Several lines of evidence indicate that transcriptional activation is coupled with DNA replication initiation, but the nature of initiation of DNA replication in mammalian cells is unclear. Polyoma virus replicon is an excellent system to analyze the initiation of DNA replication in murine cells because its replication requires an enhancer, and all components of replication machinery, except for DNA helicase large T antigen, are supplied by host cells. This system was used to examine the role of signal transducer and activator of transcription (STAT5) in replication initiation of polyoma replicon in the mouse lymphoid cell line BA/F3. The plasmid with tandem repeats of consensus STAT5 binding sites followed by polyoma replication origin was replicated by stimulation with human granulocyte-macrophage colony-stimulating factor (hGM-CSF) in the presence of polyoma large T antigen in BA/F3 cells. Mutation analysis of the hGM-CSF receptor β subunit revealed that only the box1 region is essential, and the C-terminal tyrosine residues are dispensable for the activity. Addition of the tyrosine kinase inhibitor genistein suppressed this replication without affecting transcriptional activation of STAT5. Because deletion analysis of STAT5 indicates the importance of the C-terminal transcriptional activation domain of STAT5 for the initiation of replication, the role of this region in the activation of replication was examined with a GAL4–STAT5 fusion protein. GAL4–STAT5 activated replication of the plasmid containing tandem repeats of GAL4 binding sites and polyoma replication origin in BA/F3 cells. Mutation analysis of GAL4–STAT5 indicated that multiple serine residues coordinately have a role in activating replication. This is the first direct evidence indicating the potential involvement of STAT5 in replication.
Introduction
Roles of Janus kinase (JAK) and signal transducer and activator of transcription (STAT) in cytokine signal transduction were first identified in interferon (IFN) signaling pathways, and it was revealed that all members of the cytokine receptor superfamily activate JAK and STAT.1,2 To date, 7 members of the STAT family with similar structural features, STAT1 to STAT6, have been identified.3 A DNA binding domain is located in the amino-terminal half, and linker and Src homology 2 (SH2) domains followed by the transactivation domain are in the carboxyl-terminal half.2 A conserved Y residue (single-letter amino acid code) is located in the C-terminal region, and phosphorylation of this residue plays an essential role in the dimerization and nuclear translocation of STAT. S residues in the more C-terminal region of the Y residues are phosphorylated by extracellular signal regulated kinase (Erk), p38 mitogen activated protein kinase (p38 MAPK), or Jun N terminal kinase (JNK), which is implicated in the transcriptional activity of STAT1 and STAT3.4 Mechanisms related to the contribution of phosphorylated S residues in transcriptional activation are not well understood. As observed initially in the IFN system, accumulating evidence suggests that STAT proteins are involved in the activation of cytokine-specific genes,5-8 and knockout studies of various STATs strongly support this evidence.9-15
Receptors of granulocyte-macrophage colony-stimulating factor (GM-CSFR) consist of 2 subunits, α and β, both of which are members of the cytokine receptor superfamily.16,17 The α subunit is specific for each cytokine, and the β subunit (βc) is shared by interleukin 3 (IL-3), GM-CSF, and IL-5 receptors.17 GM-CSF induces tyrosine phosphorylation of βc and various cellular proteins and activates early-response genes and cell proliferation in hematopoietic cells and in fibroblasts.18 The βc contains conserved box1 and box2 regions and 8 Y residues located in the cytoplasmic region.19 We and others have analyzed biologic activities of various mutants of βc20-23 and found that multiple distinct signaling pathways that use different receptor domains or Y residues are activated by hGM-CSFR.24-26 Apparently only the box1 region is essential for cell proliferation, whereas signaling cascades such as MAPK and phosphatidylinositol 3-kinase (PI3K), which are transduced through C-terminus Y residues, are dispensable. Because box1 is assumed to bind JAK2, it is likely that activation of JAK2 is sufficient for promotion of proliferation, and this notion was supported by the experiments using chimeric JAK2 protein, which can induce BA/F3 cell proliferation without the activation of MAPK or PI3K cascades.27
GM-CSF activates JAK2 and STAT5 in various hematopoietic cells.28,29STAT5A and STAT5Bgenes encode proteins that are approximately 95% identical in amino acid sequence.30 Mutation analyses of hGM-CSFR showed that STAT5 activation is not required for GM-CSF–dependent antiapoptosis or for proliferation. Although STAT5 is activated by various cytokines such as erythropoietin (EPO), IL-3/IL-5/GM-CSF, prolactin, growth hormone, and thrombopoietin (TPO), STAT5A and/or STAT5B knockout mice showed an essential and a redundant role for physiologic responses associated with growth hormone and prolactin.31 Because other STATs may play pivotal roles for cell differentiation and function, these results suggest that STAT5A and STAT5B are obligate mediators of mammopoietic and lactogenic signaling rather than cell proliferation.32
Because cytokines are strong proliferation-promoting factors for various hematopoietic cells, attempts were made to clarify the role of STATs in cell proliferation. Mutation analyses of the receptor domains of IL-4, GM-CSF, and EPO receptors showed a lack of correlation between cell growth and STAT6 (by IL-4) and STAT5 (by GM-CSF and EPO).33,34 In contrast, dominant-negative STAT5 partially suppressed IL-3–induced proliferation.35 Retardation of colony formation by IL-3, GM-CSF, or IL-5 of bone marrow cells derived from STAT5A/B-deficient mice has been reported.31Activation of hematopoietic cell proliferation through gp130 was shown to depend on the activation of both STAT3 and SHP-2.36 The requirement of STAT3 in Src-induced cell transformation is also indicated,37 which means that STAT3 is probably involved in cell proliferation.
We analyzed the direct role of STAT5 in the initiation of DNA replication in BA/F3 cells using the polyoma (Py) replicon as a model system. This system is widely used to analyze DNA replication of mammalian cells because Py DNA replication makes use of host DNA replication machinery, except for large T antigen (LTag), a viral-encoded DNA helicase. In addition, it is an excellent system to analyze the roles of transcription factors in DNA replication because the Py origin of replication contains an enhancer, which is an essential module in addition to the core sequence of the origin.38 The enhancer sequence can be replaced with multiple copies of a binding site for a single transcription factor.39 Using this system, we examined the ability of STAT5 to activate Py DNA replication. We found that STAT5 can activate DNA replication in response to GM-CSF stimulation and that this activation relies on the C-terminal transactivation domain of STAT.
