Abstract
Signal-regulatory proteins (SIRPs) represent a new family of inhibitory/activating receptor pairs. They consist of 3 highly homologous immunoglobulin (Ig)–like domains in their extracellular regions, but differ in their cytoplasmic regions by the presence (SIRPα) or absence (SIRPβ) of immunoreceptor tyrosine-based inhibitory motifs (ITIMs). To analyze the differential expression on hematopoietic cells, function and ligand binding capacity of SIRPα and SIRPβ molecules, soluble fusion proteins consisting of the extracellular domains of SIRPα1, SIRPα2, and SIRPβ1, as well as SIRPα/β-specific and SIRPβ-specific monoclonal antibodies (MoAbs) were generated. In contrast to SIRPα1 and SIRPα2, no adhesion of SIRPβ1 to CD47 could be detected by cell attachment assays and flow cytometry. Using deletion constructs of SIRPα1, the epitope responsible for SIRPα1 binding to CD47 could be confined to the N-terminal Ig-like loop. Flow cytometry analysis with SIRPα/β- and SIRPβ-specific MoAbs revealed that SIRPα but not SIRPβ is expressed on CD34+CD38− hematopoietic cells. In addition, a strong SIRPα expression was also observed on primary myeloid dendritic cells (DCs) from peripheral blood as well as on in vitro generated DCs. Analysis of the T-cell stimulatory capacity of in vitro generated DCs in the presence of soluble SIRPα1 fusion proteins as well as SIRPα/β-specific and CD47-specific MoAbs revealed a significant reduction of T-cell proliferation in mixed lymphocyte reaction and inhibition of induction of primary T-cell responses under these conditions. In contrast, soluble SIRPα or SIRPβ-specific antibodies had no effect. The data suggest that the interaction of SIRPα with CD47 plays an important role during T-cell activation and induction of antigen-specific cytotoxic T-lymphocyte responses by DCs.
Introduction
Signal-regulatory proteins (SIRPs) comprise a novel transmembrane glycoprotein family involved in receptor tyrosine kinase-coupled signaling pathways.1 These molecules are also called SHPS-1 (src homology 2 domain–containing phosphatase substrate-1),2 BIT (brain immunoglobulin [Ig]–like molecule with a tyrosine-based activation motif),3P84,4 and MFR (macrophage fusion receptor).5Structurally, all SIRP members share a large extracellular region with 3 Ig-like loops.1 The cytoplasmic domains of SIRPα subfamily members display 2 immunoreceptor tyrosine-based inhibitory motifs (ITIMs).1 These ITIM regions, which are also present on other inhibitory receptor molecules, recruit src homology 2 domain–containing phosphatases SHP-21,2 and SHP-16,7 and negatively regulate signal transduction cascades. Although SIRPα1 is known to inhibit receptor tyrosine kinase-coupled signaling pathways,1 it can also positively regulate the mitogen-activated protein kinase (MAPK) pathway in response to insulin and potentiate integrin-induced MAPK activation.8 9 Thus, under certain circumstances SIRPα1 is involved in activating rather than in inhibiting processes.
In contrast to SIRPα molecules, members of the SIRPβ subfamily express only short cytoplasmic domains and lack inhibitory ITIM regions.1 SIRPβ molecules are characterized by a single basic lysine residue within the hydrophobic transmembrane domain, a feature that is reminiscent of a group of activating receptors expressed on natural killer cells. These receptors interact with small intracellular adapter proteins that transduce activating signals via their immunoreceptor tyrosine-based activation motif (ITAM).10,11 Very recently, the DAP12 protein was identified as the ITAM-containing adapter protein for SIRPβ1.12 13
Several reports describe the expression and function of SIRP in neuronal tissues and on myeloid cells.3,5,14-17 Recently, we have identified CD47, an integrin-associated protein (IAP), as an extracellular ligand for human SIRPα1 and SIRPα2.17 In addition, an interaction of IAP with P84 was described in the rat.18 SIRP is involved in adhesive processes, like the extension of neurites on SIRP-coated substrate,3 the fusion of macrophages,5 the binding of SIRP+dendritic cells (DCs) to CD4+ T cells,14 and the attachment of hematopoietic cells to immobilized SIRPα1 protein.17 As a result of cell adhesion, SIRPα1 phosphorylation was observed in macrophages as well as in nonhematopoietic cells.19 20
Because the extracellular domains of SIRPα and SIRPβ molecules are highly homologous, it was plausible that both subfamily members interact with the same extracellular ligand. However, cell attachment and flow cytometric assays revealed that SIRPβ1 does not bind at high affinity to CD47 at detectable levels. Using SIRPβ-specific monoclonal antibodies (MoAbs) we also analyzed differential expression of SIRPα and SIRPβ on hematopoietic cell subsets and show that SIRPα but not SIRPβ1 is expressed on early CD34+CD133+ hematopoietic stem/progenitor cells. Finally, we show the involvement of SIRPα-CD47 interactions in the activation of T cells by DCs.
Materials and methods
Cells
Bone marrow (BM) and peripheral blood (PB) cells from healthy donors and patients with acute or chronic myeloid leukemia (AML or CML) were obtained after informed consent according to the guidelines of the local ethics committee in Tübingen. Buffy coat PB cells from normal volunteers were obtained from the Transfusion Department, Tübingen, Germany, according to institutional guidelines. Mononuclear cells were isolated on a Ficoll-Hypaque density gradient (1.077 g/mL) by collecting the interphase cells. For immunofluorescence labeling of PB lymphocytes, monocytes, and granulocytes, lysing reagent without fixative from Immunotech (Marseilles, France) was used to remove erythrocytes.
The human leukemic cell lines HL60, KG1a, K562, M07e, 207, Daudi, CCRF-CEM, Jurkat, and Molt-4 were obtained from the American Type Culture Collection (Rockville, MD). The murine myeloma cell line, SP2/0, and the human leukemic cell lines EM2, U937, and LAMA-84, were obtained from the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany). All cell lines were grown in RPMI 1640 culture medium (Gibco-BRL, Eggenstein, Germany) supplemented with 10% fetal calf serum (FCS) and antibiotics. Cells were cultured at 37°C and 5% CO2.
The NIH-3T3 cells were transfected with the complete coding sequence of the human SIRPα1 complementary (cDNA) (NIH-3T3/huSIRP1) as described previously.1 The complete DNA sequence coding for SIRPβ1 was transfected into 293E cells by the calcium phosphate method.21 Cells were grown and selected in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FCS and 1 mg/mL G418 (Sigma, Deisenhofen, Germany) for 2 weeks. A single high-expressing 293E/huSIRPβ1 clone was chosen for the screening of MoAb hybridoma supernatants.
