Zinc protoporphyrin (ZnPP), a naturally occurring molecule, is increased in iron deficiency and lead intoxication. ZnPP can also induce heme oxygenase (HO-1), the enzyme it competitively inhibits. In cultured cells (HA-1), ZnPP was the strongest HO-1 inducer of any metalloporphyrin (MP) tested. This was not due to increased oxidative stress, enhanced binding at metal response element, nor increased binding at activator protein-1 (AP-1) or SP-1 sites on HO-1. Only ZnPP, however, increased binding of nuclear proteins to early growth response-1 (Egr-1) protein consensus sequence. Pretreatment of HA-1 with cycloheximide inhibited ZnPP-induced HO-1 messenger RNA (mRNA) by 55%. Incubation with antisense Egr-1 oligomers decreased ZnPP-induced HO-1 expression by 47%. Furthermore, the level of HO-1 mRNA induction by ZnPP was 2-fold less in Egr-1–deficient fibroblasts than in wild-type cells. Because no Egr-1 binding site was previously identified on the HO-1 promoter, HA-1 cells were transfected with HO-1 CAT constructs containing segments of a 12.5-kb enhancer region of HO-1. A 196-bp fragment (RH) located approximately 9.5 kb upstream of the transcription start site mediated HO-1 induction by ZnPP alone. DNase I footprinting analysis further revealed that nuclear proteins bound to a 50-bp sequence in the RH. Within this sequence, a novel 9-bp region with 78% homology to the Egr-1 consensus sequence was identified further suggesting that Egr-1 partially mediates HO-1 induction by ZnPP. Lastly, increased apoptosis and nuclear localization were only seen with ZnPP, suggesting that increased ZnPP in disease states may serve as a cellular signaling mechanism.
Introduction
Zinc protoporphyrin (ZnPP) is an endogenous metalloporphyrin (MP) found at higher concentrations in anemia of chronic disorders,1 iron deficiency,2 and lead poisoning, 3 clinical circumstances where bone marrow cell proliferation is altered. Conversely, synthesis of ZnPP appeared diminished in states of increased cell proliferation such as in leukemia.4 Furthermore, zinc porphyrins are potent inhibitors of hematopoiesis in animal and human bone marrow.5 These observations suggest that ZnPP could serve as a signaling molecule in cell cycle regulation. One mechanism by which ZnPP could alter cell cycle is via heme oxygenase (HO-1), because ZnPP is a potent inducer of the enzyme that it competitively inhibits.6 In fact, in an in vitro model, ZnPP resulted in the highest induction of HO-1 among many MPs tested (W. S. Zhang and colleagues, oral communication, October 2000). Because increased HO-1 is associated with increased cell proliferation in several models,7,8 it may be that ZnPP-mediated HO-1 induction leads to alterations of cell cycle regulation or that ZnPP mediates cell cycle regulation to result in HO-1 induction. Nonetheless, the ability to induce HO-1 is not unique to ZnPP. For example, in chick embryo liver cells, different MPs resulted in varying levels of HO-1 induction.9 More recent studies in murine NIH 3T3 cells and transgenic mice expressing the HO-1 promoter linked to a luciferase reporter demonstrated HO-1 induction by various MPs (W. A. Zhang and colleagues, oral communication, October 2000). However, the greater ability of ZnPP to induce HO-1 raises the question as to whether unique mechanisms contribute to ZnPP-mediated HO-1 induction.
Although the induction of HO-1 by heme, the natural substrate, is believed to occur through a complex mechanism involving activator protein-1 (AP-1),10 the mechanism of HO-1 induction by MPs is not clear. Diverse agents can lead to HO-1 induction, but the mechanism of the HO-1 induction that has been best described involves stress activation. The HO-1 induction by heavy metals on the mouseHO-1 gene, for example, is mediated via the stress response elements (SRE) and not the metal regulatory element (MRE). The multimerized SRE located on the distal enhancer 1 (DE1) and 2 (DE2) regions of the mouse HO-1 gene containing the core heptad AP-1 consensus sequence was also demonstrated to be responsible for HO-1 induction by heme.10 Other inducer-response elements such as SP-1, a zinc finger protein, have also been identified on the DE2 region of the mouse HO-1 gene.10Although the role of SP-1 has not been extensively studied in induction of HO-1, it represents a class of transcription factors that requires zinc to form zinc finger motifs. These then bind to the SP-1 consensus sequence and lead to gene transcription. Another zinc finger protein that has similar motifs to SP-1 is early growth response-1 protein (Egr-1). To date, no Egr-1 response element has been identified on the mouse HO-1 gene, but Egr-1 and SP-1 can compete for binding to regions containing an overlapping SP-1 and Egr-1 binding sequence such as on the human adenosine deaminase (ADA) gene.11 The importance of SP-1 and Egr-1 lies in their effect on cell cycle regulation. SP-1 is usually an activating factor that up-regulates genes involved in cell cycle regulation,12-14 whereas Egr-1 can either transactivate target promoter sequences or in most cases inhibit gene transcription.15,16 Moreover, in tumor cells, endogenous levels of Egr-1 act to impede cell proliferation.17 18 If MPs mediate HO-1 induction via SP-1 or Egr-1 binding sites, altered cell cycle regulation could be expected.
We therefore investigated the mechanism by which MPs induce HO-1 and identified a unique Egr-1–mediated induction pathway of ZnPP as well as effects of ZnPP on cell cycle regulation.
Materials and methods
Cell culture conditions
Hamster fibroblasts (HA-1) were grown in Eagle minimum essential media (EMEM; Sigma, St Louis, MO) supplemented with 10% fetal bovine serum (FBS), glutamine (2 μmol), penicillin (100 U/mL), and streptomycin (100 μg/mL) (Life Technologies, Grand Island, NY). The cells were grown as previously described19 for experiments. Mouse embryonic fibroblast cells from homozygous Egr-1 mutant mice (Egr-1−/−) or from heterozygous Egr-1 mutant mice (Egr-1+/−)20 21 (gifts of Dr J. Milbrandt, Washington University School of Medicine, St Louis, MO) and mouse fibroblast NIH 3T3 cells (ATCC, Manassas, VA) were maintained in EMEM with 10% FBS, 1% antibiotic-antimycotic, and 1% nonessential amino acid (Life Technologies).