Materials and methods
Chemicals, media, and cytokines
Fetal calf serum (FCS) was from Sera-Tech Zellbiologische Produkte (St. Salvatol, Germany). RPMI 1640 and Dulbecco modified Eagle medium (DMEM) were from Nikken Biomedical Laboratories (Kyoto, Japan). Recombinant human (h) GM-CSF and G418 were kindly provided by Schering-Plough (Madison, NJ). Mouse (m) IL-3 produced by silkworm,Bombyx mori, was purified as described.40Plasmids encoding wild-type STAT5A and B were gifts from Dr A. Miyajima (Tokyo University, Tokyo, Japan).
Plasmid construction
Four tandem repeats of the proximal STAT5 binding site of β-casein promoter8 were used as an enhancer of plasmids to analyze replication (4XSTOICAT) and luciferase (4XST-Luc). pPyOICAT contains Py fragment (nt 5267 to nt 152), which includes the replication origin core sequence (nt 5267 to nt 56) and early gene promoter but lacks the enhancer region, and chloramphenicol acetyltransferase (CAT) coding sequence.41,42pGL3-promoter (Promega, Madison, WI) has the luciferase coding region and SV40 minimal promoter, but lacks the enhancer. The oligonucleotide containing the proximal STAT5 binding site of β-casein promoter, 5′-GATCTAGATTTCTAGGAATTCAAATCG-3′, 5′-GATCCGATTTGAATTCCTAGAAATCTA-3′, is designed to place the BglII site at the 5′ end and theBamHI site at the 3′ end. Both strands were annealed and phosphorylated by T4 polynucleotide kinase, ligated with the Takara ligation kit version II (Takara Biomedicals, Osaka, Japan), and digested with BamHI and BglII to digest the head to head-ligated product. Four-tandem oligomer was isolated by gel electrophoresis and inserted into the BglII site of pGL3-promoter or pPyOICAT. pPyOICAT contains 4 tandem repeats of the mutant STAT5 binding site30 and was constructed using oligonucleotides 5′-GATCTAGATTTATTTTAATTCAAATCG-3′, 5′-GATCCGATTTGAATTAAAATAAATCTA-5′. pPyG5OICAT,43 which contains 5 tandem repeats of the GAL4 binding site fused with the polyoma replication origin, was kindly provided by Dr Y. Ito (Kyoto University, Kyoto, Japan).
STAT5B-Δ68330 was kindly provided by Dr A. Miyajima (Tokyo University, Tokyo, Japan). Truncation mutants STAT5B-Δ781 and STAT5B-Δ721 were constructed by deletion of downstreamBamHI and NarI sites, respectively. Either theBamHI or NarI site was blunt-ended and ligated to the blunted NotI site of pME18S vector, which contains SRα promoter.44 STAT5B-Y699F was constructed by introduction of a mutation in Y699 to F (TTC) by polymerase chain reaction (PCR) mutagenesis. The AgeI-NotI region of STAT5B was replaced with the AgeI-NotI digested 2-round PCR product, which contains a point mutation within Y699. Primers used for PCR were 5′-GGCATCACCATTGCTTGGAAG (sense) and 5′-TGGCTTCACGAATCCGTCAGCTGCTT (antisense) and 5′-CTGACGGATTCGTGAAGCCACAGATCA (sense) and antisense primer of pME18S vector for the first round. For the second round of PCR, we used 5′-CTGACGGATTCGTGAAGCCACAGATCA (sense) and antisense primer of pME18S vector. Construction of GAL4–STAT5A, GAL4–STAT5B, and their mutants will be described elsewhere (Itoh, Watanabe et al, manuscript in preparation).
Cell lines and culture methods
An mIL-3–dependent pro–B-cell line, BA/F3 was maintained in RPMI 1640 medium containing 5% FCS, 1 ng/mL mIL-3, 100 U/mL penicillin, and 100 μg/mL streptomycin. Stable transformants of BA/F3 cells expressing hGM-CSFR were grown in the same type of medium but supplemented with 500 μg/mL G418. Expression levels of α and β subunits of these cell lines were examined using fluorescence-activated cell sorter analysis.22 Cells with almost equivalent levels of the subunits were used.
COS7 cells were maintained in DMEM containing 10% FCS, 100 U/mL penicillin, and 100 μg/mL streptomycin.
Gel shift analysis
The proximal STAT5 binding site of the β-casein promoter (5′-AGATTTCTAGGAATTCAATCC-3′) served as a probe. The nuclear extract was prepared as described,26 and 5 μg protein was incubated in 12 μL binding buffer (10 mM Tris-HCl, pH 8.0, 100 mM KCl, 5 mM MgCl2, 1 mM dithiothreitol, 10% glycerol, 0.1 mg/mL poly dI-dC, 0.5 mg/mL bovine serum albumin) for 30 minutes at room temperature. Samples were subjected to electrophoresis through 5% polyacrylamide gel in 0.25 × TBE buffer (22.5 mM Tris-borate, 0.5 mM ethylenediamine-N, N, N′, N′-tetraacetic acid [EDTA]) and visualized using a Fuji Image analyzer (model BAS-2000, Tokyo, Japan).