The DCs were generated in vitro as described.22 Briefly, monocytes were isolated from mononuclear PB cells by adhesion to culture dishes for 2 hours at 37°C. After removing nonadherent cells, RPMI 1640 medium supplemented with 10% FCS, antibiotics, interleukin (IL)-4 (1000 IU/mL; Genzyme, Cambridge, MA) and granulocyte-macrophage colony-stimulating factor (GM-CSF) (Leukomax, 10 ng/mL; Novartis, Basel, Switzerland) was added. Cell cultures were fed with fresh medium and cytokines every second day, and dendritic cells were collected after 7 days of culture.
Recombinant SIRPα1, SIRPα2, and SIRPβ1 proteins
GST fusion proteins of SIRPα1, SIRPα2, and SIRPβ1 were produced as previously described.17 Primers used for the amplification of SIRPβ extracellular domain were as follows: Forward primer: 5′-GCGGAATTCGCCACCATGCCCGTGCCAGCCTCC-3′, reverse primer: 5′-CCGCTCGAGAGCAGTAGGAGCCAGCGC-3′. Briefly, cDNAs coding for the extracellular domains of SIRPα1, SIRPα 2, and SIRPβ1 were cloned into the pSj26(mod) vector, which was derived from the pCDNA3 cloning vector (Invitrogen, Groningen, The Netherlands). The resultant expression plasmids were transfected into 293E cells by the calcium phosphate method.21 Cells were grown and selected in DMEM supplemented with 10% FCS and 1 mg/mL G418 (Sigma) for 2 weeks. Surviving clones were tested for expression and secretion of fusion protein by Western blotting. High-expressing clones were used to produce SIRPα1ex-, SIRPα2ex-, and SIRPβ1ex-GST fusion proteins, which were purified from cell supernatants with glutathione Sepharose (Pharmacia Biotech, Freiburg, Germany).
Cell adhesion assay
Adhesion of leukemic cell lines and primary hematopoietic cells to SIRPα1ex, SIRPα2ex, and SIRPβ1ex was performed as described previously.23 Briefly, the protein solutions were immobilized onto nitrocellulose-coated plastic dishes (35-mm diameter) by air-drying at room temperature. Nonspecific binding of cells to nitrocellulose was prevented by blocking with 1% bovine serum albumin (BSA) solution in phosphate-buffered saline (PBS). A total of 3 × 106 hematopoietic cells in serum-free RPMI 1640 medium was allowed to adhere to the immobilized protein for 1 hour at 37°C. Nonadherent cells were removed by gently rinsing the dishes with warm PBS. Specific cell binding was evaluated under a Zeiss Axiovert microscope (Carl Zeiss, Göttingen, Germany). Photographs of representative fields were taken.
To inhibit cell adhesion, immobilized SIRP fusion proteins were preincubated with different SIRPα/β-reactive MoAbs for 30 minutes at 37°C, before cell attachment in the presence of the antibodies was performed.
Transient overexpression of SIRPα1 deletion constructs in 293E cells
For the production of the deletion constructs SIRPα1Ig1 and SIRPα1Ig2-3 the following primers were used:
Forward primers: SP6: 5′-CATACGATTTAGGTGACACTATAG-3′; J6: 5′-GGATCCGCCCCCGTGGTATCGGGCCC-3′; J3: 5′-CGCGGATCCGCCCACCCGAAGGAGCAGGGC-3′.
Reverse primers: J1: 5′-CCGATTCGCCGCTCGAGTCACTGCCTCGGGACCTGG-3′; J2: 5′-GGATCCAGCTGCAACTGATACGGAC-3′; J7: 5′-GGATCCCGATACCACGGGGGCAGAGG-3′.
Sequence corresponding to the signal peptide was amplified by polymerase chain reaction (PCR) from the SIRPα1 cDNA in pRK5RS1 using primers SP6 and J2 and cloned into pCDNA3. A single fragment containing the sequence corresponding to Ig-like domains 2 and 3 and remaining downstream coding region were amplified with J6 and J1 and cloned into pCDNA3 behind the signal peptide insert to give SIRPα1Ig2-3. Sequence corresponding to the signal peptide and first Ig-like domain was amplified from SIRPα1 cDNA with primers SP6 and J7 and cloned into pCDNA3. The portion of the cDNA coding for downstream sequence beginning at the transmembrane domain was amplified with primers J6 and J1 and cloned into pCDNA3 behind the Ig1 insert to give SIRPα1Ig1.
The deletion plasmids SIRPα1Ig1 and SIRPα1Ig2-3 were transfected into 293E cells by the calcium phosphate method.21 Cells were harvested after 2 days and immunofluorescence labeling with SIRP-reactive MoAbs was performed.
Immunization and hybridoma production
The MoAbs SE5A5, SE7C2, SE8A3, SE11A6, SE12B6, SE12C3, and P3C4 were generated by immunization with SIRPα1 fusion proteins, as described previously.17 MoAbs B1D5 and B4B6 were raised in a 4- to 8-week-old female Balb/c mouse by immunization with a recombinant GST fusion protein containing the whole extracellular domain of SIRPβ1. Fifty micrograms protein diluted 1:2 in ABM-2 adjuvant solution (Pansystems, Aidenbach, Germany) was applied intramuscularly 3 times in 14-day intervals. The spleen was removed 4 days after the last injection for fusion with the SP2/0 myeloma cell line. The resulting hybridoma cells were grown in RPMI 1640 culture medium containing 10% FCS, antibiotics, and hypoxanthine, aminopterin, and thymidine (HAT; Sigma). Culture supernatants were screened by flow cytometric analysis on 293E/huSIRPβ1 cells, and positive hybridoma cells secreting antibodies selectively recognizing the SIRPβ1 transfectant cell line, but not the parental 293E cells, were cloned by limiting dilution. The selected hybridoma cells were further screened for cross-reactivity with SIRPα1. Two of 13 selected clones exclusively recognized 293E/huSIRPβ1 cells, but not NIH-3T3/huSIRPα1 cells. These 2 clones, B1D5 and B4B6, were cultured in Integra CL1000 culture flasks (Integra Biosciences, Fernwald, Germany) and antibodies were purified from supernatants using protein G Sepharose columns (Pharmacia Biotech). The isotypes of the MoAbs were determined by flow cytometry analysis using phycoerythrin (PE)-conjugated isotype-specific secondary antisera for staining (Southern Biotechnology, Birmingham, AL).
MoAbs were biotinylated by adding 6-((6-((biotinoyl)amino)hexanoyl)amino)hexanoic acid, sulfosuccinimidyl ester, sodium salt (Mobitec, Göttingen, Germany) to the protein solution in 0.1 M sodium bicarbonate buffer, pH 8.3, at a molar ratio of 1:100 (MoAb/biotin). After 2 hours of incubation, unbound biotin was separated from the biotinylated antibodies by gel filtration on a Sephadex G25 column (Pharmacia Biotech).
Immunofluorescence labeling and flow cytometry analysis
Indirect staining of cells.