Preparation of MPs
Solutions of zinc, tin, and chromium protoporphyrins (ZnPP, SnPP, and CrPP, respectively) (Porphyrin Products, Logan, UT) were prepared as described previously.22
Incubation with MPs
Cell cultures at 80% confluency were incubated with 10 μM of either ZnPP, SnPP, or CrPP for 24 hours at 37°C. Cells incubated in the absence of MPs served as controls. To elucidate whether ZnPP induction of HO-1 was related to the presence of the Zn++ions, HA-1 cells were incubated with either 100 μM ZnCl2or 10 μM ZnPP for 6 hours; thereafter, 10 μM ZnPP or 100 μM ZnCl2 was added to the cell media, respectively, for an additional 18 hours of incubation. In another experiment, HA-1 cells were incubated with various concentrations of ZnPP to evaluate whether HO-1 messenger RNA (mRNA) induction by ZnPP was dose dependent. After incubation, cells were rinsed twice with 1 × cold phosphate-buffered saline (PBS) then harvested for analyses.
Preincubation with cycloheximide
To evaluate if ZnPP-induced HO-1 mRNA required de novo protein synthesis, up to 2 mM cycloheximide was added to the cell culture media for 15 minutes prior to ZnPP incubation. Cells were then collected for Northern analysis as described below.
Egr-1 antisense transfection
To evaluate whether Egr-1 was required for HO-1 transcription, antisense experiments were conducted. The phosphorothioate-capped antisense oligodeoxynucleotide (5′-GsCsGGGGTGCAGGGGCACAsCsT-3′) (PAN Facility, Beckman Center, Stanford University Medical Center, Stanford, CA) targeted against the homologous sequence of the 5′ untranslated region of murine Egr-1 mRNA (120-101 bases upstream of the initial translation site)23 was used to block Egr-1 protein induction. Control sense oligodeoxynucleotides consisted of the complementary sense strand of the murine antisense oligomer similarly modified.24 HA-1 cells were grown to 50% confluence with 10% FBS and then the media changed to 0.4% FBS for 3 hours. Lyophilized oligomers were resuspended in PBS and added to the media at a final concentration of 40 μM. The cells were then incubated with oligomers for 12 hours. The medium was then replaced with new medium (1% FBS) containing the same concentration of oligomers and 10 μM ZnPP. After 24 hours the cells were collected for Western and Northern analyses as described below.
Evaluation of HO-1 expression
Antibodies.
Western analysis.
For detection of HO-1 immunoreactive protein, aliquots of cell sonicates were electrophoresed on a 12% polyacrylamide gel.27 Antigen-antibody complexes were visualized with the horseradish peroxidase (HRP) chemiluminescence system according to the manufacturer's instructions (Bio-Rad, Hercules, CA). Quantification was performed using Molecular Analyst image analysis software (Bio-Rad).
Cell protein content.
Sonicates were analyzed for protein content by known methods.28
Northern analysis.
Evaluation of oxidative stress
Because HO-1 induction is a generalized response to oxidative stress, multiple oxidative stress markers were measured to determine whether MP mediated HO-1 induction via oxidative signaling: (1) lactic dehydrogenase (LDH) release was determined by collecting effluent media from cell cultures and analyzing it by spectrophotometry using a standard enzyme solution30; (2) total glutathione content of cells was measured by the method of Anderson31; (3) generation of thiobarbituric acid-reactive substances (TBA-RS) was detected at 535 nm and values were determined using the extinction coefficient of 1.55 × 10−3/M per cm32; (4) protein oxidation was determined using Western analysis, and sonicates were reacted with 10 mM 2,4-dinitriphenylhydrazine and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described.33
Analysis of DNA binding of transcription factors
To identify possible transcription factors involved in HO-1 induction in HA-1 cells by MPs, electrophoresis mobility gel shift assay (EMSA) was performed after incubation with various MPs.
Oligonucleotides.
An oligonucleotide containing the core AP-1 binding site TGAGTCA from the HO-1 promoter34(5′-CTAGTGATGAGTCAGCCGGATC-3′) was obtained from Operon Technologies (Alameda, CA). The mouse MRE consensus sequence from HO-1 containing a core heptanucleotide, TGCACTC (5′-GATCCGGGTGCTGCACTCCATGAG-3′)35 was synthesized by the PAN Facility. Egr-1 (5′-GGATCCAGCGGGGGCGAGCGGGGGCGA-3′) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and the SP-1 consensus sequence from HO-1 (5′-ATTCGATCGGGGCGGGGCGAGC-3′) was synthesized by the PAN facility.
SP-1/Egr-1 (S/E) oligonucleotide probe.
It is known that SP-1 and Egr-1 can competitively bind to a common S/E site and result in opposing effects on gene transcription.11 To further define a possible role of SP-1 and Egr-1 in MP-mediated HO-1 induction, a 12-bp oligomer from the mouse HO-1 gene was chosen as an S/E probe. A sequence located in the DE2 region between base pairs 353 and 364 (GenBank, accession no. U70472)10 identified with a computer program (Wisconsin Package Version 10, Genetics Computer Group, Madison, WI) had 85% similarity to the reported S/E overlapping binding site in theADA gene.11 This consensus sequence, 5′-CGTAGTGGGCGGGGGGCTGTG-3′ (S/E) (PAN facility) on the DE2 region of the mouse HO-1 contains the core of SP-1 sequence (underlined) and Egr-1 sequence (boxed).
Preparation of cell nuclear extracts.
Cells were washed with cold PBS and pelleted in PBS at 3000gfor 5 minutes and processed using the standard method.36The nuclear extracts were dialyzed against 100 mM KCl using Slide-A-Lyzer Dialysis Cassette (Pierce, Rockford, IL) for 1 hour at 4°C and then stored at −80°C. Protein content of the nuclear extracts and cytoplasmic extracts were then determined.28
EMSA.
Nuclear protein (2 μg) was mixed with the 32P-labeled oligonucleotide probes in a buffer containing 10 mM Hepes (pH 7.9), 1 mM EDTA, 80 mM KCl, 1 μg poly[dIdC][dIdC], and 4% Ficoll. The reaction mixture was incubated at room temperature for 30 minutes and electrophoresed on a 6% polyacrylamide gel. To detect nonspecific binding, competition reactions were performed by adding 100-fold excess nonradiolabeled probe to the binding reaction mixture prior to electrophoresis.
