Abstract
The spatial distribution of subpopulations of hemopoietic progenitor cells following syngeneic transplantation was investigated at the single-cell level. The location of infused hemopoietic progenitor cells within the femoral bone marrow of nonablated recipients was determined by 5-(and-6)-carboxyfluorescein diacetate succinimidyl ester labeling of cells and in situ fixation by perfusion. Analysis performed over 15 hours after infusion demonstrated that the spatial distribution of transplanted marrow cells is not a random process. Although the majority of cells enter the bone marrow from the central marrow vessels, the subsequent localization within the bone marrow varied according to their phenotype. Candidate “stem cells” demonstrated selective redistribution and were significantly enriched within the endosteal region, whereas mature terminally differentiated and lineage-committed cells selectively redistributed away from the endosteal region and were predominantly in the central marrow region. Together, these data strongly support historical evidence of the presence of endosteal hemopoietic stem cell niches.
Introduction
In the bone marrow (BM), hemopoiesis takes place in the extravascular spaces between the marrow sinuses. Within these hemopoietic cords maturing hemopoietic cells exhibit distinctive and consistent lineage-specific spatial locations. For example, megakaryocytes develop on the adventitial surface of the vascular sinuses, granulocytic cells are consistently associated with alkaline phosphatase-positive reticular cells, and erythroblasts undergo synchronous maturation around a central macrophage-forming erythroblastic islands.1-3 Furthermore, the highest concentration of pre-B cells is in the subendosteal area, which gradually declines toward the center of the marrow.4 In contrast, mature B cells are uniformly distributed.4
Under steady-state conditions, hemopoietic BM cells develop in intimate association with a highly organized 3-dimensional microenvironment. This structural scaffolding is comprised of many cell types, including a phenotypically and probably functionally diverse population of stromal cells,5-7 together with their associated biosynthetic products, including extracellular matrix (ECM) components and hemopoietic growth factors. Much of the data regarding ECM proteins was derived from the analysis of long-term BM cultures,8which were shown to contain the ECM components fibronectin9; collagen Types I, III, IV, and V10; laminin; and various proteoglycans.11 We mapped 5 key ECM proteins in situ, showing each to have specific localization, suggesting different structural roles in the regulatory aspects of hemopoiesis.12
At present, the inability to identify hemopoietic stem cells (HSCs) in situ makes it impossible to analyze the spatial distribution of these cells in the BM and the molecules that regulate this process. However, previous studies in the mouse by Lord13 and colleagues have established that primitive hemopoietic cells (spleen colony-forming-units [CFU-s]) and all the major hemopoietic progenitor cell types conform to a well-defined spatial distribution across the longitudinal axis of the femur.14-18 Of note, CFU-s were shown to be enriched in a region of the marrow adjacent to bone.14 These data therefore provide circumstantial evidence for the presence of hemopoietic stem cell “niches” in close association with the endosteum.
The control of stem cell proliferation is affected in the locality of the HSCs themselves as initially demonstrated by the elegant experiments of Croizat et al19 and Gidali and Lajtha.20 Encapsulating the concept of highly specific, local interactions regulating hemopoiesis, Schofield21formulated the niche hypothesis in which it was suggested that the most primitive hemopoietic cell isolated at the time, the CFU-s, did not represent the ultimate hemopoietic stem cell. Rather, true HSCs were proposed to exist in association with one or more other supporting cells and would therefore, in essence, be fixed tissue cells. These microenvironmental cells were postulated to form a specific niche that, when in close association with the HSC, confer on it the HSC attribute of indefinite self-renewal capacity, while effectively inhibiting differentiation and maturation of the cell. To this day the niche hypothesis remains largely unchallenged and continues to provide an important conceptual basis for studies that analyze the role of the hemopoietic microenvironment in the regulation of the HSC. However, a more detailed description of the hypothesized niches in terms of their precise location within the BM and of their cellular and molecular composition is currently lacking.
