenitor cell input. Sorted MHC class II+ CD11c+ cells displayed typical DC morphology after 12-hour incubation in complete media (not shown). At 2 weeks after transplantation, approximately 70% of CLP-derived and approximately 65% of host-derived splenic DCs showed CD8α expression. Four weeks after reconstitution, total numbers of CLP-derived DCs decreased significantly. On day 28, approximately 80% of CLP-derived splenic DCs were CD8α+. In contrast, only approximately 35% of host-derived DCs were CD8α+ at this time point (data not shown).
CLPs give rise to DCs in vivo.
CLPs were competitively transplanted into lethally irradiated CD45 congenic animals, together with host-type whole bone marrow (A). CD45 expression was used to gate on donor-type (CD45.1) and host-type (CD45.2) CD11c+-enriched, live splenocytes on day 15 after transplantation (B). Both CLP (upper panel) and host-derived (lower panel) cells were analyzed for DC phenotype by MHC class II+ and CD11c+ expression (C, contour plots). CLP-derived MHC class II+CD11c+ cells account for approximately 10% of donor-derived MHC class II+CD11c+ cells in a CD11c+-enriched sample. MHC class II+CD11c+ cells were further analyzed for CD8α, CD40, CD80, and CD86 expression (C). Closed hisograms represent specific staining, and open histograms represent isotype-matched controls of the MHC class II+CD11c+ cells. On day 15, 69% of CLP-derived DCs and 63% of host-derived DCs were CD8α+. Both CLP and host-derived DCs expressed low to intermediate levels of CD40, CD80, and CD86. Details are given in “Materials and methods.”
CLPs give rise to DCs in vivo.
CLPs were competitively transplanted into lethally irradiated CD45 congenic animals, together with host-type whole bone marrow (A). CD45 expression was used to gate on donor-type (CD45.1) and host-type (CD45.2) CD11c+-enriched, live splenocytes on day 15 after transplantation (B). Both CLP (upper panel) and host-derived (lower panel) cells were analyzed for DC phenotype by MHC class II+ and CD11c+ expression (C, contour plots). CLP-derived MHC class II+CD11c+ cells account for approximately 10% of donor-derived MHC class II+CD11c+ cells in a CD11c+-enriched sample. MHC class II+CD11c+ cells were further analyzed for CD8α, CD40, CD80, and CD86 expression (C). Closed hisograms represent specific staining, and open histograms represent isotype-matched controls of the MHC class II+CD11c+ cells. On day 15, 69% of CLP-derived DCs and 63% of host-derived DCs were CD8α+. Both CLP and host-derived DCs expressed low to intermediate levels of CD40, CD80, and CD86. Details are given in “Materials and methods.”
We have shown that the earliest myeloid-committed progenitors, CMPs, can produce functional DCs of both CD8α+ and CD8α− phenotype after transplantation in vivo.24 Similarly, the downstream GMPs are capable of giving rise to both CD8α+ and CD8α− DCs in spleen and thymus, though with substantially lower efficiency than CMPs (Table 3 and not shown). To compare the contributions of lymphoid-committed and myeloid-committed progenitors to the entire DC compartment, we transplanted prospectively purified CLPs, TPs, pro-T cells, CMPs, and GMPs, along with host CD45 congenic bone marrow cells, into lethally irradiated hosts. Table 3 shows the mean number of donor-derived DC progeny (MHC class II+CD11c+). On a per cell basis, transplanted CLPs are more efficient than the other progenitors in giving rise to thymic DCs. CLPs and CMPs are roughly equivalent, and both are far more efficient than TPs, pro-T cells, and GMPs in producing splenic DCs. CLPs represent 0.02%, CMPs 0.2%, and GMPs 0.4% of total nucleated cells in the bone marrow; approximately 2 × 104 CLPs, 20 × 104 CMPs, and 40 × 104 GMPs should reside in the femurs and tibias of both legs of a 6- to 8-week-old animal. Therefore, the transplanted 1 × 104 CLPs, 2 × 104 CMPs, and 4 × 104 GMPs represented approximately 50% the bone marrow equivalent of CLPs and approximately 10% the bone marrow equivalent of CMPs and GMPs in these bones, respectively. Similarly, approximately 3 × 104CD25−c-Kit+CD4loCD8−TP and 6 × 104CD25+c-Kit+CD4−CD8−pro-T cells exist in a 6- to 8-week-old thymus (each population accounted for 0.02% and 0.04% of all thymic cells, respectively, in our hands (data not shown). Hence, the transplanted 2 × 104 TPs and 3 × 104 pro-T cells represented 66% and 50% thymus equivalent, respectively. By transplanting CLPs and CMPs in increasing numbers, an approximate linear increase of progenitor-derived DCs was seen in spleen and thymus (data not shown). If their behavior on transplantation reflects the endogenous production of DCs by these progenitors, and if all DCs are produced through either CLPs or CMPs, we calculate that approximately 90% of splenic DCs and approximately 50% of thymic DCs are derived from CMPs and that approximately 10% splenic DCs and approximately 50% thymic DCs are of CLP origin (Table 3).