Plasmid construsts, transfection, and enzyme assays
Construction of the CAT reporter plasmids (gift of Dr J. Alam, Alton Ochsner Medical Foundation, New Orleans, LA) shown in Figure 8and subfragments of fragment EH in Figure 9 have been described previously.10,35,37 38 A mixture of DNA containing 10 μg of the CAT expression plasmid and 1 μg pGL3luc were transiently transfected into HA-1 cells using Lipofectin Reagent (Life Technologies) according to the manufacturer's instruction. After 8 hours, the cells were incubated with 10 μM ZnPP, 100 μM ZnCl2, or media alone (controls) for 24 hours. The cells were washed 3 times with cold PBS (Mg++ and Ca++ free) and collected in a reporter lysis buffer from the CAT enzyme assay system kit (Promega, Madison, WI). CAT activity in each cell lysate was measured in duplicate following the manufacturer's protocol (Promega) and the specific CAT activity was calculated using a standard curve from a pure CAT enzyme supplied in the kit. Aliquots of the cell lysates were detected for luciferase activity using a luciferase activity assay kit (Promega). Background activity without the CAT reporter was then subtracted from the specific CAT activity and values were normalized to the luciferase activity.
DNase I protection assay
Nuclear protein extracts from HA-1 cells were prepared using the NE-PER Kit (Pierce). The noncoding strand RH probes were generated similarly as described previously10 with modifications. Plasmid pMHO1CATΔ-33 + RH was linearized and liberated by digestion with BamHI and SacI (which cleave at vector-encoded sequences on either side of the RH fragment). The digestion mixture was electrophoresed on a 6% polyacrylamide gel and the ∼200-bp fragment was purified using a QIAquick spin column (QIAquick Gel Extraction Kit; Qiagen, Valencia, CA). The purified fragment was labeled by the “end-fill” reaction using the Klenow fragment of E coli DNA polymerase I and a dNTP mixture containing [α-32P]dGTP and [α-32P]dATP. The labeled probe was further digested with NotI to generate a singly labeled probe and purified on a polyacrylamide column (Bio-Spin 30 column; Bio-Rad). Protein-binding reactions were carried out in a total volume of 25 μL reaction mixture containing a labeled fragment (10 000 cpm), 15 μg nuclear extract, and binding buffer at a final concentration of 10 mM Hepes/KOH (pH 7.9), 40 mM KCl, 1 mM MgCl2, 1 mM EDTA, 1 mM dithiothreitol, 1 μg poly[d(I-C)], and 10% glycerol. The mixture was incubated at room temperature for 30 minutes and diluted to a final volume of 50 μL with the reaction buffer. DNase I digestion was performed using the SureTrack Foot Printing Kit (Amersham Pharmacia Biotech, Piscataway, NJ).
Evaluation of apoptosis
Determination of immunoreactive p53.
Cell cytoplasm (20 μg) was analyzed for p53 immunoreactive protein using polyclonal rabbit anti-p53 antibody (Santa Cruz Biotechnology) by Western analysis as described above.
Visualization of annexin V, p53, and MPs.
Immunoreactive annexin V and p53 were also detected by immunohistochemistry using previously described methods.33A 1:150 dilution of rabbit antirat annexin IgG or 1:150 rabbit anti-p53 antibody was used. Slides were viewed with a Nikon fluorescent microscope (Japan) using a confocal laser scanning unit (Model 2010; Molecular Dynamics, Sunnyvale, CA) with excitation set at 488 nm and emission at 515 to 545 nm.
Because ZnPP and SnPP have endogenous fluorescence they could also be visualized within cells by setting the excitation at 568 nm and the emission at greater than 590 nm. To localize MP distribution in cells, dual images of the fluorescent signal from p53 and MPs were processed as anaglyphs on a SGI computer system (Molecular Dynamics). In this case, p53 was used only as a marker for the nucleus; therefore, the p53 (fluorescein isothiocyanate [FITC]) signal was further enhanced in the SnPP to allow for visualization of p53 in the nucleus because it was considerably less abundant in this circumstance than with ZnPP incubation.
Statistical analysis
For comparison between treatment groups, the null hypothesis that there was no difference between treatment means was tested by a single-factor analysis of variance (ANOVA) for multiple groups or unpaired t test for 2 groups (Statview 4.02; Abacus Concepts, Berkeley, CA). Statistical significance (P < .05) between groups was determined by means of the Fisher method of multiple comparisons.
Results
Effect of MPs on HO-1 expression
At 24 hours both HO-1 mRNA and HO-1 protein were highest in HA-1 cells incubated with 10 μM ZnPP compared to those incubated with SnPP and CrPP (Figure 1). Quantitative analysis of HO-1 protein showed 5.7-, 1.5-, and 4-fold increases after incubation with ZnPP, SnPP, and CrPP, respectively, compared to the untreated controls. Additionally, evaluation of HO activity revealed a 59.0% ± 2.0%, 69.7% ± 6.0%, and 75.0% ± 2.5% inhibition after incubation with ZnPP, SnPP, and CrPP, respectively, compared to untreated controls (n = 5 in each group; values represent mean ± SE).
HO-1 expression in HA-1 cells incubated with 10 μM MPs for 24 hours.
Upper panel shows a representative example of 3 Northern blots of HO-1 mRNA. Lower panel shows a representative example of 3 Western blots of HO-1 immunoreactive protein. Lane HO-1 is HO-1 standard obtained from rat livers injected with COCl2; lane C, untreated control; lane ZP, cells incubated with ZnPP; lane SP, cells incubated with SnPP; lane CP, cells incubated with CrPP.
HO-1 expression in HA-1 cells incubated with 10 μM MPs for 24 hours.
Upper panel shows a representative example of 3 Northern blots of HO-1 mRNA. Lower panel shows a representative example of 3 Western blots of HO-1 immunoreactive protein. Lane HO-1 is HO-1 standard obtained from rat livers injected with COCl2; lane C, untreated control; lane ZP, cells incubated with ZnPP; lane SP, cells incubated with SnPP; lane CP, cells incubated with CrPP.
Markers of oxidative stress with MP incubation
Because induction of HO-1 is a generalized marker of oxidative stress, we evaluated LDH release, glutathione depletion, TBA-RS formation, and protein oxidation as markers of oxidative stress in HA-1 cells incubated with MPs. There were no significant changes in total glutathione (Table 1), TBA-RS formation (Table 1), or protein oxidation (data not shown) with ZnPP, SnPP, or CrPP. However, incubation with CrPP, the MP resulting in the lowest HO-1 induction, was associated with a significant increase in LDH release compared with ZnPP and SnPP (Table 1). Overall, these data indicate that oxidative stress was not likely to mediate HO-1 induction by MPs in the HA-1 cells. These cells are not particularly resistant to oxidative stress and therefore represent a valid model.