Cells from growing cell lines or primary mononuclear cells from BM and PB were washed in PBS supplemented with 0.1% BSA and 0.1% sodium azide (FACS buffer). In the next step, cells were incubated with 20% human AB serum for 10 minutes at 4°C to prevent unspecific binding of mouse antibodies. Cells were then incubated with 10 μg/mL of the primary antibody for 30 minutes on ice. After washing 2 times with FACS buffer, cells were stained with PE-conjugated goat antimouse IgG1 or IgG2a antiserum (Southern Biotechnology) for 30 minutes at 4°C. After washing twice, cells were suspended in FACS buffer and analyzed on a FACSCalibur flow cytometer (Becton Dickinson, Heidelberg, Germany).
Two-color staining of cultured DCs and PB and BM cells.
Mononuclear PB and BM cells of healthy donors or patients with AML, as well as cultured DCs, were labeled with MoAb P3C4 or B1D5 (both IgG2a) and fluorescein isothiocyanate (FITC)- or PE-conjugated goat antimouse IgG2a-specific antiserum (Caltag, San Francisco, CA) as described above. In addition, FITC-conjugated antibodies against CD1a (WM35) (Peli Cluster, Amsterdam, The Netherlands), CD14 (MΦP9), and CD45 (2D1) (Becton Dickinson), or PE-conjugated antibodies against CD19, (4G7) CD33 (P67.6), and CD34 (8G12) (Becton Dickinson), as well as the MoAb AC133-PE (Miltenyi Biotec, Bergisch Gladbach, Germany) were used for fluorescence labeling of the cells. The stained cells were analyzed on a FACSCalibur flow cytometer using the Cellquest software (Becton Dickinson).
Four-color staining of BM cells.
To analyze SIRP and CD133 expression on CD34+CD38− BM cells, mononuclear cells from healthy donors were stained with anti–CD34-PerCP (8G12), anti–CD38-APC (HB7; Becton Dickinson), CD133-PE (W6B3; our laboratory; http://gryphon.jr2.ox.ac.uk/cdlistings.htm), and either P3C4 (anti-SIRPα/β), or B1D5 (anti-SIRPβ). In the next step cell-bound anti-SIRP antibodies (both IgG2a) were stained with anti-IgG2a–FITC antiserum. After washing, cells were analyzed on a FACSCalibur flow cytometer.
Four-color staining of PB cells.
To detect primary DC subsets in peripheral blood, 4-color analysis was performed on total blood as previously reported.24Briefly, blood cells were stained with anti-ILT1 antibody,24 followed by FITC-labeled multiple adsorbed goat antirat antibody (Pharmingen, San Diego, CA). After washing, cells were labeled with PCy5-conjugated anti-CD3 (UCHT1), -CD14 (RMO52), -CD16 (3G8), -CD19 (B9E6), and -CD56 MoAb (N901) (Immunotech), PE-conjugated anti-ILT3 MoAb (ZM3.8) (Immunotech), as well as with biotinylated MoAbs SE5A5 and B1D5. In the final step, streptavidin-allophycocyanin (Molecular Probes, Eugene, OR) was added. Red blood cells were lysed with FACS Lysing Solution (Becton Dickinson) and analyzed on a FACSCalibur flow cytometer.
Immunoprecipitation and Western blot analysis
To determine the specificity of our SIRP-reactive MoAbs, 1 μg SIRPα1ex or SIRPβ1ex protein was incubated with 5 μg antibody for 1 hour at 4°C. Immunoprecipitation was performed overnight at 4°C using 100 μL protein A Sepharose solution (Sigma) for each MoAb. The antibody-Sepharose complexes were washed 6 times with Tris-buffered saline (TBS: 10 mmol/L TrisHCl, pH 7.5, 100 mmol/L NaCl), and bound proteins were eluted with reducing Laemmli sample buffer.25 Western blot analysis was performed as described previously.17 Briefly, eluted proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose. After blocking the membrane with 3% BSA in TTBS (TBS + 0.1% Tween 20), a polyclonal SIRP-reactive antiserum1 was used as primary antibody, followed by incubation with alkaline phosphatase-conjugated goat antirabbit antiserum (Sigma). Detection of immunoprecipitated SIRP proteins was performed with BCIP/NBT (5-bromo-4-chloro-3-indolylphosphate p-toluidine salt/nitro blue tetrazolium chloride) Sigma Fast Tablets.
Mixed lymphocyte reaction assay
Responding cells (1.5 × 105) from allogeneic PB mononuclear cells were cultured in flat-bottomed 96-well microplates with 5 × 102 DCs. Control and inhibitory antibodies (anti-CD47, anti-SIRPα/β, anti-SIRPβ) were added to the cultures at concentrations of 100 μg/mL. Soluble SIRPα1ex and SIRPβ1ex proteins were used at 25 μg/mL. Thymidine incorporation was measured on day 5 by a 16-hour pulse with 3H-thymidine (0.5 μCi/well; Amersham Life Science, Buckingham, United Kingdom).
Induction of antigen-specific cytotoxic T lymphocyte response using HLA-A2 restricted synthetic peptides
The HLA-A2 binding peptides E75 (Her-2/neu, KIGSFLAFL, used for cytotoxic T lymphocyte (CTL) inductions) and the MUC1 peptide M1.1 (amino acids 950-958: STAPPVHNV, control peptide) were synthesized using standard Fmoc chemistry on a peptide synthesizer (432A, Applied Biosystems, Weiterstadt, Germany) and analyzed by reverse-phase high-performance liquid chromatography (HPLC) and mass spectrometry. For CTL induction 5 × 105 DCs were pulsed with 50 μg/mL of the synthetic Her-2/neu-peptide E75 for 2 hours, washed, and incubated with 2.5 × 106 autologous PB mononuclear cells in RPMI medium supplemented with 10% FCS. Control and inhibitory antibodies (anti-CD47 and anti-SIRPα/β) were added to the cultures at concentrations of 100 μg/mL. Soluble SIRPα1ex, SIRPβ1ex, and control GST proteins were used at 25 μg/mL. Cells were restimulated after 7 days of culture and 1 ng/mL human recombinant IL-2 (Genzyme) was added every second day.26 The cytolytic activity of induced CTL was analyzed on day 5 after the last restimulation in a standard 51Cr-release assay.
CTL assay
The standard 51Cr-release assay was performed with some modifications as described.26 Target cells (Croft cells, an Epstein-Barr virus [EBV] immortalized B-cell line, kindly provided by O. J. Finn, University of Pittsburgh, Pittsburgh, PA) were pulsed with 25 μg/mL peptide (E75 as the cognate peptide and M1.1 as an irrelevant control peptide) for 2 hours and labeled with [51Cr]-sodium chromate in RP10 medium for 1 hour at 37°C. Cells (104) were transferred to a well of a round-bottomed 96-well plate. Varying numbers of CTL were added to give a final volume of 200 μL and incubated for 4 hours at 37°C. At the end of the assay supernatants (50 μL/well) were harvested and counted in a microbeta counter. The percent specific lysis was calculated as: 100 × (experimental release − spontaneous release/maximal release − spontaneous release). Spontaneous and maximal release were determined in the presence of either medium or 1% Triton X-100, respectively.