We have recently developed a novel approach based on the use of BM transplantation to track cells at the individual cell level as they lodge in the BM of nonablated recipients. Although transplantation into myeloblated recipients represents the standard means by which patients are given a graft of HSC, the most appropriate method for analyzing the spatial distribution of cells within the BM, and consequently the factors that regulate this process, is one in which the hemopoietic microenvironment has not been altered by preparative ablation. We have analyzed the spatial distribution of various hemopoietic progenitor cell-enriched BM subpopulations within the femur over the first 15 hours following intravenous infusion. This brief time period allows the investigation of the initial phases of donor cell homing with minimal added complexity from their proliferation.22Transplantations of different BM subpopulations demonstrated that, although the majority of cells entered the BM from the central marrow vessels, the subsequent localization within the BM varied according to their phenotype. HSC-enriched populations exhibited selective lodgment in the endosteal region. In contrast, hemopoietic cells expressing surface markers associated with lineage commitment redistributed away from the endosteal region and demonstrated high selectivity for the central marrow region. Therefore, the distribution of transplanted hemopoietic cells within the BM is not random but closely reflects that previously defined for the different subpopulations in steady-state adult mouse BM.14-18 More importantly, these data demonstrate for the first time that the discrete spatial localization of transplanted primitive and mature hemopoietic cells within the BM appears to be the result of specific, hierarchically dependent patterns of migration that culminate in the retention of these populations at anatomically distinct marrow sites.
Materials and Methods
Mice
Specific pathogen-free 6- to 10-week-old BALB/c H-2D mice were purchased from the Animal Resources Centre (Canning Vale, WA, Australia) and housed in a conventional clean facility for at least 1 week prior to experimental use. All mice received mouse chow (Barastok, St Arnaud, Victoria, Australia) and acidified water ad libitum.
Isolation of hemopoietic cell suspensions
Mice were killed by cervical dislocation. BM was routinely collected from femurs, tibiae, and iliac crests. These bones were thoroughly ground in phosphate-buffered saline (PBS) supplemented with 2% heat-inactivated (HI) fetal calf serum (FCS; Hyclone, Logan, UT). The bone fragments were washed multiple times, and the supernatant cell suspension and wash fractions were filtered through a 40-μm filter (Becton Dickinson, Franklin Lakes, NJ) to remove large bone particles. The marrow was centrifuged (400g for 5 minutes) and resuspended in fresh buffer. The cell supernatant was refiltered through a 40-μm filter and diluted to 107 cells/mL PBS supplemented with 2% HI FCS (buffer).
Hemopoietic cell enrichment strategies
Lineage-positive cells (Lin+).
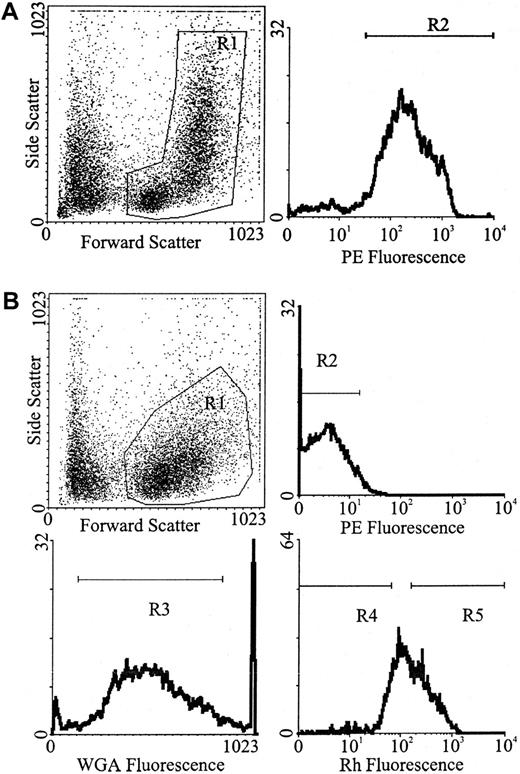
Whole BM was labeled with a cocktail of primary rat antimouse antibodies: anti-B220 (CD45R; B cells),23 anti–Mac-1 (CD11b; macrophages),24 anti–Gr-1 (Ly-6G; neutrophils),25 anti–Lyt-2 (CD8), and anti-L3T4 (CD4; T cells)26 (Hybridoma supernatants). Each batch of antibody was evaluated by flow cytometric analysis for the concentration that resulted in the greatest shift in mean channel fluorescence and/or the percentage of positive cells detected. Double-strength antibody cocktail (50 μL) was added per 5 × 106 cells suspended in an equal volume of buffer, and the cell/antibody suspension was incubated on ice for 20 minutes. The cells were washed in buffer, resuspended at the same cell concentration, and incubated with a final concentration of 1:80 (6.8 μg/mL) goat antirat phycoerythrin (PE)-conjugated secondary antibody (Biosource International, Camarillo, CA) on ice in the dark for a further 20 minutes. Finally, the cells were washed in buffer, resuspended at 5 × 106 cells/mL buffer, and stored on ice prior to Lin+ cells being isolated by fluorescence activated-cell sorting (FACS) (Figure1A).