Effect of metal ion on HO-1 expression with ZnPP incubation
Another means of HO-1 induction is via heavy metals. Incubation with 10 μM ZnPP, the concentration causing maximum HO-1 mRNA induction (Figure 2, insert), followed by incubation with 100 μM ZnCl2 further increased HO-1 mRNA by 1.5-fold compared to cells incubated with ZnPP alone or 2-fold with ZnCl2 alone (Figure 2). Similarly by reversing the incubation order, that is, ZnCl2 followed by ZnPP, HO-1 mRNA increased by 1.7-fold compared to cells incubated with ZnPP alone or 2.4-fold with ZnCl2 alone (Figure 2). These results indicated that the induction of HO-1 by ZnPP was not associated with the metal effect of the Zn++, but rather pointed to an independent mechanism. This was further corroborated by the lack of increased MRE binding in the nuclear extract of cells treated with ZnPP or ZnCl2 (data not shown).
Densitometric evaluation of HO-1 mRNA in HA1 cells incubated with ZnPP or ZnCl2.
HO-1 mRNA was measured by Northern analysis and normalized to the β-actin mRNA obtained from the same blot. Lane C is untreated control; lane ZP, cells incubated with 10 μM ZnPP for 24 hours; lane ZC, cells incubated with 100 μM ZnCl2 for 24 hours; lane ZP/ZC, cells incubated with 10 μM ZnPP for 6 hours then with 100 μM ZnCl2 for an additional 18 hours; lane ZC/ZP, cells incubated with 100 μM ZnCl2 for 6 hours then with 10 μM ZnPP for 18 hours. Each bar represents the mean ± SE of 3 experiments. *P < .05 versus controls; †P < .05 versus ZnPP- or ZnCl2-treated cells. Insert: Determining the maximum HO-1 mRNA induction with ZnPP incubation. Densitometric evaluation of HO-1 mRNA normalized to the β-actin mRNA. Lane C is untreated control; lane 1, cells incubated with 1 μM ZnPP for 24 hours; lane 10, cells incubated with 10 μM ZnPP for 24 hours; lane 50, cells incubated with 50 μM ZnPP for 24 hours. Maximal induction was achieved with 10 μM ZnPP.
Densitometric evaluation of HO-1 mRNA in HA1 cells incubated with ZnPP or ZnCl2.
HO-1 mRNA was measured by Northern analysis and normalized to the β-actin mRNA obtained from the same blot. Lane C is untreated control; lane ZP, cells incubated with 10 μM ZnPP for 24 hours; lane ZC, cells incubated with 100 μM ZnCl2 for 24 hours; lane ZP/ZC, cells incubated with 10 μM ZnPP for 6 hours then with 100 μM ZnCl2 for an additional 18 hours; lane ZC/ZP, cells incubated with 100 μM ZnCl2 for 6 hours then with 10 μM ZnPP for 18 hours. Each bar represents the mean ± SE of 3 experiments. *P < .05 versus controls; †P < .05 versus ZnPP- or ZnCl2-treated cells. Insert: Determining the maximum HO-1 mRNA induction with ZnPP incubation. Densitometric evaluation of HO-1 mRNA normalized to the β-actin mRNA. Lane C is untreated control; lane 1, cells incubated with 1 μM ZnPP for 24 hours; lane 10, cells incubated with 10 μM ZnPP for 24 hours; lane 50, cells incubated with 50 μM ZnPP for 24 hours. Maximal induction was achieved with 10 μM ZnPP.
Effect of inhibition of de novo protein synthesis on ZnPP-induced HO-1 expression
Addition of cycloheximide before ZnPP incubation decreased HO-1 mRNA induction in HA-1 cells in a dose-dependent manner (Figure3). At 300 μM, the maximal inhibition on ZnPP-mediated HO-1 induction was observed (55%) (Figure 3). This suggests that new protein synthesis is required only in part for HO-1 induction and that there may be other nonprotein factors mediating HO-1 induction by ZnPP.
Effect of cycloheximide on HO-1 mRNA induction by ZnPP.
HA-1 cells were preincubated with various doses of cycloheximide for 15 minutes prior to incubation with 10 μM ZnPP. HO-1 mRNA was evaluated by Northern analysis and normalized to the β-actin mRNA obtained from the same blot. Lane Cyc, cells incubated with 2 mM cycloheximide alone for 15 minutes as control; lane ZP, cells incubated with 10 μM ZnPP for 24 hours. Lanes 0.05, 0.1, 0.3, 1, and 2 show cells preincubated with 0.05, 0.1, 0.3, 1, or 2 mM cycloheximide and then incubated with ZnPP for 24 hours.
Effect of cycloheximide on HO-1 mRNA induction by ZnPP.
HA-1 cells were preincubated with various doses of cycloheximide for 15 minutes prior to incubation with 10 μM ZnPP. HO-1 mRNA was evaluated by Northern analysis and normalized to the β-actin mRNA obtained from the same blot. Lane Cyc, cells incubated with 2 mM cycloheximide alone for 15 minutes as control; lane ZP, cells incubated with 10 μM ZnPP for 24 hours. Lanes 0.05, 0.1, 0.3, 1, and 2 show cells preincubated with 0.05, 0.1, 0.3, 1, or 2 mM cycloheximide and then incubated with ZnPP for 24 hours.
Effect of transcription factors on HO-1 expression
Because binding at the MRE region did not account for the HO-1 induction with ZnPP, binding at other regulatory elements, such as AP-1, SP-1, and Egr-1 was also investigated with ZnPP. After 24 hours of incubation with ZnPP, cells did not show increased AP-1 binding, unlike cells similarly incubated with ZnCl2, SnPP, and CrPP (Figure 4). Furthermore ZnPP- and CrPP-treated cells differed from cells treated with SnPP or ZnCl2 in that they did not show increased SP-1 binding compared to controls (Figure 4B). In contrast enhanced Egr-1 binding was noted only with ZnPP-treated cells (Figure 4C).
AP-1, SP-1, and Egr-1 binding in nuclear extracts from HA-1 cells incubated with 100 μM ZnCl2 or 10 μM MPs for 24 hours.
Each panel represents an electrophoretic mobility shift gel from 3 separate experiments. (A) AP-1 binding. Lane F, free probe; lane C; untreated control; lane ZC, cells incubated with ZnCl2; lane ZP, cells incubated with ZnPP; lane SP, cells incubated with SnPP; lane CP, cells incubated with CrPP; lane ZC+AP-1, ZnCl2treated cells incubated with a 100-fold excess of cold AP-1 probe to demonstrate specificity of binding. (B) SP-1 binding. Lane F, free probe; lane C, untreated control; lane ZC, cells incubated with ZnCl2; lane ZP, cells incubated with ZnPP; lane CP, cells incubated with CrPP; lane SP, cells incubated with SnPP; lane ZC+SP-1, ZnCl2 treated cells incubated with a 100-fold excess of cold SP-1 probe to demonstrate specificity of binding. (C) Egr-1 binding. Lane F, free probe; lane C, untreated control; lane ZP, cells incubated with ZnPP; lane ZC, cells incubated with ZnCl2; lane CP, cells incubated with CrPP; lane SP, cells incubated with SnPP; lane ZP+Egr-1, ZnPP-treated cells incubated with a 100-fold excess of cold Egr-1 probe to demonstrate specificity of binding. In each gel, cold competition was performed with the compound leading to the highest nuclear protein binding.