Transient transfection, Py replication assay, and luciferase assay
DNA replication of the transfected plasmid was assayed byDpnI analysis,25,41 and transcription activity was monitored according to luciferase activity. Plasmids were introduced into semiconfluent BA/F3 cells (2 × 106 cells per sample) by electroporation, as described.26 Cells resuspended in factor-depleted media were incubated for 5 hours and then stimulated with 5 ng/mL hGM-CSF. After 24 hours of incubation, the cells were harvested and used for either replication assay or luciferase assay. For replication assay, low-molecular-weight DNA was isolated by the Hirt extraction method, as described.25,45Ten microliters of DNA solution was digested with HindIII (4XSTOICAT) or BamHI (pPyG5OICAT), which linearizes template plasmid, and DpnI. Because DpnI digests only methylated or hemimethylated recognition sites of DNA, newly synthesized DNA is resistant to DpnI digestion. DNA was separated by electrophoresis and transferred to Hybond-N+ (Amersham Pharmacia Biotech Limited, Buckinghamshire, England) by alkaline blotting.46 DNA blots were hybridized with denaturedHindIII-digested template plasmid labeled with32P by a random priming kit (United States Biochemical, Cleveland, OH) with the use of QuikHyb rapid hybridization solution (Stratagene, La Jolla, CA). Blots were visualized and quantified using a Fuji Image Analyzer (model BAS-2000).
For the luciferase assay, proteins were extracted by freezing and thawing of the cells18 and the assay was done, as described, using a luciferase assay substrate (Promega) and a luminometer (model LB9501; Berthold Lumat, Tokyo, Japan). Transfection efficiency was normalized by the alkaline phosphatase activity of the cotransfected CMV–alkaline phosphatase plasmid.
Isolation of chromatin fraction of BA/F3 cells
Chromatin fractions were isolated as described.47Briefly, cells (2 × 107 per sample) treated with or without genistein (20 μg/mL) were incubated for 30 minutes and then stimulated with hGM-CSF (10 ng/mL) for 15 minutes. The harvested cells were suspended in 1 mL cytoskeleton buffer (100 mM NaCl, 300 mM sucrose, 10 mM 1,4-peperazinebis (ethanesulfonic acid) (PIPES), pH 6.8, 3 mM MgCl2, 1 mM ethylenebis (oxyethylenenitrilo) tetraacetic acid (EGTA), 0.5% Triton X-100, and 1.2 mM phenylmethylsulfonyl fluoride [PMSF]) and incubated for 10 minutes on ice. The suspensions were centrifuged and the supernatants were stocked as soluble fractions. Precipitates were resuspended in 500 μL digestion buffer (50 mM NaCl, 400 mM sucrose, 1 mM PIPES, pH 6.8, 3 mM MgCl2, 1 mM EGTA, 0.5% Triton X-100, 1.2 mM PMSF, 100 μg/mL DNase I, and 50 μg/mL RNase A) and incubated for 20 minutes at room temperature. Next, ammonium sulfate (final concentration 250 mM) was added; the supernatants were retained as the chromatin fraction and precipitates were referred to as nuclear matrix fractions.
Transfection to COS7 cells and preparation of cytoplasmic and nuclear fractions
Transfection of plasmids to COS7 cells was done by electroporation, and nuclear fractions were isolated as described.26 Briefly, cells were electroshocked and cultured in DMEM (10% FCS) for 48 hours. Cells were stimulated with hGM-CSF (10 ng/mL) for 30 minutes and harvested. The cells were incubated in buffer (10 mM N-2-Hydroxyethylpiperazine-N′-ethanesulphonic acid (HEPES), pH 7.9, 10 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol, 0.1 mM EDTA, and 0.1 mM PMSF) for 15 minutes on ice, and NP40 was added at a final concentration of 1%. Cells were mixed vigorously for 15 seconds and centrifuged. The supernatant was stored as the cytosol fraction. Nuclear proteins were extracted as described.26
Results
Induction of DNA binding activity of STAT5 through the hGM-CSF receptor in BA/F3 cells does not require βc tyrosine residues
Human GM-CSF activates STAT5 in BA/F3 cells, and we earlier reported the detailed region requirement of hGM-CSFR βc for STAT5 tyrosine phosphorylation by receptor mutation analysis.22Y-series mutants of βc contain only a single intact Y residue, with the remaining Ys mutated to F. Fall mutant has substitutions of all 8 Y residues together. Using these mutants, we found that the level of STAT5 tyrosine phosphorylation was dramatically decreased with lack of all of the βc Ys (Fall), but was increased when any one (or 2, in the case of Y12) Y was added back.22 In the present work, we examined the role of βc cytoplasmic Y residues in induction of STAT5 DNA binding activity, using gel shift analysis and the proximal STAT5 binding site of β-casein promoter8 as a probe. Although the extent of binding activity was much less than that seen with the wild-type receptor, DNA binding activity was clearly induced by the addition of GM-CSF through the Fall mutant, and any one of the Y-series mutants also enhanced DNA binding of STAT5 by GM-CSF stimulation (Figure 1A). The extent of binding strength through Y-series mutants correlates with that observed with tyrosine phosphorylation of STAT5 through these mutants.22We analyzed the transcriptional activation of STAT5 through these mutant receptors using 4XST-Luc, which contains 4 tandem repeats of the proximal STAT5 binding site of the β-casein promoter, followed by the luciferase coding region. As expected, Fall can activate luciferase activity of 4XST-Luc, and adding back of any Y residues enhanced the activity (data not shown). These results suggest that DNA binding activity and luciferase activity through mutant βc are correlated. We also examined the possible requirement of box1 and box2 motifs, which are conserved among cytokine receptors, using internal deletion mutant of βc, which lacks either box1 (Δbox1) or box2 (Δbox2).48 As shown in Figure 1A, Δbox1 did not activate DNA binding activity of STAT5, but Δbox2 did transduce signals for DNA binding of STAT5 in BA/F3 cells.
Activation of STAT5 DNA binding and STAT-dependent Py origin replication in BA/F3 cells through a series of mutants of hGM-CSFR.