Statistical analysis
To determine the statistical significance of the results,t tests were performed.
Results
CD47+ hematopoietic cells adhere to SIRPα1ex and SIRPα2ex, but not to SIRPβ1ex
We have previously shown that hematopoietic cells adhere to the extracellular domains of SIRPα1 and SIRPα2 via the transmembrane IAP CD47.17 To test whether SIRPβ molecules show the same adhesive capacity, a GST fusion protein containing the whole extracellular domain of SIRPβ1 was generated. Cell attachment assays with this protein revealed however, that the CD47brightcell line Jurkat (Figure 1) and all the other tested CD47+ hematopoietic cell lines do not bind SIRPβ1ex. This result was confirmed by FACS analysis with biotinylated SIRPβ1ex protein, followed by streptavidin-PE staining. Whereas biotinylated SIRPα1ex and SIRPα2ex stained all tested CD47+ hematopoietic cell lines, no binding of biotinylated SIRPβ1ex was observed (data not shown). Because the extracellular Ig-like loops of SIRPα1, SIRPα2, and SIRPβ1 are highly homologous, single amino acid residues within these regions seem to be critical for CD47 binding. The fact that none of the tested CD47+ hematopoietic cell lines bind to SIRPβ1ex supports the hypothesis that SIRPβ molecules do either not interact with CD47 or only at very low affinity below the detection level of our assays.
Cell adhesion to SIRPα1ex and SIRPα2ex, but not to SIRPβ1ex.
SIRP fusion proteins (2 μL of 20 μg/mL solutions) were immobilized on nitrocellulose-coated Petri dishes by air-drying. Adhesion of Jurkat cells to SIRPα1ex, SIRPα2ex, and SIRPβ1ex was analyzed by incubating the cells for 1 hour at 37°C, as described.17Specific cell binding is shown in the round protein drop and was evaluated and photographed using a Zeiss Axiovert microscope (original magnification × 40; n = 3). SIRPα adhered to all tested CD47+ cell lines (HL-60, KG1a, K562, MO7e, 207, Daudi, CCRF-CEM, Molt-4, EM2, U937, and LAMA-84); adhesion of SIRPβ was always negative.
Cell adhesion to SIRPα1ex and SIRPα2ex, but not to SIRPβ1ex.
SIRP fusion proteins (2 μL of 20 μg/mL solutions) were immobilized on nitrocellulose-coated Petri dishes by air-drying. Adhesion of Jurkat cells to SIRPα1ex, SIRPα2ex, and SIRPβ1ex was analyzed by incubating the cells for 1 hour at 37°C, as described.17Specific cell binding is shown in the round protein drop and was evaluated and photographed using a Zeiss Axiovert microscope (original magnification × 40; n = 3). SIRPα adhered to all tested CD47+ cell lines (HL-60, KG1a, K562, MO7e, 207, Daudi, CCRF-CEM, Molt-4, EM2, U937, and LAMA-84); adhesion of SIRPβ was always negative.
SIRPα molecules interact with CD47 via their N-terminal immunoglobulinlike domain
To identify functional epitopes within the extracellular SIRPα domains, responsible for the interaction with CD47, deletion constructs containing either the N-terminal Ig-like domain of SIRPα1 (SIRPα1Ig1) or the second and third Ig-like loop (SIRPα1Ig2-3), respectively, were transiently overexpressed in 293E cells. Transfected cells were used for immunofluorescence analysis with 7 previously described SIRPα/β-reactive MoAbs.17 Five of these antibodies (SE5A5, SE7C2, SE11A6, SE12C3, and P3C4) recognized only 293E cells transfected with the SIRPα1Ig1 construct, whereas the remaining 2 MoAbs (SE8A3 and SE12B6) exclusively bound to the SIRPα1Ig2-3 transfectants (summarized in Table1). In previous studies we could show that 3 of the SIRPα1Ig1-specific antibodies blocked cell adhesion to immobilized SIRPα1ex protein. Two of these antibodies also inhibited cell binding to SIRPα2ex (SE5A5 and SE12C3), whereas MoAb SE7C2 exclusively inhibited adhesion to SIRPα1ex. Interestingly, the 2 SIRPα1Ig2-3–specific MoAbs SE8A3 and SE12B6 selectively inhibited the CD47-SIRPα2 interaction, whereas binding of SIRPα1ex to CD47 remained unaltered. Thus, most likely SIRPα1 exclusively utilizes the N-terminal Ig-like loop for binding to CD47, whereas SIRPα2 additionally requires the second and/or third Ig-like domains.
Generation of SIRPβ-specific MoAbs
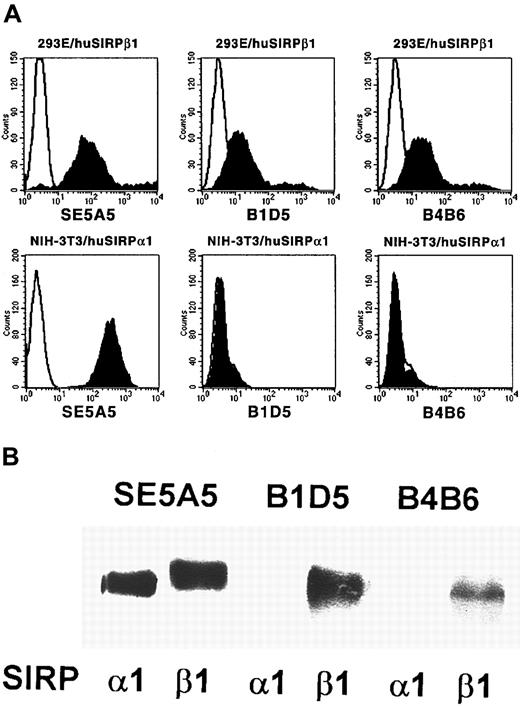
To study cell surface expression of human SIRPα and SIRPβ molecules, SIRPβ-specific MoAbs were raised by immunization of a Balb/c mouse with the recombinant GST fusion protein SIRPβ1ex. Two antibodies, B1D5 and B4B6, were selected because of their specific reactivity with 293E cells transfected with SIRPβ1, but not with NIH-3T3/huSIRPα1 transfectants, as determined by flow cytometric analysis (Figure 2A). The specificity of these antibodies was further confirmed by immunoprecipitation experiments followed by Western blotting with a SIRP-reactive polyclonal antiserum. Whereas all of the previously described SIRP-reactive MoAbs precipitated SIRPα1ex and SIRPβ1ex (Figure 2B; SE5A5 is shown as an example), the 2 SIRPβ-specific antibodies B1D5 and B4B6 exclusively immunoprecipitated SIRPβ1 protein.
Specificity of MoAbs B1D5 and B4B6.