AP-1, SP-1, and Egr-1 binding in nuclear extracts from HA-1 cells incubated with 100 μM ZnCl2 or 10 μM MPs for 24 hours.
Each panel represents an electrophoretic mobility shift gel from 3 separate experiments. (A) AP-1 binding. Lane F, free probe; lane C; untreated control; lane ZC, cells incubated with ZnCl2; lane ZP, cells incubated with ZnPP; lane SP, cells incubated with SnPP; lane CP, cells incubated with CrPP; lane ZC+AP-1, ZnCl2treated cells incubated with a 100-fold excess of cold AP-1 probe to demonstrate specificity of binding. (B) SP-1 binding. Lane F, free probe; lane C, untreated control; lane ZC, cells incubated with ZnCl2; lane ZP, cells incubated with ZnPP; lane CP, cells incubated with CrPP; lane SP, cells incubated with SnPP; lane ZC+SP-1, ZnCl2 treated cells incubated with a 100-fold excess of cold SP-1 probe to demonstrate specificity of binding. (C) Egr-1 binding. Lane F, free probe; lane C, untreated control; lane ZP, cells incubated with ZnPP; lane ZC, cells incubated with ZnCl2; lane CP, cells incubated with CrPP; lane SP, cells incubated with SnPP; lane ZP+Egr-1, ZnPP-treated cells incubated with a 100-fold excess of cold Egr-1 probe to demonstrate specificity of binding. In each gel, cold competition was performed with the compound leading to the highest nuclear protein binding.
Effect of Egr-1 protein on HO-1 mRNA induction
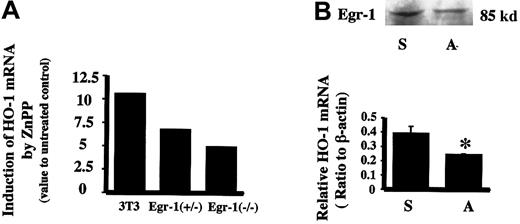
To further verify if Egr-1 protein could mediate the induction of HO-1 by ZnPP, Egr-1−/− and Egr-1+/− mouse fibroblast cells were examined for ZnPP-mediated HO-1 inducibility. Cells deficient in Egr-1 showed a lesser ability to induce HO-1 mRNA by ZnPP as compared to wild-type fibroblast cells (Figure5A). Furthermore, transfection of HA-1 cells with an Egr-1 antisense oligomer (40 μM) before incubation with ZnPP decreased Egr-1 protein by 21% and resulted in a 47% inhibition of both HO-1 mRNA and protein (Figure 5B).
Effect of Egr-1 on HO-1 mRNA induction by ZnPP.
(A) Relative HO-1 mRNA induction by ZnPP in Egr-1−/−mouse fibroblast cells. The HO-1 mRNA value for nontreated cell controls was assigned as 1. 3T3 indicates wild-type mouse fibroblast cells (NIH 3T3) incubated with 10 μM ZnPP for 24 hours. Egr+/− indicates Egr-1 heterozygote mouse fibroblasts incubated with 10 μM ZnPP for 24 hours. Egr−/−indicates Egr-1 homozygote null mutant mouse fibroblasts incubated with 10 μM ZnPP for 24 hours. (B) Effect of Egr-1 antisense oligonucleotides on ZnPP-induced HO-1 mRNA in HA-1 cells. Upper panel shows Egr-1 immunoreactive protein levels after incubation with Egr-1 antisense oligomer. Lower panel is the densitometric evaluation of HO-1 mRNA normalized to β-actin mRNA. Lane S, cells treated with 40 μM Egr-1 sense oligomers prior to incubation with ZnPP; lane A, cells treated with 40 μM Egr-1 antisense oligomers prior to incubation with ZnPP. Values represent the mean ± SE of 2 experiments; *P < .05 versus sense.
Effect of Egr-1 on HO-1 mRNA induction by ZnPP.
(A) Relative HO-1 mRNA induction by ZnPP in Egr-1−/−mouse fibroblast cells. The HO-1 mRNA value for nontreated cell controls was assigned as 1. 3T3 indicates wild-type mouse fibroblast cells (NIH 3T3) incubated with 10 μM ZnPP for 24 hours. Egr+/− indicates Egr-1 heterozygote mouse fibroblasts incubated with 10 μM ZnPP for 24 hours. Egr−/−indicates Egr-1 homozygote null mutant mouse fibroblasts incubated with 10 μM ZnPP for 24 hours. (B) Effect of Egr-1 antisense oligonucleotides on ZnPP-induced HO-1 mRNA in HA-1 cells. Upper panel shows Egr-1 immunoreactive protein levels after incubation with Egr-1 antisense oligomer. Lower panel is the densitometric evaluation of HO-1 mRNA normalized to β-actin mRNA. Lane S, cells treated with 40 μM Egr-1 sense oligomers prior to incubation with ZnPP; lane A, cells treated with 40 μM Egr-1 antisense oligomers prior to incubation with ZnPP. Values represent the mean ± SE of 2 experiments; *P < .05 versus sense.
Searching for Egr-1–like binding sites on the mouseHO-1 gene
Despite the above observations, no Egr-1 binding site had been previously identified on the HO-1 gene. However, a region of homology to an S/E overlapping binding site, as seen in theADA gene,11 was identified on the mouse HO-1 DE2 region. To determine whether binding occurred at this site in the ZnPP-treated cells, we used a synthesized oligomer containing this S/E sequence. Binding at the S/E was observed with ZnCl2-, SnPP-, and CrPP-treated cells but not with those treated with ZnPP (Figure 6). This does not preclude ZnPP-mediated binding to other sequences on HO-1.
S/E binding in nuclear extracts from HA-1 cells incubated with 100 μM ZnCl2 or 10 μM MPs for 24 hours.