(A) BA/F3 cells expressing wild-type α subunit and various β mutants were depleted of mIL-3 for 5 hours and restimulated with hGM-CSF (10 ng/mL) for 15 minutes. Gel shift assay was done using an oligonucleotide corresponding to the STAT binding site. Arrow indicates specific STAT binding complex to the probe. (B) Plasmid containing 4 tandem repeats of STAT5 binding sites or mutant STAT5 binding sites fused to Py virus replication origin were transfected to Ba/F-wild cells and left for 5 hours in depletion media. Cells were restimulated with hGM-CSF (10 ng/mL) and harvested after 24 hours of incubation, followed by replication assay. Transfected plasmids were extracted using Hirt solution and digested with DpnI, which selectively degrades an unreplicated plasmid, and then the plasmids were separated through an agarose gel and analyzed by Southern blotting. (C) Plasmid containing wild-type STAT5 binding site followed by Py virus replication origin was transfected to BA/F3 cells expressing various hGM-CSFR mutants, and replication induced by hGM-CSF was analyzed as described above. Arrow indicatesDpnI-resistant replicated bands.
Activation of STAT5 DNA binding and STAT-dependent Py origin replication in BA/F3 cells through a series of mutants of hGM-CSFR.
(A) BA/F3 cells expressing wild-type α subunit and various β mutants were depleted of mIL-3 for 5 hours and restimulated with hGM-CSF (10 ng/mL) for 15 minutes. Gel shift assay was done using an oligonucleotide corresponding to the STAT binding site. Arrow indicates specific STAT binding complex to the probe. (B) Plasmid containing 4 tandem repeats of STAT5 binding sites or mutant STAT5 binding sites fused to Py virus replication origin were transfected to Ba/F-wild cells and left for 5 hours in depletion media. Cells were restimulated with hGM-CSF (10 ng/mL) and harvested after 24 hours of incubation, followed by replication assay. Transfected plasmids were extracted using Hirt solution and digested with DpnI, which selectively degrades an unreplicated plasmid, and then the plasmids were separated through an agarose gel and analyzed by Southern blotting. (C) Plasmid containing wild-type STAT5 binding site followed by Py virus replication origin was transfected to BA/F3 cells expressing various hGM-CSFR mutants, and replication induced by hGM-CSF was analyzed as described above. Arrow indicatesDpnI-resistant replicated bands.
Activation of STAT leads to initiation of polyoma replicon DNA replication
Because transcription factors are apparently involved in regulating DNA replication in eukaryotic cells, we evaluated whether STAT5 would activate the initiation of DNA replication with the use of polyoma replicon. Plasmids containing 4 tandem repeats of STAT binding sites followed by polyoma replication origin (4XSTOICAT) were transfected to BA/F3 GM-CSFR cells and incubated with or without hGM-CSF (10 ng/mL). After 24 hours of culture, DpnI assay was done as described in “Materials and methods.” As shown in Figure 1B, replication of the 4XSTOICAT was induced by stimulation of hGM-CSF. When we used plasmids containing 4 tandem repeats of mutant STAT5 binding sites followed by polyoma replication origin, no replication was induced with the addition of hGM-CSF to BA/F-wild cells. Because the mutant site cannot bind to STAT530 (our unpublished results), the essential role of STAT5 and its binding site for initiation of polyoma origin-dependent replication was suggested. We then analyzed activities of STAT-dependent DNA replication with various hGM-CSF receptor mutants in BA/F3 cells. A stable line of BA/F3 cells expressing hGM-CSF α subunit was transfected with various mutants of hGM-CSFR and 4XSTOICAT, and DpnI assay was done. Δbox2, but not Δbox1, induced replication, indicating that box1 is essential but box2 is dispensable for replication initiation (lanes 3, 4). Any one of the Y-series mutants induced DNA replication of 4XSTOICAT. When we examined levels of activation of DNA replication, no correlation was observed with that of DNA binding activity. However, because Fall induced DNA replication, Y residues of βc may not be essential, and the requirement of the βc region for Py DNA replication seems the same as that for the STAT5 DNA binding activity induced by hGM-CSF.
Genistein inhibits Py replication, but not STAT5 chromatin localization
We earlier found that adding the tyrosine kinase inhibitor genistein suppressed proliferation promotion by hGM-CSF, but not cell survival or MAPK cascade activation.24 25 Therefore, genistein may be a specific inhibitor of signaling pathways for cell proliferation. Here, we tested the effects of genistein on replication and transcription activation by STAT5. Both 4XST-Luc and 4XSTOICAT were transfected to BA/F-wild cells, and then the cells were stimulated with hGM-CSF in the presence of the indicated doses of genistein. After 24 hours of culture, the cells were harvested and divided into 2 samples for luciferase and replication analyses. The addition of genistein completely suppressed DNA replication of 4XSTOICAT (Figure 2A). In contrast, the addition of genistein induced the luciferase activity of 4XST-Luc to a great extent in response to hGM-CSF (Figure 2B).
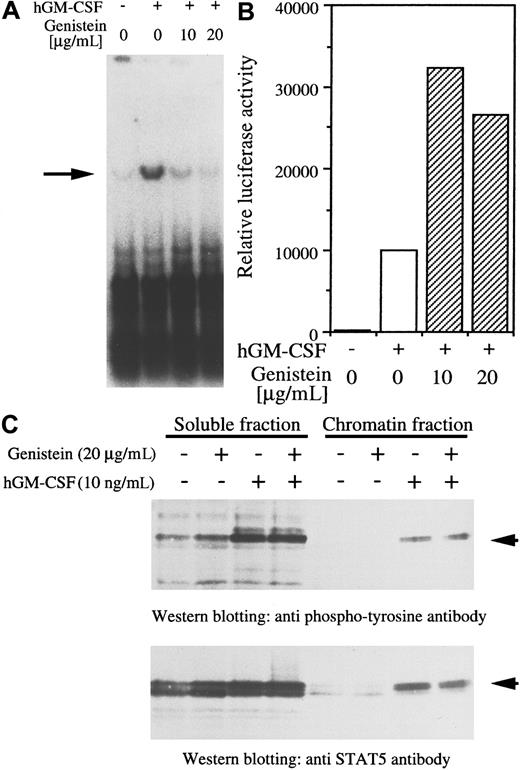
Characterization of Py origin-dependent replication by activation of STAT5.