(A) 293E/huSIRPβ1 and NIH/3T3/huSIRPα1 cells were immunolabeled with MoAbs SE5A5, B1D5, and B4B6 and stained with PE-conjugated goat antimouse IgG1 or IgG2a antiserum (filled histograms). Nonbinding IgG1 and IgG2a antibodies were used as negative controls (black lines). Cells were analyzed on a FACSCalibur flow cytometer (n = 3). (B) Recombinant SIRPα1ex and SIRPβ1ex protein was immunoprecipitated with MoAbs SE5A5, B1D5, and B4B6. Precipitated protein was separated by 12% SDS-PAGE and immunoblotted with a polyclonal antibody against SIRP (n = 3).
Specificity of MoAbs B1D5 and B4B6.
(A) 293E/huSIRPβ1 and NIH/3T3/huSIRPα1 cells were immunolabeled with MoAbs SE5A5, B1D5, and B4B6 and stained with PE-conjugated goat antimouse IgG1 or IgG2a antiserum (filled histograms). Nonbinding IgG1 and IgG2a antibodies were used as negative controls (black lines). Cells were analyzed on a FACSCalibur flow cytometer (n = 3). (B) Recombinant SIRPα1ex and SIRPβ1ex protein was immunoprecipitated with MoAbs SE5A5, B1D5, and B4B6. Precipitated protein was separated by 12% SDS-PAGE and immunoblotted with a polyclonal antibody against SIRP (n = 3).
SIRPα and SIRPβ are differentially expressed on PB and BM mononuclear cells
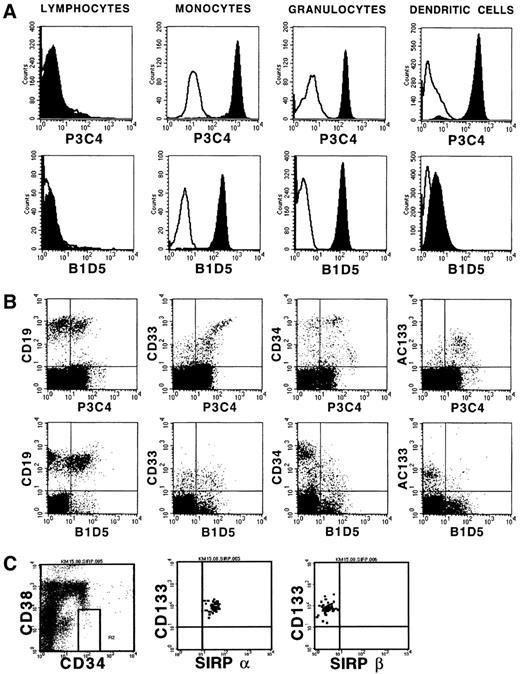
Previously, we showed differential SIRP expression on hematopoietic cells using MoAbs recognizing both SIRPα and SIRPβ molecules.17 In this study we extended the analysis and differentiated between SIRPα and SIRPβ expression using SIRPβ-specific antibodies. Figure 3A shows that PB cells show a very similar expression pattern of both SIRP subfamily members, with a strong expression on monocytes and granulocytes, and almost no expression on lymphocytes. In contrast, the SIRPαbright myeloid dendritic cells generated in vitro show only a very weak SIRPβ signal. Two-color analysis of BM cells with SIRPα/β- and SIRPβ-specific MoAbs shows coexpression of both SIRP subfamily members on a subset of CD19+ B-cell precursors and on CD33+ myeloid progenitor cells (Figure3B). However, immature CD34++ and CD133+hematopoietic cells almost exclusively express inhibitory SIRPα molecules. A similar selective expression of SIRPα was also observed on immature CD34+CD38−CD133+hematopoietic progenitor cells (Figure 3C). This indicates that SIRPα but not SIRPβ may play an important role in the regulation of early hematopoiesis.
Expression of SIRPα and SIRPβ on hematopoietic cells.
(A) PB cells and in vitro–generated DCs were immunolabeled with MoAbs P3C4 (IgG2a) or B1D5 (IgG2a) and PE-conjugated goat antimouse IgG2a-specific antiserum (filled histograms). Nonbinding IgG2a antibody was used as negative control (black line). Cultured DCs were further labeled with FITC-conjugated CD1a-specific MoAb. Gated lymphocytes, monocytes, and granulocytes, as well as CD1a+ DCs were analyzed on a FACSCalibur flow cytometer (n = 3). (B) Ficoll- isolated BM cells were labeled with MoAbs P3C4 or B1D5 and stained with FITC-conjugated goat antimouse IgG2a-specific antiserum, as well as with PE-conjugated MoAbs against CD19, CD33, CD34, and AC133 antigen. Gated mononuclear cells were analyzed on a FACSCalibur flow cytometer (n = 3). (C) Expression of SIRP on CD34+CD38− BM cells. Cells were stained with CD34-FITC, CD38-APC, CD133-PE, and either P3C4 (SIRPα) or B1D5 (SIRPβ) plus anti-IgG2a-FITC, and analyzed on a FACSCalibur flow cytometer. The plots show coexpression of CD133 and SIRP on gated CD34+CD38− cells.
Expression of SIRPα and SIRPβ on hematopoietic cells.
(A) PB cells and in vitro–generated DCs were immunolabeled with MoAbs P3C4 (IgG2a) or B1D5 (IgG2a) and PE-conjugated goat antimouse IgG2a-specific antiserum (filled histograms). Nonbinding IgG2a antibody was used as negative control (black line). Cultured DCs were further labeled with FITC-conjugated CD1a-specific MoAb. Gated lymphocytes, monocytes, and granulocytes, as well as CD1a+ DCs were analyzed on a FACSCalibur flow cytometer (n = 3). (B) Ficoll- isolated BM cells were labeled with MoAbs P3C4 or B1D5 and stained with FITC-conjugated goat antimouse IgG2a-specific antiserum, as well as with PE-conjugated MoAbs against CD19, CD33, CD34, and AC133 antigen. Gated mononuclear cells were analyzed on a FACSCalibur flow cytometer (n = 3). (C) Expression of SIRP on CD34+CD38− BM cells. Cells were stained with CD34-FITC, CD38-APC, CD133-PE, and either P3C4 (SIRPα) or B1D5 (SIRPβ) plus anti-IgG2a-FITC, and analyzed on a FACSCalibur flow cytometer. The plots show coexpression of CD133 and SIRP on gated CD34+CD38− cells.
SIRPα and SIRPβ expression is reduced on primary leukemic blasts
Compared to myeloid cells from normal PB and BM, the majority of myeloid leukemic blasts are either negative for SIRPα/β or express it at highly reduced levels.17 Using the new SIRPβ-specific MoAbs, a weak SIRPβ expression was observed on only 2 of 10 analyzed acute myeloid leukemia samples (Figure4). Both SIRPβ+ probes were from patients with AML of the FAB (French-American-British) classification type M4/M5, and were also strongly positive for SIRPα. All the other analyzed AML samples of FAB types M0-M3 were negative for SIRPβ. These cells showed also a negative or reduced SIRPα expression. Together these data show, that not only the expression of inhibitory SIRPα molecules,21 but also of activating SIRPβ proteins is reduced on myeloid leukemic blasts.