The 12-bp oligomer (S/E) from the mouse HO-1 gene DE2 enhancer region has 85% similarity to the consensus sequence for an SP-1 and Egr-1 overlapping binding site. Lane F, free probe; lane ZC, cells incubated with ZnCl2; lane ZP, cells incubated with ZnPP; lane SP, cells incubated with SnPP; lane CP, cells incubated with CrPP.
S/E binding in nuclear extracts from HA-1 cells incubated with 100 μM ZnCl2 or 10 μM MPs for 24 hours.
The 12-bp oligomer (S/E) from the mouse HO-1 gene DE2 enhancer region has 85% similarity to the consensus sequence for an SP-1 and Egr-1 overlapping binding site. Lane F, free probe; lane ZC, cells incubated with ZnCl2; lane ZP, cells incubated with ZnPP; lane SP, cells incubated with SnPP; lane CP, cells incubated with CrPP.
Identification of the ZnPP-responsive elements in the mouseHO-1 gene
To localize other DNA sequences that mediate HO-1 gene activation in response to ZnPP, HA-1 cells were transiently transfected with plasmids containing various segments of the 5′-flanking region of the mouse HO-1 gene linked to the E coli CATgene. Transfected cells were incubated with 10 μM ZnPP or 100 μM ZnCl2 for 24 hours. ZnPP-dependent fusion gene regulation was assessed by measuring CAT activity in cellular extracts and then normalized to luciferase activity. Because ZnPP was found to inhibit luciferase activity in a dose-dependent manner (data not shown), the transfection efficiency measured by the luciferase activity was corrected to the level found in cells not treated with ZnPP. Analysis of approximately 12.5 kb of the HO-1 5′-flanking region indicated that the ZnPP responsive element resides within a 900-bp fragment (EH) located 9 kb upstream of the transcription initiation site (Figure7). By analyzing subfragments of EH in a manner analogous to that described above, the ZnPP responsive element was localized to a 196-bp RsaI/HindIII fragment, RH (Figure 8). Interestingly, neither the AB1 enhancer (Figure 8) nor the SX2 enhancer (Figure 7), which were previously shown to mediate transcriptional activity of theHO-1 gene in response to various agents including heme and cadmium, were responsive to ZnPP in HA-1 cells.
ZnPP and ZnCl2 responsive elements in the mouseHO-1 gene transfected to HA-1 cells.
A partial restriction map and structural organization of the mouseHO-1 gene and the 5′-flanking region are shown. Position +1 represents the transcription initiation site. The location of the EH and SX2 enhancer elements are indicated by solid bars. B indicates BamHI; E, EcoRI; H, HindIII; X, XhoI. HA-1 cells were transiently transfected with the indicated CAT constructs and incubated with 10 μM ZnPP or 100 μM ZnCl2 for 24 hours. The CAT assay was carried out as described under “Materials and methods.” The data represent the average value from 3 independent experiments and the mean fold induction of CAT activity in extracts of cells incubated with ZnPP or ZnCl2 compared to that in extracts of cells incubated with the constructs alone.
ZnPP and ZnCl2 responsive elements in the mouseHO-1 gene transfected to HA-1 cells.
A partial restriction map and structural organization of the mouseHO-1 gene and the 5′-flanking region are shown. Position +1 represents the transcription initiation site. The location of the EH and SX2 enhancer elements are indicated by solid bars. B indicates BamHI; E, EcoRI; H, HindIII; X, XhoI. HA-1 cells were transiently transfected with the indicated CAT constructs and incubated with 10 μM ZnPP or 100 μM ZnCl2 for 24 hours. The CAT assay was carried out as described under “Materials and methods.” The data represent the average value from 3 independent experiments and the mean fold induction of CAT activity in extracts of cells incubated with ZnPP or ZnCl2 compared to that in extracts of cells incubated with the constructs alone.
Localization of transcription enhancer activity of the EH subfragment in HA-1 cells.
The indicated fragments of EH were subcloned into the vector pMHO1CATΔ-33, which contains a minimal HO-1 promoter (−33 to +73). E indicates EcoRI; A, AflII; B, BsrBI; T, TaqI; R, RsaI; H,HindIII. HA-1 cells were transiently transfected with the indicated CAT constructs and incubated with 10 μM ZnPP or 100 μM ZnCl2 for 24 hours. The CAT assay was carried out as described under “Materials and methods.” The data represent the average value from 3 independent experiments and the mean fold induction of CAT activity in extracts of cells incubated with ZnPP or ZnCl2 compared to that in extracts of cells incubated with the constructs alone.
Localization of transcription enhancer activity of the EH subfragment in HA-1 cells.
The indicated fragments of EH were subcloned into the vector pMHO1CATΔ-33, which contains a minimal HO-1 promoter (−33 to +73). E indicates EcoRI; A, AflII; B, BsrBI; T, TaqI; R, RsaI; H,HindIII. HA-1 cells were transiently transfected with the indicated CAT constructs and incubated with 10 μM ZnPP or 100 μM ZnCl2 for 24 hours. The CAT assay was carried out as described under “Materials and methods.” The data represent the average value from 3 independent experiments and the mean fold induction of CAT activity in extracts of cells incubated with ZnPP or ZnCl2 compared to that in extracts of cells incubated with the constructs alone.
DNase I footprint analysis of the RH fragment
To determine specific sequences interacting with nuclear factors after incubation with ZnPP, an end-labeled RH fragment was incubated with nuclear extracts from HA-1 cells incubated with ZnPP, ZnCl2, or other MPs. The DNase I digestion products were electrophoresed on a denaturing polyacrylamide gel. The nuclear proteins from ZnPP-treated cells bound to a 50-bp region located between 12 and 62 bp on the 5′ end of the RH fragment (Figure9), whereas in ZnCl2- and SnPP-treated cells, this region was not protected by nuclear proteins (Figure 9). This newly identified GC-rich region contained a 9-bp sequence (GAGGGGTCG) that had a 78% homology to the known Egr-1 transcription factor consensus sequences (GGCGGGGGCG).
Nuclear protein binding analysis of the RH enhancer fragment by DNase I protection assays.
The noncoding strand of the RH fragment was labeled with32P at the 3′ end by an end-fill reaction. DNase I footprint analysis was carried out using 15 μg nuclear extract from HA-1 cells incubated with MPs. The DNase I digestion products were coelectrophoresed with the G+A chemical sequencing ladder of the RH probe on a denaturing 6% polyacrylamide gel and autoradiographed for 48 hours. Lane G+A is G+A chemical sequencing ladder; lane RH, RH probe alone digested with DNase I; lane BSA, RH probe incubated with BSA prior to DNase I digestion; lane ZC, nuclear extract from ZnCl2-treated cells incubated with RH probe prior to DNase I digestion; lane SP, nuclear extract from SnPP-treated cells incubated with RH probe prior to DNase I digestion; lane ZP, nuclear extract from ZnPP-treated cells incubated with RH probe prior to DNase I digestion. The region protected from DNase I digestion is indicated by the arrows and the 9-bp sequence, which has 78% homology with Egr-1 is indicated with the parentheses.