Plasmids containing 4 tandem repeats of STAT5 binding sites fused to Py virus replication origin (A) or luciferase (B) were transfected to BA/F-wild cells. Cells were restimulated with hGM-CSF (10 ng/mL) and harvested after 24 hours of incubation. The tyrosine kinase inhibitor genistein (10 or 20 μg/mL) was added 30 minutes before restimulation. Either DpnI (A) or luciferase assay (B) was done as described in “Materials and methods.” Arrow in (A) indicates the replicated plasmid. (C) Subcellular translocation of STAT5 by hGM-CSF in the presence or absence of hGM-CSF. Cells were stimulated with hGM-CSF (10 ng/mL) in the presence or absence of genistein (20 μg/mL) and separated into soluble and chromatin fractions. Recovery of STAT5 in these fractions was analyzed by Western blotting with anti-STAT5 or phosphotyrosine antibodies.
Characterization of Py origin-dependent replication by activation of STAT5.
Plasmids containing 4 tandem repeats of STAT5 binding sites fused to Py virus replication origin (A) or luciferase (B) were transfected to BA/F-wild cells. Cells were restimulated with hGM-CSF (10 ng/mL) and harvested after 24 hours of incubation. The tyrosine kinase inhibitor genistein (10 or 20 μg/mL) was added 30 minutes before restimulation. Either DpnI (A) or luciferase assay (B) was done as described in “Materials and methods.” Arrow in (A) indicates the replicated plasmid. (C) Subcellular translocation of STAT5 by hGM-CSF in the presence or absence of hGM-CSF. Cells were stimulated with hGM-CSF (10 ng/mL) in the presence or absence of genistein (20 μg/mL) and separated into soluble and chromatin fractions. Recovery of STAT5 in these fractions was analyzed by Western blotting with anti-STAT5 or phosphotyrosine antibodies.
To clarify the target of genistein, we examined the subcellular localization of STAT5 of the fractionated cells. Cells were stimulated with hGM-CSF in the presence or absence of genistein, and soluble chromatin fractions were separated. Both fractions were subjected to polyacrylamide gel electrophoresis, and Western blotting was done using anti-STAT5 (Transduction Laboratories, Lexington, KY) or antiphosphotyrosine (4G10; Upstate Biotechnology, Lake Placid, NY) antibodies. As shown in Figure 2C (lower panel), in response to hGM-CSF stimulation, recovery of STAT5B in chromatin fractions was dramatically increased and these fractions were tyrosine phosphorylated (Figure 2C, upper panel), which means that tyrosine-phosphorylated STAT5 was moved to the chromatin fraction after the stimulation. In contrast, slight decreases in the soluble fractions were observed. When cells were treated with genistein, no change in STAT5B recovery of chromatin, soluble fractions was observed, and the presence of genistein did not affect the tyrosine phosphorylation of STAT5. The findings suggest that genistein suppressed replication through specific inhibition of the replication machinery. Further experiments of immunoprecipitation of STAT5 followed by antiphosphotyrosine antibody Western blotting and gel shift analysis indicated that tyrosine phosphorylation and DNA binding activity of STAT5 were not suppressed by genistein (data not shown); hence, the target of genistein seems to be events after chromatin opening by STAT5.
C terminus of STAT5 is required for DNA replication initiation
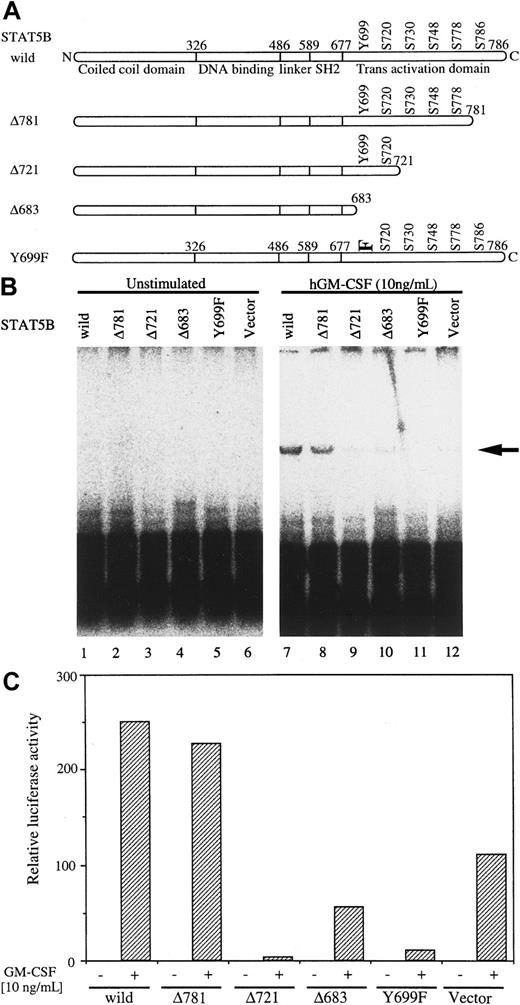
STAT family proteins have common structural features, including a C-terminal transcriptional activation domain, tyrosine residue, and SH2 region.49 To determine the STAT5B region required for replication initiation, we constructed STAT5B mutants and analyzed their activity in BA/F3 cells. Wild-type as well as mutant STAT5B, shown schematically in Figure 3A, were transfected together with 4XSTOICAT and SRα-LTag. Cells were cultured in the presence or absence of hGM-CSF for 24 hours, and DpnI assay was done. Without hGM-CSF stimulation, no replicated band was observed (Figure 3B, lanes 1-6). Because endogenous STAT5 exists, a replicated band was observed in the vector control sample (lane 12), but cotransfection of STAT5B dramatically enhanced the intensity of the band (lane 7). When we transfected mutant STAT5B instead of wild-type STAT5B, deletion up to amino acid 781 did not affect replication activity, but further deletion up to 721 resulted in loss of the activity (lanes 8, 9). Further deletion up to 683 confirmed this result (lane 10). The C-terminal Y residue of STAT5B is phosphorylated upon cytokine stimulation and is thought to be essential for dimerization through the SH2 region of STAT. When we mutated STAT5B Y699 to F (Y699F), DNA replication was abrogated (lane 11), and complete loss of the replicated band suggests that this mutant acts in a dominant-negative fashion to endogenous STAT5B. We also analyzed the transcriptional activation potential of these STAT5B mutants using the 4XST-Luc plasmid (Figure 3C). As shown in Figure 3C, mutant Δ781 can induce transcription of 4XST-Luc, but further deletion up to 721 resulted in loss of the activity. Mutation of Y699 also resulted in a complete loss of luciferase activation. In both transcription and replication, Δ683, which lacks Y residue, showed weaker dominant-negative effects than those observed with Y699F. We speculate that this is caused by differences in expression levels of Δ683 and Y699F, which can be deduced from expression levels of these mutants in COS7 cells. These results indicate that Y699 and the C-terminal transactivation domain of STAT5B are essential for both transcription and replication activation by hGM-CSF in BA/F3 cells.