Flow cytometric separation strategy.
(A) For the isolation of Lin+ white blood cells were gated from whole BM (R1) followed by the selection of PE-positive cells (R2). (B) For the isolation of Lin− cells, white blood cells were gated from Lin− MACS-depleted marrow (R1), followed by the selection of PE-negative cells (R2). For the isolation of Lin−WGAdim-medRhdim or bright, cells were gated from Lin− MACS-depleted marrow (R1), followed by the selection of PE-negative cells (R2), followed by the selection of WGAdim-med-cells (R3), followed by the selection of either Rhdim- (R4) or Rhbright-cells (R5).
Flow cytometric separation strategy.
(A) For the isolation of Lin+ white blood cells were gated from whole BM (R1) followed by the selection of PE-positive cells (R2). (B) For the isolation of Lin− cells, white blood cells were gated from Lin− MACS-depleted marrow (R1), followed by the selection of PE-negative cells (R2). For the isolation of Lin−WGAdim-medRhdim or bright, cells were gated from Lin− MACS-depleted marrow (R1), followed by the selection of PE-negative cells (R2), followed by the selection of WGAdim-med-cells (R3), followed by the selection of either Rhdim- (R4) or Rhbright-cells (R5).
Lineage-negative cells (Lin−).
BM mononuclear cells of low density (< 1.0777 g/cm3) were isolated by discontinuous density centrifugation, using Nycoprep for animals (Accurate Chemical and Scientific Corporation, Westbury, NY). The isolated cells were washed in buffer prior to further manipulation.
Lin− cells were isolated in a similar manner to that previously described by Bertoncello et al.27 Briefly, low-density cells were labeled with the same cocktail of primary rat antimouse antibodies as described above. Lin+ cells were removed by immunomagnetic selection, using the MACS system (Miltenyi Biotec, Bergisch, Gladbach, Germany). Washed, antibody-labeled cells were incubated with goat antirat immunoglobulin G microbeads (Miltenyi Biotec) at 4°C for 15 minutes by adding 15 μL of beads to 85 μL of cell suspension (containing 107 cells in buffer) and agitating regularly. The cells were washed in buffer and resuspended in 1.5 mL PBS, 5 mM EDTA, and 1% bovine serum albumin (BSA)/108 cells. Up to 2 mL of the cells were then added to the column (C column, maximum capacity 2 × 108 cells, generally not run at more than half the maximum stated capacity), run into the mesh, and left to magnetize for 5 minutes. The Lin− cells (nonmagnetic fraction) were collected by eluting the cells through a 22-gauge needle with 25 mL PBS, 5 mM EDTA, and 1% BSA. When Lin− cells were to be transplanted, the cells were washed with buffer and then incubated with the goat antirat PE-conjugated secondary antibody as described above. Washed cells were resuspended at 5 × 106 cells/mL buffer and stored on ice prior to Lin− cells being isolated by FACS (Figure 1B).
Isolation of populations enriched in stem cells (Lin−, wheat germ agglutinin (WGA)dim-medium, rhodamine (Rh) 123dull cells), and progenitors (Lin−, WGAdim-medium, Rh 123bright).
Immunomagnetically enriched Lin− cell suspensions resuspended at 106 cells/mL buffer were incubated in a final Rh (Molecular Probes, Eugene, OR) concentration of 0.1 μg/mL (diluted in buffer) for 20 minutes at 37°C in the dark. The cells were centrifuged, resuspended at 106 cells/mL in buffer, effluxed for 15 minutes at 37°C in the dark, centrifuged, and resuspended at 108 cells/mL in PBS 0.5% HI FCS. The cells were labeled with a final concentration of 0.25 μg/mL biotinylated WGA (Vector Laboratories, Burlingame, CA) (diluted in PBS) at room temperature for 15 minutes in the dark, washed with PBS 0.5% HI FCS, and resuspended at 106 cells/mL PBS 0.5% HI FCS. The cells were labeled with a fluorochrome cocktail, containing a final dilution of 1:160 streptavidin-Red 670 (Caltag, San Francisco, CA) and 1:80 goat antirat PE for 20 minutes on ice, washed in buffer, resuspended at 5 × 106 cells/mL buffer, and held on ice for FACS (Figure 1B).