SIRPα and SIRPβ expression on AML blasts.
Ficoll-isolated PB or BM cells from patients with AML were immunolabeled with MoAb P3C4 or B1D5, followed by PE-conjugated goat antimouse IgG2a-specific antiserum (filled histograms). Nonbinding IgG2a antibody was used as negative control (black line). Cells were further stained with FITC-conjugated CD45-specific MoAb and gated CD45low leukemic blasts were analyzed. AML samples were divided into 3 groups: (1) normal SIRPα/β expression; (2) reduced SIRPα, no SIRPβ expression; (3) no SIRPα/β expression. The histograms show representative examples of each group. The number of analyzed samples is given in parentheses.
SIRPα and SIRPβ expression on AML blasts.
Ficoll-isolated PB or BM cells from patients with AML were immunolabeled with MoAb P3C4 or B1D5, followed by PE-conjugated goat antimouse IgG2a-specific antiserum (filled histograms). Nonbinding IgG2a antibody was used as negative control (black line). Cells were further stained with FITC-conjugated CD45-specific MoAb and gated CD45low leukemic blasts were analyzed. AML samples were divided into 3 groups: (1) normal SIRPα/β expression; (2) reduced SIRPα, no SIRPβ expression; (3) no SIRPα/β expression. The histograms show representative examples of each group. The number of analyzed samples is given in parentheses.
Differential expression of SIRPα and SIRPβ on primary PB DCs
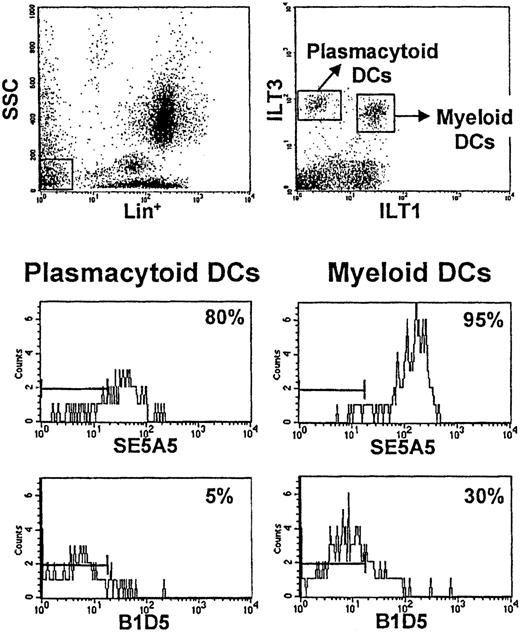
To determine whether primary DCs express SIRPα and/or SIRPβ, we stained lineage-negative PB cells with anti-ILT3 and anti-ILT1 antibodies, which allow us to identify plasmacytoid DCs (lin−/ILT1−/ILT3+) and myeloid DCs (lin−/ILT1+/ILT3+) (Figure5).24 SIRPα was expressed on both DC subsets, myeloid DCs having a higher level of expression. SIRPβ was expressed on a very small fraction of plasmacytoid DCs (∼5%-10%), and on a significant percentage of myeloid DCs, varying from 30% to 70% in different individuals. The strong expression of SIRPα on primary and cultured myeloid DCs indicated a possible role for this surface receptor in the generation of immune responses.
Differential expression of SIRPα and SIRPβ on distinct subsets of primary DCs.
Cells from whole blood were stained with anti-ILT1 antibody followed by FITC-labeled goat antirat antibody. After washing, cells were labeled with PCy5-conjugated anti-CD3, -CD14, -CD16, -CD19, and -CD56 MoAb; PE-conjugated anti-ILT3 MoAb; and biotinylated MoAbs SE5A5 and B1D5 or a biotinylated control antibody. In the final step, streptavidin-allophycocyanin was added. Primary DCs in human PB are included within lineage (lin)−/low side-scatter cells (top left). Differential expression of ILT1 and ILT3 receptors on these cells allows the identification of 2 distinct subsets of primary DCs (top right). ILT3+ILT1− cells correspond to plasmacytoid dendritic cells. ILT3+ILT1+ cells correspond to myeloid DCs. SIRPα (analyzed with MoAb SE5A5) is expressed on both subsets and is particularly bright on ILT3+ILT1+ DCs. SIRPβ (analyzed with MoAb B1D5) is expressed on a significant percentage of ILT3+ILT1+ DCs (30%-70% in different individuals) and only on few ILT3+ILT1− DCs (5%-10%). Bars on the left of the histograms indicate the region in which 99% of the control cells stained with isotype-matched control antibodies were found.
Differential expression of SIRPα and SIRPβ on distinct subsets of primary DCs.
Cells from whole blood were stained with anti-ILT1 antibody followed by FITC-labeled goat antirat antibody. After washing, cells were labeled with PCy5-conjugated anti-CD3, -CD14, -CD16, -CD19, and -CD56 MoAb; PE-conjugated anti-ILT3 MoAb; and biotinylated MoAbs SE5A5 and B1D5 or a biotinylated control antibody. In the final step, streptavidin-allophycocyanin was added. Primary DCs in human PB are included within lineage (lin)−/low side-scatter cells (top left). Differential expression of ILT1 and ILT3 receptors on these cells allows the identification of 2 distinct subsets of primary DCs (top right). ILT3+ILT1− cells correspond to plasmacytoid dendritic cells. ILT3+ILT1+ cells correspond to myeloid DCs. SIRPα (analyzed with MoAb SE5A5) is expressed on both subsets and is particularly bright on ILT3+ILT1+ DCs. SIRPβ (analyzed with MoAb B1D5) is expressed on a significant percentage of ILT3+ILT1+ DCs (30%-70% in different individuals) and only on few ILT3+ILT1− DCs (5%-10%). Bars on the left of the histograms indicate the region in which 99% of the control cells stained with isotype-matched control antibodies were found.
SIRPα is involved in DC-mediated T-cell activation
To determine whether SIRPα/CD47 interactions play a role in mixed lymphocyte reactions, we analyzed the influence of inhibitory SIRPα/β- and CD47-reactive MoAbs on the capacity of cultured DCs to stimulate alloreactive T cells. Both, SIRPα/β-reactive (P = .0119) and CD47-reactive antibodies (P = .0035) significantly reduced T-cell proliferation (Figure 6). Soluble SIRPα1ex protein at a concentration of 25 μg/mL reduced T-cell proliferation to less than 50% (P = .001), whereas anti-SIRPβ antibody B1D5 (P = .9653) and the soluble SIRPβ1ex protein (P = .4828) had no effect on T-cell proliferation in the mixed lymphocyte reaction (MLR). These data show SIRPα and CD47 are involved in T-cell activation induced by DC.