Nuclear protein binding analysis of the RH enhancer fragment by DNase I protection assays.
The noncoding strand of the RH fragment was labeled with32P at the 3′ end by an end-fill reaction. DNase I footprint analysis was carried out using 15 μg nuclear extract from HA-1 cells incubated with MPs. The DNase I digestion products were coelectrophoresed with the G+A chemical sequencing ladder of the RH probe on a denaturing 6% polyacrylamide gel and autoradiographed for 48 hours. Lane G+A is G+A chemical sequencing ladder; lane RH, RH probe alone digested with DNase I; lane BSA, RH probe incubated with BSA prior to DNase I digestion; lane ZC, nuclear extract from ZnCl2-treated cells incubated with RH probe prior to DNase I digestion; lane SP, nuclear extract from SnPP-treated cells incubated with RH probe prior to DNase I digestion; lane ZP, nuclear extract from ZnPP-treated cells incubated with RH probe prior to DNase I digestion. The region protected from DNase I digestion is indicated by the arrows and the 9-bp sequence, which has 78% homology with Egr-1 is indicated with the parentheses.
Effect of ZnPP on apoptosis
It is known that Egr-1 mediates cell cycle regulation and, in some models, initiates apoptosis by p53-mediated events.16Furthermore, HO-1 induction is associated with changes in cell cycle and cell proliferation.7 8 Therefore, it was important to understand whether increased Egr-1 binding associated with ZnPP also affected the cell cycle. Markers of apoptosis were assessed in the ZnPP-treated cells and these were compared to cells incubated with other MPs or ZnCl2.
After 24 hours of incubation, ZnPP-treated cells showed an increase in annexin V immunoreactive protein compared to CrPP- or SnPP-treated cells or untreated cell controls as determined by immunohistochemistry (Figure 10A). In addition, p53 protein levels in the cytoplasm were increased in ZnPP-treated cells as compared to cells treated with ZnCl2, SnPP, or CrPP or untreated controls (Figure 10B). The induction of p53 protein was dose dependent on ZnPP concentration (Figure 10B).
Markers of apoptosis in MP-incubated cells.
(A) A representative slide of immunoreactive annexin V detection in cultured HA-1 cells incubated with 10 μM MPs for 24 hours. Four slides in each group were incubated with annexin V antibody and FITC as described in “Materials and methods.” All images were obtained at the same intensity to allow for comparison. Lane C, untreated controls; lane CP, cells incubated with CrPP; lane SP, cells incubated with SnPP; lane ZP, cells incubated with ZnPP. (B) Upper panel shows representative example of 2 Western analyses for immunoreactive p53 protein content in HA-1 cells after 10 μM MP incubation. Lane p53, p53 protein standard obtained from human 293 cells; lane C, untreated controls; lane ZC, cells incubated with 100 μM ZnCl2; lane ZP, cells incubated with ZnPP;lane SP, cells incubated with SnPP; lane CP, cells incubated with CrPP. Lower panel indicates that p53 protein induction was dose dependent on ZnPP concentration. Lane p53, p53 protein standard obtained from human 293 cells; lane C, untreated controls; lane 1, cells incubated with 1 μM ZnPP; lane 10, cells incubated with 10 μM ZnPP; lane 50, cells incubated with 50 μM ZnPP.
Markers of apoptosis in MP-incubated cells.
(A) A representative slide of immunoreactive annexin V detection in cultured HA-1 cells incubated with 10 μM MPs for 24 hours. Four slides in each group were incubated with annexin V antibody and FITC as described in “Materials and methods.” All images were obtained at the same intensity to allow for comparison. Lane C, untreated controls; lane CP, cells incubated with CrPP; lane SP, cells incubated with SnPP; lane ZP, cells incubated with ZnPP. (B) Upper panel shows representative example of 2 Western analyses for immunoreactive p53 protein content in HA-1 cells after 10 μM MP incubation. Lane p53, p53 protein standard obtained from human 293 cells; lane C, untreated controls; lane ZC, cells incubated with 100 μM ZnCl2; lane ZP, cells incubated with ZnPP;lane SP, cells incubated with SnPP; lane CP, cells incubated with CrPP. Lower panel indicates that p53 protein induction was dose dependent on ZnPP concentration. Lane p53, p53 protein standard obtained from human 293 cells; lane C, untreated controls; lane 1, cells incubated with 1 μM ZnPP; lane 10, cells incubated with 10 μM ZnPP; lane 50, cells incubated with 50 μM ZnPP.
Nuclear localization of ZnPP
Because new protein synthesis did not completely account for HO-1 induction and Egr-1 antisense oligonucleotides did not completely suppress HO-1 induction, we examined the cellular localization of the ZnPP. Only ZnPP-treated cells demonstrated translocation to the nucleus after 24 hours of incubation, suggesting a direct cellular signaling mechanism of this MP (Figure 11).
Nuclear localization of ZnPP and SnPP in HA-1 cells.
Representative pseudoimages of fluorescent signal for MPs and immunoreactive p53. Three slides in each group were incubated with ZnPP or SnPP then incubated with p53 antibody and FITC-labeled secondary antibody as described in “Materials and methods” and analyzed at various time points. The MPs were visualized using their endogenous fluorescence by setting excitation at 568 nm and emission at more than 590 nm, and FITC was detected by setting the excitation at 488 nm and the emission at 515 to 545 nm. Upper panel is ZnPP-incubated HA-1 cells. Lower panel shows SnPP-incubated HA-1 cells. Lane 1h, 1-hour incubation with MP; lane 4h, 4-hour incubation with MP; lane 12h, 12-hour incubation with MP; lane 24h, 24-hour incubation with MP. The yellow arrows represent colocalization of MPs and p53; the orange arrow represents the endogenous fluorescence of the MPs; the green arrow represents the p53 signal. In the SnPP-treated cells, the FITC signal was further enhanced to allow for visualization of p53 within the nucleus. Note the nuclear colocalization of ZnPP and p53 and the lack of nuclear colocalization of SnPP and p53. CrPP could not be visualized due to a lack of endogenous fluorescence.
Nuclear localization of ZnPP and SnPP in HA-1 cells.