Flow cytometry
Labeled cells were sorted on a FACStarplus cell sorter equipped with a 5-watt argon ion laser (Coherent Innova 90, Palo Alto, CA) emitting 488 nm light at 200 mW, and a Spectra-Physics ultraviolet (UV) laser (Mountain View, CA) emitting 350/360 nm light at 50 mW. Light-scatter signals were collected through a 488-nm band pass 10 filter and a 1-decade logarithmic neutral density filter in the forward light-scatter path. Rh-emitted green fluorescence pulses were collected through an FITC 530-nm band pass 15 filter. Orange fluorescence pulses emitted following excitation PE were reflected through a 440-dichroic short pass mirror and collected through a 575-nm band pass dichroic 26 filter. Pulses emitted following the excitation of Red 670 were collected through a long-pass RG655 filter.
Although progenitors such as CFU-s at day 12 have bright WGA labeling, the stem cell subset has been characterized as having dim to medium WGA labeling28 and low Rh retention.29-31 For an enriched stem cell subset we isolated Lin− (PE negative) cells with low to medium WGA RED-670 fluorescence and then low Rh fluorescence (WGAdim-med/Rhdull).32 Cells for sorting were kept chilled, sorted at a rate of approximately 3000 cells/second, and collected in serum-coated tubes. Overlap of the Rh, PE, and RED-670 emission spectra was compensated for electronically.
5-(and-6)-carboxyfluorescein diacetate succinimidyl ester labeling
Cells to be transplanted were labeled with the fluorescent dye 5-(and-6)-carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, OR) by using a method adapted from Nordon et al.33 Briefly, cell populations of varying enrichment for hemopoietic progenitor cells were washed in PBS 0.5% HI FCS to remove high levels of serum. Washed cells were resuspended in PBS 0.5% HI FCS at a density of 106 cells/mL and preincubated at 37°C for 2 minutes. CFSE was diluted to 5 mM in dimethyl sulfoxide and then to 5 μM in PBS. CFSE was added to the cells to give a final concentration of 0.5 μM, and the dye solution/cell mixture was incubated at 37°C for a further 10 minutes. Staining was stopped by adding 10 times the dye solution/cell volume of ice-cold PBS containing 20% FCS. Finally, the cells were washed in PBS and resuspended for injection in up to 0.3 mL PBS per recipient. The toxicity of CFSE labeling has been studied both previously33 and in house, and no toxic effects have been found at the dose of CFSE used.
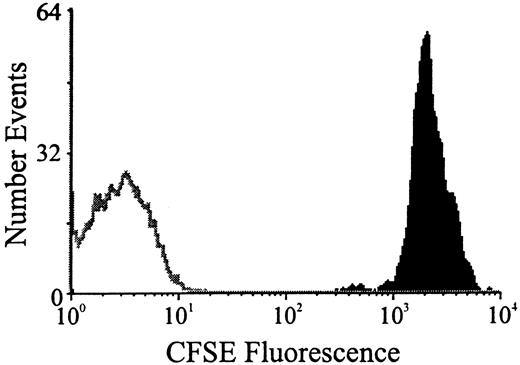
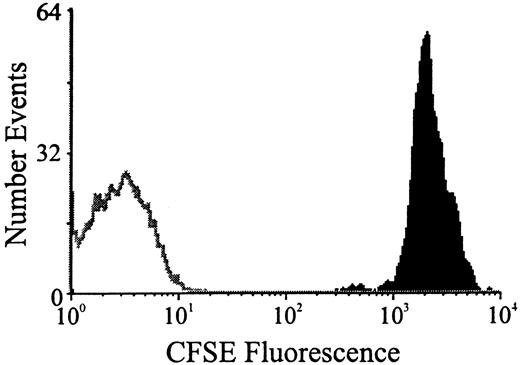
The CFSE labeling and viability of an aliquot of each transplanted cell population was analyzed by FACS. Green fluorescence pulses caused by the excitation of CFSE were collected through an FITC 530-nm band pass 15 filter. Transplanted CFSE-labeled cells were always detected at more than 3 logs above background (Figure 2).
Fluorescence histogram.
CFSE-stained BM cells are solid black; the single black line is an overlay of unlabeled BM.
Fluorescence histogram.
CFSE-stained BM cells are solid black; the single black line is an overlay of unlabeled BM.
More than 98.5% of transplanted cells were viable as determined by using the viability dye Fluoro-Gold (hydroxystilbamidine methanesulfonate; Molecular Probes) as previously described.34 Fluoro-Gold was excited by UV, and emissions were collected through a long-pass RG630 filter.