CLP- and CMP-derived DCs are able to present allogeneic antigens efficiently
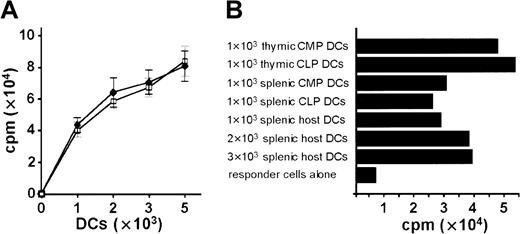
We evaluated the antigen-presenting capacity of progenitor-derived DCs by a mixed lymphocyte reaction (MLR) assay. DCs developed in vitro from C57BL (H-2b) CLPs and from lineage-depleted bone marrow cells were irradiated on day 5 of culture and seeded in graded numbers. CLP-derived DCs were as efficient as bone marrow–derived DCs in their capacity to stimulate allogeneic (BALB/c: H-2d) lymph node cells (Figure 5A). CLP-derived MHC class II+CD11c+ DCs in thymus and spleen on day 15 after transplantation are as potent as CMP-derived and host-derived DCs to induce MLR (Figure 5B).
DCs derived from CLPs and CMPs were functionally active in allogeneic mixed leukocyte reactions.
(A) Stimulation of 105 allogeneic BALB/c lymph node cells by graded numbers (x-axis) of in vitro CLP-derived DCs (day 5 of culture) (closed diamonds) and by bone marrow (Lin−)-derived DCs (day 5 of culture) (open squares). Cultures were grown and MLR was performed as described in “Materials and methods.” Results are the means ± SE each with 3 to 6 wells per point. (B) MLR of in vivo CLP-, CMP-, and host-derived DCs sorted by high expression of CD11c and MHC class II. Sorted DCs were cultured in numbers as indicated for 12 hours in complete media without cytokines, and 2 × 105 nucleated BALB/c splenocytes were added. MLR was performed as described in “Materials and methods.” (A, B) Results of 3 representative experiments are shown.
DCs derived from CLPs and CMPs were functionally active in allogeneic mixed leukocyte reactions.
(A) Stimulation of 105 allogeneic BALB/c lymph node cells by graded numbers (x-axis) of in vitro CLP-derived DCs (day 5 of culture) (closed diamonds) and by bone marrow (Lin−)-derived DCs (day 5 of culture) (open squares). Cultures were grown and MLR was performed as described in “Materials and methods.” Results are the means ± SE each with 3 to 6 wells per point. (B) MLR of in vivo CLP-, CMP-, and host-derived DCs sorted by high expression of CD11c and MHC class II. Sorted DCs were cultured in numbers as indicated for 12 hours in complete media without cytokines, and 2 × 105 nucleated BALB/c splenocytes were added. MLR was performed as described in “Materials and methods.” (A, B) Results of 3 representative experiments are shown.