Requirement of a region of STAT5 for Py origin-dependent replication.
(A) Mutants with deletion or Y-residue point mutation are shown schematically. The mutant STAT5 was transfected with 4XSTOICAT (B) or 4XST-Luc (C) into BA/F-wild cells, and the cells were depleted of mIL-3 for 5 hours. hGM-CSF (10 ng/mL) was added and culture was continued for another 24 hours. Cells were harvested, and DpnI (B) or luciferase (C) assay was done as described in “Materials and methods.” Arrow in (B) indicates the replicated plasmid.
Requirement of a region of STAT5 for Py origin-dependent replication.
(A) Mutants with deletion or Y-residue point mutation are shown schematically. The mutant STAT5 was transfected with 4XSTOICAT (B) or 4XST-Luc (C) into BA/F-wild cells, and the cells were depleted of mIL-3 for 5 hours. hGM-CSF (10 ng/mL) was added and culture was continued for another 24 hours. Cells were harvested, and DpnI (B) or luciferase (C) assay was done as described in “Materials and methods.” Arrow in (B) indicates the replicated plasmid.
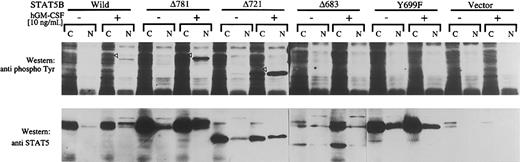
To analyze the mechanism of lack of replication and transcription-stimulating activity of these mutants (Δ683, Δ721, and Y699F), we examined tyrosine phosphorylation and nuclear translocation of these mutants with a transient expression system in COS7 cells. Mutants of STAT5 were transfected to COS7 cells with hGM-CSFR α and βc plasmids and cultured for 2 days. Cells were stimulated for 30 minutes with hGM-CSF (10 ng/mL), and total cell lysates of nuclear and cytoplasmic fractions were analyzed by Western blotting with antiphosphotyrosine and anti-STAT5 antibodies. As shown in Figure 4 (upper panel), wild type, Δ781, and Δ721 were tyrosine phosphorylated in response to hGM-CSF (open arrowhead). Because no tyrosine phosphorylation was observed with Δ683 or Y699F, the results suggest that Y699 is a major phosphorylation site of STAT5B. When we examined nuclear translocation by blotting with α STAT5 antibody, wild type as well as Δ781 and Δ721, but not Δ683 or Y699F, were translocated to the nucleus. A weak Y699F band was observed, but this band is assumed to be caused by an incomplete purity of fractionation because a trace amount of SHP-2, which is assumed to localize exclusively in the cytoplasm, was also observed in the nuclear fraction. Exclusive localization of Y699F and Δ683 in the cytoplasm was also supported by immunostaining with anti-STAT5 antibody (data not shown). These results suggest that failure of activation of DNA replication by Δ683 and Y699F was caused by a lack of nuclear translocation, whereas requirement of the transcriptional activation domain for DNA replication was suggested in the case of Δ721.
Characterization of mutant STAT5B.
Wild-type as well as mutant STAT5B with hGM-CSFR α and βc subunits were transfected to COS7 cells. After 2 days of culture, the cells were stimulated with hGM-CSF (10 ng/mL) for 30 minutes and harvested. Cells were separated into soluble and nuclear fractions, and phosphorylation and translocation of STAT5 were analyzed by Western blotting.
Characterization of mutant STAT5B.
Wild-type as well as mutant STAT5B with hGM-CSFR α and βc subunits were transfected to COS7 cells. After 2 days of culture, the cells were stimulated with hGM-CSF (10 ng/mL) for 30 minutes and harvested. Cells were separated into soluble and nuclear fractions, and phosphorylation and translocation of STAT5 were analyzed by Western blotting.