Transplants
Cell populations of varying enrichment for hemopoietic progenitor and stem cells were transplanted into nonablated recipients by injection at the lateral tail vein. The actual numbers of cells injected were 2.2 to 7.3 × 105 Lin+ cells, 2.0 to 2.2 × 105 Lin− cells, 1.3 to 1.7 × 105Lin−/WGAdim-med/Rhbright cells, and 0.8 to 3.3 × 105Lin−/WGAdim-med/Rhdull cells.
Cells were allowed to home for up to 15 hours. This time point was chosen on the basis of observations,22 35 demonstrating that transplanted cells quickly progress into cell cycle and that their progeny increasingly contribute to levels of donor cells detected after 24 hours. Therefore, at time points earlier than 24 hours after transplantation , the percentages of donor cells found in the marrow will more accurately reflect the fate of transplanted cells rather than their progeny.
Transplantation analysis
At the end of the transplantation period the femoral BM was fixed by perfusing 2% paraformaldehyde, 0.05% glutaraldehyde through the descending aorta at physiologic pressure as previously described.36 Femurs were removed, and further immersion fixed for up to 24 hours, prior to the bones being decalcified in 10% EDTA for 2 to 3 weeks. Bones were then dehydrated in graded ethanol and embedded in paraffin as previously described.36Longitudinal sections (3.5 μm) of each femur were cut, dewaxed, and brought to water. Sections were washed in PBS prior to mounting in antifade mounting medium (Vectashield, Vector Laboratories). All sections were analyzed under a fluorescence microscope (Zeiss, Camperdown, NSW, Australia), using an FITC and Texas red dual filter set (green excitation at 578 nm and red excitation at 610 nm). This filter set was specifically chosen because the short emission bandwidths clearly allow CFSE-positive BM cells to be easily distinguished from host marrow cells.
Statistical analysis
Analysis involved determining if there was a specific pattern of spatial distribution at the different time points analyzed and if there was a change in this pattern with either time or purification of the transplant population. To determine if the spatial distributions observed were random, analysis of the distribution of donor cells from the different marrow subpopulations at each time point was done by using the following formula:
OENi, EENi, OCNi, ECNi were the observed and expected number of cells in the endosteal and central marrow regions for mousei, respectively. This statistic was then compared with an χ2 distribution with n degrees of freedom. All tests were 2 sided, and a P value < .05 was considered to be significant. Analysis was then done to determine which of the following factors influenced the percentage of cells at the endosteum: the subpopulation of donor cells infused, the time after transplantation , and the number of cells infused. This analysis was done by using a multiple linear regression model that used a forward selection procedure. After adjusting for the subpopulation transplanted and the time after transplantation , the number of cells infused was not significant (P = .29). Hence, the final analysis only included the other 2 factors. The interaction between the time after transplantation and the marrow subpopulation was then examined by using the following regression model weighted by the number of cells counted for each mouse:Yijk = μ + αI + β1itj + β2it + εijk. Yijk is the percentage of cells at the endosteum of mouse k injected with cells of purity level i and killed at time tj after transplantation,μ is the grand mean, αi is the parameter associated with purity level i,β1i is the coefficient term fortj associated with purity level i, β2i is the coefficient term fort associated with purity level i, tj is the sacrifice time, andεijk is the overall error term. This model indicated that each purity level exhibited its own pattern over time (P < .0001; test of equalities ofβ1i and β2i).
To determine whether there was a significant change in the number of donor cells detected per section a one-way analysis of variance was done followed by a Tukey test to determine which points were statistically significant.
Results
Analysis of spatial distribution
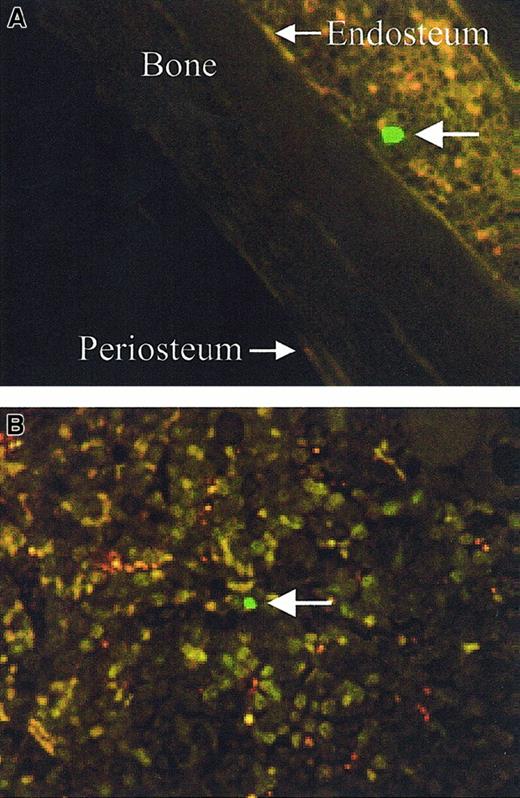
The spatial distribution of transplanted cells was determined by analyzing the location of CFSE-labeled cells (positive cells) from at least 6 longitudinal sections per transplant recipient. Central longitudinal sections were analyzed as opposed to transverse sections, as each individual section encompasses more of the entire femur. To ensure that individual cells were only analyzed once, every alternate 3.5-μm section was analyzed. The location of positive cells were designated as either endosteal (previously arbitrarily defined as within 12 cells of the endosteum37) or central (> 12 cells from either endosteum) (Figure 3).