Pro-B cells do not have detectable DC differentiation potential
It was reported that CD19+ pro-B cells from BALB/c mice develop into DCs under the same culture conditions as TPs.25 We wanted to compare pro-B cells with CLPs and TPs in their ability to generate DCs. Rigorously purified pro-B cells (either IgM−, NK1.1−, B220+, CD43+, or IgM−, CD19+, CD43+) were cultured at 103, 5 × 103, and 104 cells per well with IL-1β, IL-3, IL-4, IL-7, SLF, Flt3-L, and TNF-α or in the same cocktail without IL-4. We were unable to detect any DC progeny determined by surface phenotype and morphology on days 4 and 6 of cultures. We repeatedly transplanted 3 to 4 × 104double-sorted pro-B cells (either IgM−, NK1.1−, B220+, CD43+, or IgM−, CD19+, CD43+) and looked for DC read-out on days 8 and 15 after transplantation. Although pro-B cells generated a large amount of mature B cells, no DC progeny could be detected (not shown).
MEP do not have significant DC differentiation potential
MEPs cultured under the same conditions as CMPs and GMPs expanded approximately 5 times and only gave rise to 0.1% to 0.5% MHC class II+CD11c+ cells by day 6 of culture. However, when the MEP fraction was again divided by CD34 expression, the less-positive fraction did not give rise to DCs. It seems likely, then, that a few progenitors at the phenotype margin between CMP and MEP have some in vitro DC potential. This could be due either to a small number of CMP contaminants among the 2000 to 5000 MEPs cultured or to a few MEPs at the phenotypic margin that may be able to differentiate into DCs but not to granulocytes or macrophages. By transplanting 3 × 104 MEPs (accounting for 33% of bone marrow equivalent of those progenitors), no DC read-out could be detected on days 8 and 15 after transplantation (not shown). Therefore, MEPs have no significant, if any, DC differentiation potential.
Discussion
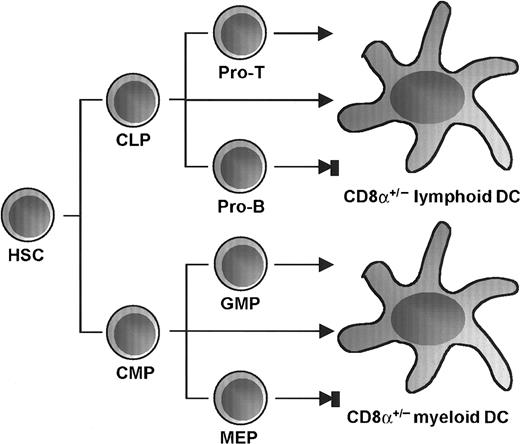
We have previously shown that CMPs efficiently give rise to both CD8α+ and CD8α− DC subsets.24Here we show that the downstream granulocyte-macrophage–restricted progenitors, but not the megakaryocyte-erythrocyte–restricted progenitors, give rise to DCs, and we compare the DC potentials of the myeloid-committed progenitors to lymphoid-committed progenitors. We show directly that CLPs, the earliest lymphoid-restricted progenitors, are capable of differentiating into both CD8α+ and CD8α− functional DCs. This differentiation activity into DCs is preserved in early T-cell progenitors, declining along with T-cell maturation, but is absent in B-cell progenitors. Thus, in addition to the DC developmental pathway from CMP, there is a DC developmental pathway initiated at the CLP, which continues to T-cell–committed progenitors (Figure 6).
Proposed differentiation pathways from HSC to DC.
The earliest CLPs and CMPs in bone marrow are capable of developing into DCs of both CD8α+ and CD8α−phenotypes. DC development potential is preserved in early T-cell progenitors and declines along with T-cell maturation as well as during granulocyte-macrophage commitment but is absent in B-cell progenitors and megakaryocyte-erythrocyte progenitors.
Proposed differentiation pathways from HSC to DC.