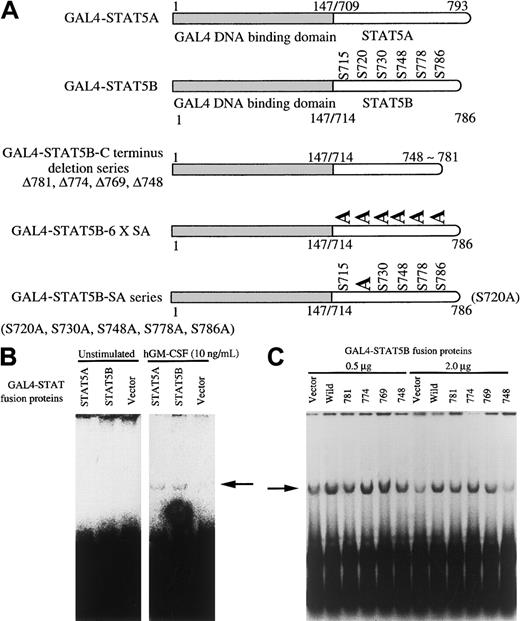
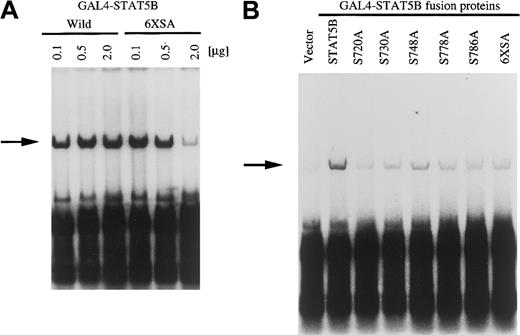
C-terminus transcriptional activation domain of STAT5 fused with GAL4 DNA binding domain can activate Py replication through the GAL4 binding sequence
We next analyzed the role of the STAT5 transcriptional activation domain using the yeast GAL4 fusion protein system. GAL4 protein and various hybrid proteins composed of the GAL4 DNA binding domain and the activating domain of other transcription factors were shown to transactivate replication of the plasmid containing the Py replicon and GAL4 binding sequence in the upstream of replication origin.50 It has been reported that fusion proteins in which the DNA binding domain of the yeast GAL4 transcription factor is linked to the transactivation domain of STAT5A can lead to transactivation of a luciferase reporter construct with a promoter that contains 3 binding sites for the GAL4 protein.51 We constructed similar plasmids, GAL4–STAT5A and GAL4–STAT5B; the mouse STAT5 C-terminal transcriptional activation domains (amino acids 709-793 of STAT5A and amino acids 714-786 of STAT5B) were fused with the C terminus of GAL4 DNA binding element (Figure 5A). These fusion proteins do not contain Y residues, which were shown to be a major phosphorylation site. As a replicon, the 5-tandem repeat of GAL4 binding domain fused with the polyoma replication origin (pPyG5OICAT) was used.43 Either GAL4–STAT5A or GAL4–STAT5B was transfected with Py LTag into BA/F-wild cells. After 24 hours of culture with or without hGM-CSF, DpnI assay was done. As shown in Figure 5B (right panel), cotransfection of either GAL4–STAT5A or GAL4–STAT5B induced DNA replication by the addition of hGM-CSF to BA/F-wild cells. Replication occurs in a GM-CSF–dependent manner because no band was observed with the nonstimulated samples (left panel). To analyze the role of the transcriptional activation domain of STAT5B, we tested the induction of replication by C-terminal deletion mutants of GAL4–STAT5B (GAL4BΔ781, 774, 769, and 748). Two different doses (0.5 μg, 2.0 μg) of GAL4–STAT5B and its mutants were transfected, and the ability to induce replication of pPyG5OICAT was analyzed. As shown in Figure 5C, transfection of 0.5 μg of any one of the mutants can induce replication of pPyG5OICAT. When we transfected 2 μg GAL4–STAT5 or its mutants, deletion up to 769 did not affect replication induction ability, but further deletion up to 748 resulted in loss of replication. These results indicate that the transcriptional activation domain between amino acids 748 and 769 is essential for replication activation.
Activation of Py replicon by GAL4–STAT fusion protein.
(A) Schematic diagram of GAL4–STAT fusion proteins used. (B) GAL4–STAT5A or GAL4–STAT5B was transfected with Py LTag and pPyG5OICAT, which contains a 5-tandem repeat of GAL4 binding site and Py replication origin, into BA/F3 cells. After 24 hours of culture, replication was analyzed by DpnI assay. The replicatedDpnI-resistant plasmid is indicated by the arrow. (C) The indicated doses of GAL4–STAT5 or its mutants were transfected with SRα-LTag and pPyG5OICAT into BA/F-wild cells. After 24 hours of stimulation, DpnI assay was done.
Activation of Py replicon by GAL4–STAT fusion protein.
(A) Schematic diagram of GAL4–STAT fusion proteins used. (B) GAL4–STAT5A or GAL4–STAT5B was transfected with Py LTag and pPyG5OICAT, which contains a 5-tandem repeat of GAL4 binding site and Py replication origin, into BA/F3 cells. After 24 hours of culture, replication was analyzed by DpnI assay. The replicatedDpnI-resistant plasmid is indicated by the arrow. (C) The indicated doses of GAL4–STAT5 or its mutants were transfected with SRα-LTag and pPyG5OICAT into BA/F-wild cells. After 24 hours of stimulation, DpnI assay was done.
Six S residues exist in the C-terminal transcriptional activation domain of STAT5B, and the role of these residues in transactivation was not clarified. To determine the requirement of S residues of the STAT5B transcriptional activation domain, we constructed a mutant carrying all the S residues substituted with A (GAL4–STAT5B-6XSA, Figure 5A). Because the effects of deletion of the transactivation domain were observed only when we transfected a high amount of GAL4–STAT5B mutants, we transfected 3 different amounts of GAL4–STAT5B or GAL4–STAT5B-6XSA with pPyG5OICAT and Py LTag into BA/F-wild cells. As shown in Figure 6A, a small or middle amount (0.1, 0.5 μg) of GAL4–STAT5B-6XSA stimulated Py replication to the same extent as did the wild-type GAL4–STAT5B, whereas a larger amount (2 μg) of transfected GAL4–STAT5B-6XSA did not induce Py replication. We then looked at the role of each S residue by constructing GAL4–STAT5B mutants with one S mutated with A and other Ss left intact (Figure 5A). The mutants (2 μg) were then transfected to BA/F-wild cells. As shown in Figure 6B, none of the mutants induced Py replication. When we transfected these mutants with a low dose (0.5 μg), all the mutants induced Py replication, as was seen with the wild type (data not shown). These results suggest that these S residues coordinately play a role in replication initiation.
Role of S residues of STAT5 for initiation of Py replication.
(A) Various amounts of GAL4–STAT5B-wild or GAL4–STAT5B-6XSA were transfected to BA/F-wild cells together with SRα-LTag and pPyG5OICAT. (B) Two micrograms of GAL4–STAT5B or GAL4–STAT5B S-residue mutants were transfected with SRα-LTag and pPyG5OICAT. Cells were cultured for 24 hours in the presence of hGM-CSF (10 ng/mL). Replication was analyzed by DpnI assay. The arrows indicate replicated plasmid.
Role of S residues of STAT5 for initiation of Py replication.
(A) Various amounts of GAL4–STAT5B-wild or GAL4–STAT5B-6XSA were transfected to BA/F-wild cells together with SRα-LTag and pPyG5OICAT. (B) Two micrograms of GAL4–STAT5B or GAL4–STAT5B S-residue mutants were transfected with SRα-LTag and pPyG5OICAT. Cells were cultured for 24 hours in the presence of hGM-CSF (10 ng/mL). Replication was analyzed by DpnI assay. The arrows indicate replicated plasmid.