MLR in the presence of SIRPα/β- or CD47-specific MoAbs or soluble SIRPα1ex.
Responding cells (1.5 × 105) from allogeneic PB mononuclear cells were cultured with stimulator cells (DCs) in the presence of control (IgG1) antibody, antibody B1D5 specific for SIRPβ inhibitory antibodies specific for CD47 (CC2C6) and SIRPα/β (SE5A5), or soluble SIRPα1ex and SIRPβ1ex protein. Thymidine incorporation was measured on day 5 by a 16-hour pulse with3H-thymidine. The results show the means and SDs from a representative experiment in cpm (n = 6 replicates).
MLR in the presence of SIRPα/β- or CD47-specific MoAbs or soluble SIRPα1ex.
Responding cells (1.5 × 105) from allogeneic PB mononuclear cells were cultured with stimulator cells (DCs) in the presence of control (IgG1) antibody, antibody B1D5 specific for SIRPβ inhibitory antibodies specific for CD47 (CC2C6) and SIRPα/β (SE5A5), or soluble SIRPα1ex and SIRPβ1ex protein. Thymidine incorporation was measured on day 5 by a 16-hour pulse with3H-thymidine. The results show the means and SDs from a representative experiment in cpm (n = 6 replicates).
To further analyze the influence of soluble SIRPα1ex and SIRPβ1ex proteins as well as inhibitory CD47-reactive and SIRPα/β-reactive antibodies on the ability of DCs to induce a primary CTL response against a tumor-associated antigen, DCs were pulsed with the synthetic E75 peptide derived from the HER-2/neu antigen and used as antigen-presenting cell (APC) for CTL induction in vitro. As demonstrated in Figure 7 the in vitro generated CTL efficiently lysed Croft cells pulsed with the cognate E75 peptide but not target cells presenting an irrelevant MUC1 peptide. The addition of an inhibitory antibody against CD47 (Figure 7G) or SIRPα/β (Figure 7E) to the cell cultures during the in vitro priming reduced the cytotoxic activity of the induced T cells. The presence of the soluble SIRPα1ex protein (Figure 7F) almost completely abolished the induction of antigen-specific CTL. In line with the results obtained from the MLR soluble SIRPβ1ex protein and mAb specific for SIRPβ had no effect on the interaction between T cells and DC.
HER-2/neu-specific CTL responses induced with peptide-pulsed DCs.
In vitro generated DCs were pulsed with the E75 synthetic peptide derived from Her-2/neu and used as APC to induce a primary MHC class I restricted CTL response in vitro. Control antibody (A), SIRPβ-specific MoAb B1D5 (C), and the inhibitory MoAbs against SIRPα/β (SE5A5) (E) and CD47 (G), as well as soluble proteins (control, B), soluble SIRPβ1ex (D), SIRPα1ex protein (F) were added to the cultures during the in vitro priming. Cytotoxic activity of induced CTL was determined after 2 restimulations in a standard51Cr-release assay using Croft cells pulsed with the cognate E75 peptide (▪) or irrelevant M1.1 peptide (■) as targets.
HER-2/neu-specific CTL responses induced with peptide-pulsed DCs.
In vitro generated DCs were pulsed with the E75 synthetic peptide derived from Her-2/neu and used as APC to induce a primary MHC class I restricted CTL response in vitro. Control antibody (A), SIRPβ-specific MoAb B1D5 (C), and the inhibitory MoAbs against SIRPα/β (SE5A5) (E) and CD47 (G), as well as soluble proteins (control, B), soluble SIRPβ1ex (D), SIRPα1ex protein (F) were added to the cultures during the in vitro priming. Cytotoxic activity of induced CTL was determined after 2 restimulations in a standard51Cr-release assay using Croft cells pulsed with the cognate E75 peptide (▪) or irrelevant M1.1 peptide (■) as targets.
Discussion
Signal-regulatory proteins were initially described as negative regulators of receptor tyrosine kinase-coupled signaling pathways.1 More detailed analysis revealed, however, that SIRPs can be classified into 2 groups: SIRPα proteins that contain ITIMs and SIRPβ proteins that lack these inhibitory motifs.1 Very recently, SIRPβ1 was identified as an activating receptor, which associates with the small transmembrane adapter protein DAP12 and transduces stimulatory signals.12,13 Thus, SIRPαand SIRPβ molecules represent another pair of inhibitory and activating receptors. Such pairs are also described for killer cell Ig-like receptors (KIRs),27,28 Ig-like transcripts (ILTs),29,30or paired Ig-like receptors (PIRs).31,32 The extracellular regions of each of these receptor pairs are highly homologous. Therefore the existence of a common ligand for both SIRPα and SIRPβ was likely. Unexpectedly, we could show that the extracellular domain of the activating receptor SIRPβ1 does either not, or only at very low affinity, interact with the previously described SIRPα ligand CD47, although a high sequence similarity between the extracellular domains of SIRPα and SIRPβ molecules has been described.1 Further studies with more sensitive methods are in progress to test whether SIRPβ binds at very low affinity to CD47, or whether additional cofactors/coligands are required for CD47 binding. In line with the observation of differential binding of SIRPs to CD47 is the fact that activating KIRs do not bind to HLA class I molecules, which are ligands for inhibitory KIRs.33 Further, the inhibitory CD94/NKG2A receptor has a higher binding affinity for HLA-E than the activating CD49/NKG2C receptor,34 although the extracellular domains of KIRs and NKG2 activating and inhibitory isoforms contain only subtle variations in their amino acid sequence. Hence, the distinct ligand recognition properties observed must depend on these single sequence variations.
Deletion constructs containing only defined regions of the extracellular SIRPα1 domain were generated to localize the epitopes responsible for the SIRPα-CD47 interaction. Interestingly, we found that the ligand binding site of SIRPα1 is localized within the N-terminal Ig-like loop, whereas the binding of SIRPα2 to CD47 additionally requires the second and/or third Ig-like domain. This suggests that different members of the SIRPα group use different epitopes to interact with CD47. For a more precise localization of the SIRPα epitopes responsible for binding to CD47, the preparation of constructs with a few amino acids deletions will be crucial.
Because SIRPα and SIRPβ molecules have opposing functional activity and distinct ligand binding properties, their differential tissue distribution may be a central issue to the understanding of their biological role. Thus, it was of particular interest to know whether both subfamily members are expressed in the same cell or whether certain cell types exclusively express SIRPα or SIRPβ. We could previously show that SIRPα/β is expressed on hematopoietic stem and/or progenitor cells, monocytes, granulocytes, and DCs. Because all antibodies used in the previous study recognize both SIRPα and SIRPβ, expression analysis of subfamily members was not possible. In the present study we have described 2 novel MoAbs which selectively bind to the extracellular domain of SIRPβ1, but not to SIRPα1 or SIRPα2. This enabled us to specifically analyze SIRPβ expression and to detect cells that exclusively express SIRPα but not SIRPβ by subtracting the specificities of SIRPβ-reactive from SIRPα/β-reactive antibodies. Flow cytometric analysis with these MoAbs revealed that monocytes and granulocytes, as well as their myeloid precursors, react with both SIRPα/β- and SIRPβ-specific antibodies showing that these cells express at least SIRPβ. Recent data additionally showed that these cells also express SIRPα, as revealed by immunoprecipitation followed by Western blotting with SIRP-reactive antibodies.12 Whether SIRPα and SIRPβ molecules mediate opposing functions in the same cell, or whether they control separate signal transduction pathways in a coordinated manner, has to be determined.