Spatial designation of CFSE-positive cells.
Large arrows indicate the CFSE-positive cells, endosteal (less than 12 cells from the endosteum) (A, original magnification × 60) and central (> 12 cells from either endosteum) (B, original magnification × 80).
Spatial designation of CFSE-positive cells.
Large arrows indicate the CFSE-positive cells, endosteal (less than 12 cells from the endosteum) (A, original magnification × 60) and central (> 12 cells from either endosteum) (B, original magnification × 80).
The endosteal region of the femur comprises an average of 13% (13.5% ± 0.7%) (mean ± SEM by analyzing 3 central sections from each of 3 randomly chosen mice, using the software analysis package Imagepro Plus 4.0 [Media Cybernetics, Silver Spring, MD]) of the cellular area, excluding the central vein. To ensure that the femurs from the transplant recipients were of equivalent size and not toward the top or the base of the femur, where less central marrow would increase the endosteal proportion and potentially bias the results, the total BM width was analyzed in 81 randomly selected central marrow sections, using the same software. This width was shown to be 877 ± 12 μm (mean ± SEM).
For the Lin+, Lin−, purified progenitors, and HSC transplants the average number of positive cells analyzed per recipient per time point was 67 ± 6, 42 ± 3, 27 ± 4, and 25 ± 4, respectively, from between 6 and 22 sections per recipient for the Lin+ and Lin− transplants and 15 and 75 sections for the purified progenitors and HSC transplants. There were between 3 and 6 recipients per group per time point. The number of donor cells detected per section analyzed and the percentage of cells in each region was then determined.
Engrafting marrow cells are not randomly distributed within the BM
If engrafting donor cells were randomly distributed within the femur, the number of donor cells in the endosteal region would be equal to the proportion of BM that this area comprises (ie, 13% of the total number of donor cells detected). However, in all transplantation groups at all time points, the spatial distribution of engrafting cells was not random (P < .0025).
One hour following a transplantation of Lin+ cells, 58% ± 3% of the donor cells were located within the central marrow region and 42% ± 3% were found within the endosteal region of the BM (Figure 4A). During the subsequent 14 hours there was a progressive and significant decrease in the proportion of donor cells located within the endosteal region associated with a corresponding progressive increase in the proportion of donor cells within the central region. As a consequence, at day 15 after transplantation only 24% ± 0.9% of the infused Lin+ cells were located in the endosteal region, whereas 76% ± 0.9% were found within the central marrow region of the BM. This distribution pattern 15 hours after transplantation was 1.9-fold greater than expected if the distribution was random, but it was the closest to a random pattern detected following a transplantation of any of the marrow cell subpopulations.
The spatial distribution of various marrow subpopulations present in the BM up to 15 hours after transplantation.
(A) Lin+ (○) and Lin− cells (●); (B) WGAdim-med/Rhbright cells (○), and WGAdim-med/Rhdull cells (●). Individual circles represent individual recipients. Each symbol joined by the line is the mean (SEM of between 3 and 6 individual animals). Where no error bar is visible, it falls within the symbol.
The spatial distribution of various marrow subpopulations present in the BM up to 15 hours after transplantation.
(A) Lin+ (○) and Lin− cells (●); (B) WGAdim-med/Rhbright cells (○), and WGAdim-med/Rhdull cells (●). Individual circles represent individual recipients. Each symbol joined by the line is the mean (SEM of between 3 and 6 individual animals). Where no error bar is visible, it falls within the symbol.