The earliest CLPs and CMPs in bone marrow are capable of developing into DCs of both CD8α+ and CD8α−phenotypes. DC development potential is preserved in early T-cell progenitors and declines along with T-cell maturation as well as during granulocyte-macrophage commitment but is absent in B-cell progenitors and megakaryocyte-erythrocyte progenitors.
It has been shown that the early thymocyte progenitor (TP) and pro-T cell populations contain cells that are capable of differentiating into CD8α+ DCs in the thymus and secondary lymphoid organs7,20 and that give rise to DCs in vitro.9 Our data indicate that approximately 80% of single CLP and approximately 50% of single TPs can differentiate into DCs. Thus, the developmental potential of DCs is efficiently maintained during the transition from CLP to TP stages.
It has been reported that pro-B cells in BALB/c mice can differentiate into DCs in vitro, though at a low frequency.25 In addition, pro-B cells from Pax5−/− mice were able to give rise to DCs in vitro.32 We did not detect DC development from purified pro-B cells from C57BL mice in vivo or in vitro, with doses as high as 104 cells. Because pro-B cells in Pax5−/− mice are capable of differentiating into myelomonocytic cells, DC differentiation from Pax5−/−pro-B cells might not reflect physiological differentiation potentials. Elsewhere we have shown that CLPs and pro-T cells, but not pro-B cells, can be redirected from lymphoid to myelomonocytic development through the introduction of transgenes encoding hIL-2R or hGM-CSFR into these cells and culturing the transfectants with the cognate human cytokine.33 It is interesting that the natural loss of DC generative capacity and the revealed loss of myeloid conversion from the lymphoid pathway occur at similar differentiation checkpoints.
An important question in DC biology has been whether CD8α expression on DCs marks lymphoid origin. Our data indicate that CLPs, as well as CMPs and GMPs, give rise to both CD8α+ and CD8α− DCs in spleen and that virtually all thymic DCs derived from either CLPs or CMPs and GMPs were CD8α+.24 Although 1 in 2780 cells with the CMP phenotype is capable of differentiating into B cells on OP9 stromal layers,23 it seems unlikely that approximately 10 CMPs with B-cell differentiation capacity within the transplanted 2 × 104 CMPs account for most CMP-derived DCs (80%-90%) that express CD8α+.24 In addition, GMPs generate CD8α+ and CD8α−DCs but not B cells, and we here show that pro-B cells do not generate DCs as discussed above. These data indicate that CD8α expression does not delineate the ontogeny of DC; rather, both types of DC can develop from either myeloid-restricted or lymphoid-restricted stages of hematopoietic differentiation. In a recent report, it was shown that TPs are capable of giving rise to splenic CD8α−DCs.34 According to the authors, these CD8α− DCs were possibly missed before because of the DC enrichment technique. In addition, it has been reported that TPs develop into CD8α−/low DCs in lymph nodes.35 We have not tested whether CD8α expression represents a stage of DC maturation or whether CD8α+ and CD8α− DCs represent 2 distinct and independent DC types. In our study, there was a gradual increase in the percentage of CLP-derived CD8α+ DC during the 2 to 4 weeks after transplantation. A similar observation was made earlier for TP-derived DCs.8,36 Therefore, CD8α expression on DCs might be maturation-stage dependent. In support of this hypothesis, it has been reported that CD8α is an activation marker of Langerhans cells after antigen uptake and migration into draining lymph nodes.37,38 Additionally, it has been shown that CD8α+ DCs reside in the T-cell areas of secondary lymphoid organs, whereas CD8α− DCs are found in the marginal zones39,40 and marginal zone DCs have been proposed to be the immature precursors of DC in the T-cell areas.41-43 On the other hand, if CD8α+ and CD8α− DCs develop independently, it is also possible that increased CD8α+ DC detection after the transplantation of committed progenitors might result from a longer half-life of CD8α+ DCs. Data are conflicting on the half-lives of these DC subsets.39 44
It is of interest to know whether DCs derived from CLPs or CMPs have diverse, specific functions. Because each progenitor can give rise to CD8α+ and CD8α− DCs, previous experiments using CD8α expression on DCs to define a “lymphoid” origin are not helpful on this point.45-52 Our data show that CLP-derived DCs are as potent as CMP-derived DCs, at least for T-cell stimulation by presenting allogeneic antigens (Figure 5). However, to test for differences between CLP- and CMP-derived DCs, more detailed functional analyses are required.