Interestingly, most of immature CD34+ and CD133+ and 100% of CD34+CD38−CD133+ hematopoietic stem/progenitor cells exclusively express SIRPα but not SIRPβ. Possibly, the inhibitory receptor SIRPα isoforms regulate proliferation during early hematopoiesis. After the onset of myeloid differentiation the cells acquire SIRPβ, and activating signals may be balanced with inhibitory signals. To date, no cell types were identified which exclusively express SIRPβ.
In previous studies we could show that SIRP expression is significantly reduced or absent on the majority of myeloid cells from different leukemias.17 Therefore, we speculated that this reduction of inhibitory SIRPα expression was either the cause or a consequence for the deregulated proliferation of these cells. The present data with SIRPβ-specific MoAbs showed that many AML cells express SIRPα but not SIRPβ. Only myeloid leukemic blasts of the M4/M5 (myelo/monocytic) FAB type, which show a strong expression of SIRPα, were also positive for SIRPβ. Thus, the absence of SIRPβ on leukemic blasts from patients with M0/M1- type AML is most probably not caused by a malignant transformation but may rather reflect the fact that these blasts express phenotypes similar to normal hematopoietic stem cells which are negative also for SIRPβ.
Although SIRPβ is up-regulated during myeloid differentiation to monocytes and granulocytes, it is down-regulated during further differentiation into myeloid dendritic cells. DCs derived from monocytes cultured in GM-CSF and IL-4 down-regulated cell surface expression of SIRPβ, although some variability was observed in different DC preparations. This is also in line with our data from primary peripheral blood DCs. Whereas SIRPα was expressed on all primary DCs, SIRPβ was found only on a variable fraction of DCs. Several reports describe the existence of at least 2 DC subsets in the blood.24,35-37 Both subsets are positive for MHC class II expression and negative for lineage-specific antigens (eg, CD3, CD14, CD16, CD19). One molecule that distinguishes the 2 DC subsets is the β2-integrin chain, CD11c. The CD11c− subset is functionally immature, requiring monocyte-derived cytokines to develop into typical DCs.35 This cell population corresponds to the recently described plasmacytoid DC of lymphoid tissue,38,39 which is mainly found in inflamed lymph nodes in and around the high endothelial venules.24 These DCs, which are also termed lymphoid DCs, based on the lack of several myeloid markers, express CD62L and CXCR3, and produce extremely large amounts of type I interferon.24,40 In contrast to lymphoid DCs, CD11c+ DCs express the activation antigen CD45RO.35 These myeloid DCs are mainly localized in the dark and light zones of germinal centers and are similar to many tissue DCs.41 Very recently, Cella and colleagues demonstrated that both DC subsets express the Ig-like transcript receptor (ILT) 3, but only CD11c+ DCs also coexpress the stimulatory receptor ILT1.24 This observation enabled us to distinguish primary DC populations from PB monocytes and lymphocytes in flow cytometry analysis. Staining of primary DC subsets with SIRPα/β- and SIRPβ-specific MoAbs revealed that SIRPα is expressed on both subsets and that SIRPβ is predominantly expressed on a variable fraction of myeloid DCs. Because both of these DC types represent mature APC with potent T-cell stimulating activity, we speculated that SIRPα, but not SIRPβ, might play an important role during this immunologic process. On the other hand, the expression of SIRPβ on a variable percentage of myeloid DCs suggests that such expression may be tightly regulated and induced or down-regulated by yet unknown stimuli.
To investigate the role of SIRP and CD47 during T-cell activation by dendritic cells, mixed lymphocyte reactions in the presence of SIRPα/β -, SIRPβ-, and CD47-specific MoAbs, as well as of soluble fusion proteins consisting of the extracellular domains of SIRPα1 and SIRPβ1 were performed. We observed a significant reduction in T-cell proliferation in the presence of inhibitory SIRPα/β- or CD47-specific MoAbs and the soluble SIRPα1 protein. As expected, soluble SIRPβ1ex or antibody specific for SIRPβ did not show any effect in this assay. In line with these results, CD47- and SIRPα/β-specific inhibitory antibodies reduced the induction of peptide-specific CTL by DCs in vitro. The addition of the soluble SIRPα1ex to the cultures during CTL priming almost completely abolished the induction of a CTL response by peptide pulsed DCs. These results suggest that the interaction between SIRPα on antigen-presenting DCs and CD47 on SIRPα− T cells is essential for effective stimulation of a T-cell response. The reason for this could be a costimulatory signal that T cells may receive by this receptor-ligand interaction. Our observations are in line with a report of Brooke and coworkers, who showed that the proliferation of resting memory T cells in response to ovalbumin-pulsed monocytes in cattle was significantly reduced in the presence of a MoAb against SIRPα.14 In addition, a costimulatory role in T-cell activation was also described for CD47.42,43 Because the binding of soluble SIRPα1ex fusion protein to CD47 on lymphocytes is not sufficient for an effective T-cell activation, one can further postulate that contact between SIRPα+ DCs and CD47+ T cells is required. We could previously show that the interaction of SIRPα with CD47 mediates cellular adhesion.17 In addition, several other reports describe an involvement of SIRPα or CD47 in adhesion-dependent processes.3,5,14,17,44,45 Moreover, Reinhold and colleagues observed that the costimulatory signal transduced by CD47 in T cells is adhesion dependent.42 Therefore, one possible role for SIRPα and CD47 during T-cell activation by DCs could be a primary induction of cell-cell adhesion, as already described for other adhesion receptors including integrins.46-48 This adhesion-enhanced cell-cell contact may then enable other molecules to transduce stimulatory or costimulatory signals into the cell.
The authors thank Yvonne Hoffmann, Stefanie Kurtz, and Sylvia Stefan for excellent assistance in DC cultures and T-cell assays, and Heike Letzkus for her help in the generation of SIRP-reactive MoAbs. We would also like to thank Dr C. Faul for the well-organized supply of bone marrow cells.
Supported by a grant from the German Josè Carreras Leukemia Foundation (DJCLS-R20) and by the Deutsche Forschungsgemeinschaft (SFB510, project A1).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hans-Jörg Bühring, Medizinische Klinik, II, Otfried-Müller-Strasse 10, 72076 Tübingen, Germany; e-mail: hans-joerg.buehring@med.uni-tuebingen.de.