Interestingly, 1 hour following a transplantation of Lin−cells, 53% ± 4% of the donor cells were located within the central marrow region, and 47% ± 4% were found within the endosteal region of the BM (Figure 4A). This distribution is equivalent to that seen following a transplantation of Lin+ cells. During the subsequent 4 hours there was a significant decrease in the proportion of donor cells located within the central region to 37% ± 4% associated with a corresponding significant increase in the proportion of donor cells within the endosteal region to 63% ± 4%. The distribution of infused cells then remained relatively unchanged for the rest of the analysis period. This distribution pattern of infused cells 15 hours after transplantation was 5.1-fold greater than expected for a random distribution.
To further analyze this stem cell-enriched Lin− fraction it was subsetted on the basis of Rh (top 15% to 20% fluorescence for Rhbright progenitor cells and bottom 15% to 20% fluorescence for Rhdull HSC). This subsetting allowed us to distinguish between a very mature and immature population with very different cloning and marrow repopulating efficiencies. The proportion of high-proliferative potential colony-forming cells in the 2 fractions are at a ratio of 0.6:170, respectively.38 The marrow repopulating ability of Rhbright versus Rhdullcells is 1:5, respectively.39
One hour following a purified progenitor transplantation 45% ± 3% of donor cells were found in the endosteal marrow region, and 55% ± 3% were located in the central marrow region (Figure 4B). Statistically there was no significant redistribution of these cells over the transplantation period.
One hour following a HSC transplantation 72% ± 3% of donor cells were found in the central marrow region, and only 28% ± 3% were located in the endosteal marrow region (Figure 4B). This proportion of cells at the endosteum was significantly lower than that following a transplantation of progenitor cells. However, in contrast to the lack of cell redistribution following a transplantation of purified progenitors, there was significant redistribution to the endosteal region following a HSC transplantation resulting in more than 60% of these cells being located in the endosteal region 15 hours after transplantation. In addition, compared to Lin− cells, the redistribution of cells over the initial 5 hours following a HSC transplantation was significantly faster (P = .0019), resulting in an equivalent proportion of cells in the endosteal region to that following a transplantation of Lin− cells 5 hours after transplantation. Similarly to that seen following a transplantation of Lin− cells, this distribution pattern then remained relatively unchanged for the rest of the analysis period.
Changes in the spatial distribution of engrafting cells over the transplantation period are not due to a change in the total number of donor cells within the BM
The average number of donor cells detected per section per 100 000 cells transplanted was analyzed to ensure that any apparent changes in the spatial distribution of engrafting cells was not the consequence of a change in the total number of cells located within the marrow (Table 1). In all transplantation groups the changes in the spatial distribution over the transplantation period cannot simply be attributed to a change in the number of donor cells within the femur. For instance, following an infusion of Lin+ cells, there were no significant changes in the number of donor cells detected within the femur over the transplantation period, and yet there was a significant increase in the proportion of donor cells in the central region (Figure 4A).
Following a transplantation of highly purified HSC, there was a significant drop in the number of donor cells in the femur between 1 and 3 hours after transplantation, but no changes occurred over the rest of the transplantation period. However, there was a significant increase in the proportion of donor cells at the endosteum over the entire transplantation period (Figure 4B). This increase also cannot be attributed to cells either being released from the marrow or cell death and an equivalent number of cells entering the marrow from the circulation. By 5 hours after transplantation, HSCs of donor origin are no longer detectable in the peripheral blood by FACS analysis (data not shown), and yet the proportion detected within the endosteal region continues to increase.
Discussion
The data strongly suggest that hierarchically distinct hemopoietic progenitor cells exhibit distinct patterns of lodgment. Although immediately (1 hour) following a transplantation of HSCs the majority of donor cells was located in the central marrow region, there was a rapid redistribution of these cells, resulting in a preferential seeding in the endosteal region. This pattern was similar to that evident following a transplantation of Lin− marrow cells, although there were significantly more donor cells initially found in the endosteal region following a transplantation of Lin−cells. In addition, the redistribution of HSCs to the endosteal region was significantly faster than that following a transplantation of Lin− cells. In contrast, hemopoietic cells expressing surface markers associated with lineage commitment preferentially redistributed away from the endosteal region and demonstrated high selectivity for the central marrow region. Similarly, purified progenitor cells, with low cloning efficiency and marrow repopulating ability, also demonstrated high selectivity for the central marrow region. These distributions were clearly not a random phenomenon but appear to be the result of specific, hierarchically dependent patterns of migration and retention of cells in anatomically distinct marrow sites. In addition, the data strongly support the presence of HSC niches within the endosteal region.