It is also important to know whether CMP- and CLP-derived DC subsets use the differentiation machinery common to myeloid and lymphoid differentiation or whether each subset requires molecular events specific for either myeloid or lymphoid lineage. Our data show that CLPs express the IL-7R and require IL-7 to differentiate into DCs. On the other hand, myeloid-related cytokines, such as GM-CSF and M-CSF, have no effect on DC development from CLP, though GM-CSF is important for DC development from myeloid progenitors, as shown here and previously.11,14,53,54 Because IL-7 is an indispensable cytokine for both T- and B-cell development,55 56 but not for myeloid development, the differentiation machinery of DCs from these lymphoid progenitors may be related to molecular events downstream of the IL-7R.
Mice deficient in the transcription factorsRelB,15PU.1,16 andIkaros17 reportedly lack CD8α−DC but not CD8α+ DC. However, because CD8α is not a marker of lymphoid derivation,24 these data no longer suggest that these transcription factors are deterministic for myeloid or lymphoid DC differentiation pathways. Furthermore, we show that bothRelB and PU.1 are expressed in purified CD8α+ and CD8α− splenic DCs and in CLP-derived DCs and that PU.1 has been already turned on at both CLP and CMP (Figure 3).23 Although these nonquantitative PCR results do not reflect the amount of transcribed mRNA, these factors may play an important role in the development of myeloid and lymphoid DC, irrespective of their CD8α expression. Indeed, another mouse strain deficient in PU.1 reportedly lacks both CD8α+ and CD8α− DC.57 Further studies are required to understand the role of these transcription factors in each pathway.
Parallel reconstitution experiments using purified CMPs and CLPs show that these populations are roughly equivalent at producing splenic DCs. Because CMPs are more frequent than CLPs, it is reasonable to propose that the contribution of CMPs to total splenic DCs is approximately 10-fold higher than CLPs; CMPs and CLPs likely contribute equally to the formation of the thymic DC compartment. Furthermore, there were no apparent differences in absolute or relative numbers of CD8α+ and CD8α− DC in the spleens of IL-7–deficient and wild-type mice (M.G.M. et al, unpublished data, November 2000). Because IL-7 is necessary for CLPs to differentiate into DCs, at least in vitro, these data support our findings that the contribution of CLPs to the splenic DC compartment is smaller than that of CMPs.
In conclusion, lymphoid-restricted progenitors can generate DCs, though CD8α expression is not a marker for “lymphoid” DCs.24 The ability to obtain DCs from purified lymphoid-restricted CLPs and myeloid-restricted CMPs should provide the means to clarify the differences in the developmental mechanisms and functions between these DC subsets of different lineal origins.
We thank S.-I. Nishikawa for the anti–IL-7Rα antibody, L. Jerabek for excellent laboratory management, V. Braunstein for antibody preparation, the Stanford FACS facility for flow cytometer maintenance, and L. Hidalgo, B. Lavarro, and D. Escoto for animal care.
Supported in part by a fellowship from Deutsche Krebshilfe, Dr Mildred-Scheel-Stiftung für Krebsforschung (M.G.M.), National Institutes of Health training grant 5T32 AI-07290 (D.T.), a Jose Carreras International Leukemia Society grant (K.A.), the Claudia Adams Barr Program (K.A.), and a United States Public Health Service grant (CA42551) (I.L.W.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Markus G. Manz, Department of Pathology and Developmental Biology, B261 Beckman Center, Stanford University School of Medicine, 279 Campus Dr, Stanford CA 94305-5428; e-mail:manz@leland.stanford.edu.