Discussion
We obtained evidence that the activation of STAT5 can lead to initiation of Py DNA replication and that the transcriptional activation domain is required for this activity. This is the first direct evidence indicating the potential involvement of STAT5 in replication.
All transcription factors with replication-enhancing activity appear to require an activation domain, in addition to the DNA binding domain. Deletion or mutation analysis of STAT showed that the region required for transcription and that for replication activation are not separable. The activation domain for replication overlaps that for transcription in many cases, such as GAL4, VP16, and c-Jun.39,50,52 However, in the case of p53 and c-Rel, no identical requirement of the region for Py DNA replication and transcription was found.43 53 Our results obtained with the mutant STAT5 Δ721 indicate that the transcriptional activation domain is essential and sufficient for the activation of Py replication. Because deletion of the transactivation domain does not affect STAT5 nuclear translocation or DNA binding, we speculate that the transactivation domain may play a role to recruit molecules to the replication machinery.
Initiation of Py DNA replication occurs when the chromatin structure around the origin opens for the binding of LTag. LTag forms a double hexamer in a manner dependent on adenosine triphosphate and induces a structural change at the origin. Unwinding of double-stranded DNA then begins in the presence of replication protein A (RP-A), and DNA polymerase α-primases initiate DNA synthesis on the unwound DNA covered with RP-A. The activation domain of VP16, GAL4, E2, and p53 binds to a single-stranded DNA binding protein, RP-A, which is essential for the initiation of Py DNA replication,54-56and c-Jun interacts with LTag.57 To examine the potential involvement of STAT5 in the recruitment of RP-A, we overexpressed VP-16 in the STAT-Py system. Because no interference was observed (data not shown), VP-16 and STAT5 may not use the same mechanism. Similarly, the target protein of polymavirus enhancer binding protein 2αB1 is probably different from that of RP-A or other VP16 binding proteins.47
Mutation of S residues of the STAT5 transactivation domain suggested the role of S residues for replication activation. Because mutation of any one of the S residues resulted in loss of replication activation, we could not define a specific role of each S residue. Among the members of the STAT family, the role of S is reported only for STAT1 and STAT3 for their transcriptional activation.58 In contrast, although phosphorylation of S730 of STAT5B by prolactin stimulation occurs, this phosphorylation is not essential for DNA binding or transcriptional activation.59
Core binding protein and p300 can interact with STAT5, and histone acetyltransferase activity may participate to maintain a transcriptionally active chromatin structure.60 Induction of germline transcription in the T-cell receptor γ locus by STAT5 has been reported.61 Therefore, it is feasible that STAT5 plays roles in both the activation of chromatin accessibility and recruitment of replication machinery.
We reported that activation of STAT5 is not essential or sufficient for proliferation promotion of BA/F3 cells. βc fused with JAK2 can promote proliferation without activation of STAT527; on the other hand, GyrB fused with JAK2 cannot sustain the survival or proliferation of BA/F3 cells, although it can lead to activation of STAT5.62 There are several possible explanations as to why STAT5 cannot induce proliferation even though it can lead to DNA replication. STAT5 is not sufficient to promote other cell-cycle machinery such as the cyclin/CDK complex. As another possibility, it should be considered that STAT5 can initiate polyoma replicon-dependent replication but not cellular DNA replication. The former possibility is of interest because it was reported that coactivation of the ras/MAPK pathway, in addition to the constitutively active STAT5, promotes proliferation of BA/F3 cells, which means that an additional signal is required to promote cell proliferation.63 Acute myelogenous leukemia 1, c-Rel, c-Jun, and c-Fos can initiate polyoma replicon DNA replication, which means that STAT5 is not the only factor related to activation of replication. GM-CSF can lead to replication of Py replicon, whose enhancer does not contain STAT binding site.25 This notion is supported by our previous results that activation of STAT5 is not essential to promote proliferation in BA/F3 cells. It also suggests that other transcription factors can activate initiation of DNA replication.
Because STAT is assumed to play a role in cytokine-specific functions, it probably is not like a major role related to the general cell proliferation. Our results are important because it seems highly likely that STAT functions as a part of the DNA replication machinery. Selection of transcription factors for initiation of DNA replication may depend on cell types and other factors.
Our previous results indicated that no specific Y residue of βc was required for tyrosine phosphorylation of STAT5 in BA/F3 cells,22 and the same requirement was observed for the initiation of replication. Therefore, STAT5 is probably activated independently of receptor Y residues. This implication is inconsistent in that other STATs are considered to recognize and bind to a specific sequence surrounding certain Y residues of cytokine receptors. In addition, we reported that the GyrB–JAK2 fusion protein can activate STAT5 without involving GM-CSF receptor activation.62Taking these data together, we speculate that STAT5 is directly phosphorylated by JAK2.
We previously found that β-casein luciferase is not activated by Fall, although Fall can induce tyrosine phosphorylation of STAT5.22 Other modifications such as serine phosphorylation may be required for transcriptional activation of STAT5, and this modification depends on Y residues. When we examined transcriptional activation of tandem repeats of STAT5 binding sites derived from the β-casein promoter, even Fall activated luciferase activity; hence the Y residue is not required for the transcriptional activation of STAT5. Because β-casein promoter contains various putative transcription factor binding sites,8 a combination of activation of multiple transcription factors may be required for full activation of β-casein promoter, and receptor Y residues may play a role in activating other transcription factors.
We thank Yukitaka Izawa for excellent technical support, Drs Yoshiaki Ito and Kosei Ito for providing materials and for helpful discussion, and Mariko Ohara for comments. T.I. is a recipient of a research fellowship from the Japan Society for the Promotion of Science for Young Scientists.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sumiko Watanabe, Department of Molecular and Developmental Biology, Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo 108-8639, Japan; e-mail: sumiko@ims.u-tokyo.ac.jp.