We previously showed that 6 weeks following a transplantation of Rh/Hoechstdull HSCs all of the cells of donor origin were located in the endosteal region.37 However, because of the inability of obtaining sufficient numbers of these cells to analyze their spatial distribution at very early time points after transplantation, it remained possible that the cells initially homed to another marrow region and migrated to the endosteal region after proliferation. In the current study we have isolated sufficient numbers of Lin−/WGAdim-med/Rhdull HSCs to be able to analyze their spatial distribution at very early time points after transplantation, demonstrating that these cells home to this area prior to proliferation.
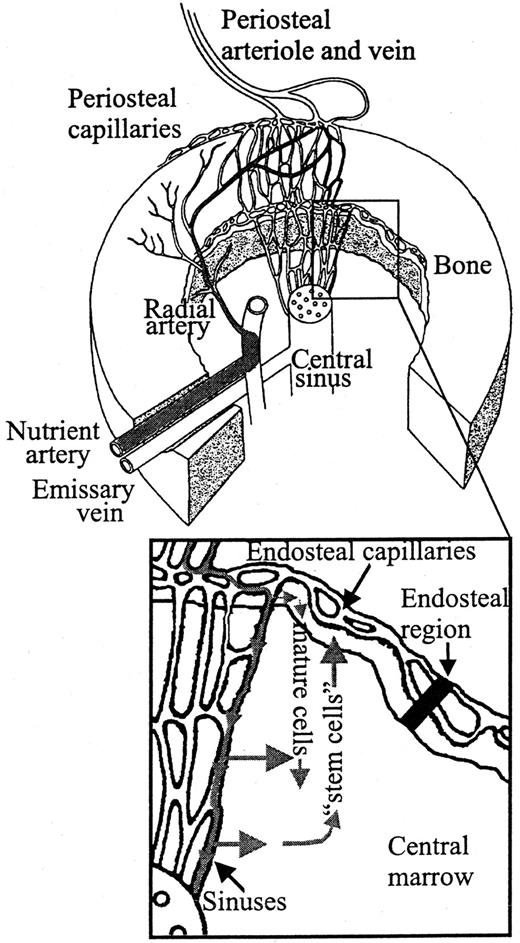
Although it has been documented that hemopoietic cells exit the BM into the circulation through the sinus walls,40 at present it is unknown exactly where engrafting transplanted cells exit the peripheral blood by transendothelially migrating into the BM. This is due to the technical difficulty in capturing very limited numbers of cells undergoing this process and to defining the exact timing at which it occurs after transplantation. The arterial blood of marrow comes from 2 major sources: the nutrient artery (primary source) and the cortical capillary system.41 The passage of blood through the marrow was originally determined by Brookes in 1971.41Together with the data presented in this study, we suggest a model in which transplanted marrow cells travel through the femoral arterial blood supply and transendothelially migrate from the femoral sinuses having passed through the bone cortex. This is schematically represented in Figure 5. We have shown that the majority of transplanted cells transendothelially migrate in the central marrow region (58%, 53%, 71%, and 72% of cells from Lin+, Lin−, purified progenitors, and HSCs, respectively, 1 hour after transplantation), with some transendothelial migration occurring in the endosteal region (42%, 47%, 29%, and 28% from Lin+, Lin−, purified progenitors, and HSCs, respectively, 1 hour after transplantation). Hemopoietic cells expressing surface markers associated with lineage commitment that entered the marrow endosteally then redistribute to the central region, and hemopoietic cells not expressing surface markers associated with lineage commitment but that have high cloning efficiencies and marrow repopulating abilities that entered the marrow centrally then redistribute to the endosteal region.
A model suggesting the flow of transplanted BM cells entering the BM.
The schematic of the marrow circulation is adapted from Lichtman.40
A model suggesting the flow of transplanted BM cells entering the BM.
The schematic of the marrow circulation is adapted from Lichtman.40
We would like to acknowledge Michelle Cook for her help with the animal work and Brenda Williams for the isolation of the different progenitor cell populations. We would like to thank Ralph Rossi for his invaluable help and advice with the flow cytometric isolation of enriched subsets of hemopoietic progenitor cells. In addition we would like to thank Kally Yuen for help with the statistical analysis of the data and Ivan Bertoncello, Paul Simmons, and David Haylock for their critical analysis of the manuscript.
S. Nilsson is a R. D. Wright Fellow, granted from the National Health and Medical Research Council, Australia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Susan Nilsson, Trescowthick Research Laboratories, Peter MacCallum Cancer Institute, Locked Bag No. 1, A'Beckett Street, Melbourne, Victoria, 3000, Australia; e-mail:s.nilsson@pmci.unimelb.edu.au.