Representative pseudoimages of fluorescent signal for MPs and immunoreactive p53. Three slides in each group were incubated with ZnPP or SnPP then incubated with p53 antibody and FITC-labeled secondary antibody as described in “Materials and methods” and analyzed at various time points. The MPs were visualized using their endogenous fluorescence by setting excitation at 568 nm and emission at more than 590 nm, and FITC was detected by setting the excitation at 488 nm and the emission at 515 to 545 nm. Upper panel is ZnPP-incubated HA-1 cells. Lower panel shows SnPP-incubated HA-1 cells. Lane 1h, 1-hour incubation with MP; lane 4h, 4-hour incubation with MP; lane 12h, 12-hour incubation with MP; lane 24h, 24-hour incubation with MP. The yellow arrows represent colocalization of MPs and p53; the orange arrow represents the endogenous fluorescence of the MPs; the green arrow represents the p53 signal. In the SnPP-treated cells, the FITC signal was further enhanced to allow for visualization of p53 within the nucleus. Note the nuclear colocalization of ZnPP and p53 and the lack of nuclear colocalization of SnPP and p53. CrPP could not be visualized due to a lack of endogenous fluorescence.
Discussion
It is known that MPs can induce HO-1, the enzyme that they are known to competitively inhibit.39-41 Perhaps accumulation of heme from inhibition of HO activity could result in increased HO-1 expression because heme is a very strong inducer of HO-1. This would predict that the most potent MPs would lead to the highest induction of HO-1. However, in this study, the highest degree of HO-1 induction was observed in ZnPP, a moderate inhibitor of HO that cannot match the potency of SnPP or CrPP.22 This high degree of HO-1 induction with ZnPP was not associated with oxidative injury, as demonstrated with other MPs,9 nor was there evidence of metal response element regulation.35 Additionally, because both Zn++ ions and ZnPP were additive in HO-1 induction, ZnPP likely mediated HO-1 induction via different mechanisms than Zn++.
If the heme accumulated by the inhibition of HO activity was responsible for later HO-1 induction with the various MPs, we would anticipate increased AP-1 binding with ZnPP incubation,10but this was not noted. Nonetheless other MP and ZnCl2 were associated with increased AP-1 binding. Because exogenous Zn++ was introduced when cells were incubated with ZnCl2 and because there is a SP-1 response element on the mouse HO-1 gene, it may be that this zinc finger protein-binding region was involved in Zn++-mediated or perhaps in ZnPP-mediated HO-1 induction. This would mean that ZnPP-incubated cells could also show increased SP-1 binding if Zn++ dissociates from the porphyrin ring as considered by others,42 although this is debatable. The increased SP-1 binding with ZnCl2 suggested that SP-1 could serve in Zn++-mediated HO-1 induction, but the lack of SP-1 binding with ZnPP demonstrated that HO-1 induction by ZnPP is not likely attributable to the Zn++ ion.
Egr-1, another zinc finger protein, binds to the same GC-rich sites as SP-1 and can displace SP-1, thereby inhibiting its activity.11 In this study, the only MP associated with increased Egr-1 binding was ZnPP. The increased Egr-1 binding and the lack of SP-1 binding seen with ZnPP incubation further corroborated a competitive relationship between Egr-1 and SP-1. Despite our observations, Egr-1 sites have not been previously documented on theHO-1 gene, and nuclear protein from ZnPP-treated cells did not bind to a sequence of homology on the mouse HO-1 gene for an overlapping S/E sequence, as previously described for the humanADA gene.11 This could indicate that the S/E overlapping sequence used in the experiments was not the putative S/E binding sequence on the HO-1 gene or that Egr-1 binding occurred on other sites that have not yet been identified. To better understand ZnPP-mediated HO-1 induction, reporter analysis was performed and a newly identified ZnPP-related binding region was found on the HO-1 promoter. This region had substantial homology to the known Egr-1 binding sequence; therefore, a novel Egr-1–like response sequence is hypothesized. This is in agreement with the ZnPP-mediated loss of HO-1 induction when Egr-1 was decreased in various models tested here.
Altered Egr-1 gene expression is also known to mediate apoptosis16,43,44; therefore, it was logical that ZnPP-mediated Egr-1 binding increased apoptosis. Others have shown an antiapoptotic effect of ZnCl; therefore, the proapoptotic effect of ZnPP is not likely to be related to the Zn++ ion, but rather to the Zn-porphyrin complex. The Egr-1 protein is an antiproliferative signal in tumor cells that is required for apoptosis because Egr-1 transactivates the promoter for thep53 gene and up-regulates p53 expression.16This was confirmed by our findings as well because p53 protein was increased with ZnPP incubation. These experiments are the first to document that ZnPP increases apoptosis.
It is known that ZnPP is a naturally occurring compound found in states of anemia1 or in lead intoxication.3 This MP is also a potent inhibitor of hematopoiesis.5 The present observations demonstrate a link between the Zn-porphyrin complex, apoptosis, and HO-1 induction. It may be that ZnPP serves as a signal for decreased cell proliferation and apoptosis in situations where hemoglobin is deficient, thereby obviating red cell formation in the absence of hemoglobin. Nonetheless, the effects of ZnPP do not appear to be related only to hematopoietic cells because the present observations were made in fibroblasts. It is not clear, however, whether this model reflects the in vivo circumstance.
Despite strong evidence to demonstrate the role of Egr-1 in HO-1 induction, we speculate that ZnPP may have a direct effect in cells because ZnPP was seen to localize and remain in the nucleus, and that ZnPP-mediated HO-1 induction was not completely dependent on new protein synthesis nor Egr-1. The nuclear localization of ZnPP suggests that it may serve as a nuclear transcription factor for theHO-1 gene. This remains to be vigorously determined.
In summary, ZnPP is a unique MP that mediates HO-1 induction in part through Egr-1 binding. This MP is also associated with apoptosis. The increased formation of ZnPP in disease states may serve as a signaling mechanism for decreased cellular proliferation.
We thank Dr Jawed Alam of the Alton Ochsner Medical Foundation for providing the HO-1 CAT constructs. We also thank Dr Jeffery Milbrandt and Dr Alexander Gorodinsky of the Washington University School of Medicine for providing the Egr−/−and Egr+/− cells. We are very grateful to Dr Eric Sibley and Ms Lynne Olds of Stanford University Department of Pediatrics for their advice and technical support with the DNase I footprint experiments.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Phyllis A. Dennery, Division of Neonatal and Developmental Medicine, Department of Pediatrics, School of Medicine, Stanford University, 750 Welch Rd, Palo Alto, CA 94304; e-mail:dennery@leland.stanford.edu.