Abstract
Clonality assessment through Southern blot (SB) analysis ofTCRB genes or polymerase chain reaction (PCR) analysis ofTCRG genes is important for diagnosing suspect mature T-cell proliferations. Clonality assessment through reverse transcription (RT)–PCR analysis of Vβ-Cβ transcripts and flow cytometry with a Vβ antibody panel covering more than 65% of Vβ domains was validated using 28 SB-defined clonal T-cell receptor (TCR)αβ+ T-ALL samples and T-cell lines. Next, the diagnostic applicability of the Vβ RT-PCR and flow cytometric clonality assays was studied in 47 mature T-cell proliferations. Clonal Vβ-Cβ RT-PCR products were detected in all 47 samples, whereas single Vβ domain usage was found in 31 (66%) of 47 patients. The suspect leukemic cell populations in the other 16 patients showed a complete lack of Vβ monoclonal antibody reactivity that was confirmed by molecular data showing the usage of Vβ gene segments not covered by the applied Vβ monoclonal antibodies. Nevertheless, this could be considered indirect evidence for the “clonal” character of these cells. Remarkably, RT-PCR revealed an oligoclonal pattern in addition to dominant Vβ-Cβ products and single Vβ domain expression in many T-LGL proliferations, providing further evidence for the hypothesis raised earlier that T-LGL derive from polyclonal and oligoclonal proliferations of antigen-activated cytotoxic T cells. It is concluded that molecular Vβ analysis serves to assess clonality in suspect T-cell proliferations. However, the faster and cheaper Vβ antibody studies can be used as a powerful screening method for the detection of single Vβ domain expression, followed by molecular studies in patients with more than 20% single Vβ domain expression or large suspect T-cell populations (more than 50%-60%) without Vβ reactivity.
Introduction
In striking contrast to the straightforward clonality assessment in most mature B-cell proliferations through single immunoglobulin light chain expression, clonality assessment in suspect T-cell proliferation is not possible through routine immunologic marker analysis. For instance, the T-cell receptor (TCR) expression pattern (TCRαβ or TCRγδ), the unusual CD4/CD8 expression pattern, or the so-called loss of T-cell markers is insufficient to establish the malignant (clonal) character of a suspect T-cell proliferation.1 As a consequence, molecular analysis of TCR gene rearrangements has developed as an important tool for clonality assessment in such proliferations. As with normal T cells in various differentiation stages, TCR genes are rearranged in most T-cell malignancies.2 The presence or absence of identically (clonally) rearranged TCR genes is therefore helpful for distinguishing between (mono)clonal and polyclonal (reactive) T-cell proliferations, respectively.
Southern blot (SB) analysis is the classical diagnostic method for clonality assessment. It is highly reliable and in principle can detect every clonal TCR gene rearrangement, provided that optimally positioned probes and appropriate restriction enzymes are used.3-6 Nevertheless, several drawbacks limit the routine application of SB analysis in diagnostic laboratories. SB is labor intensive and time consuming, especially when sequential hybridizations are required; furthermore, relatively large amounts of high-quality DNA are needed for reliable results, which precludes its application on paraffin-embedded samples. Despite these disadvantages, SB-based detection of clonal TCR rearrangements is the gold standard technique by which to validate other methods for clonality assessment in suspect T-cell proliferations. Today, polymerase chain reaction (PCR) analysis of TCR gamma (TCRG) genes is most widely applied; the relatively restricted combinatorial repertoire of TCRG genes limits the number of required PCR primers. However, this limited repertoire also results in high background amplification of similar rearrangements in normal T cells, thereby reducing the sensitivity of the assay. Other approaches focus on analysis of TCRB genes or TCRB gene products (RNA, proteins, or both), exploiting the high diversity in Vβ gene usage for clonality studies. PCR-based analysis of the Vβ repertoire mostly concerns reverse transcription (RT)–PCR amplification of Vβ-Cβ transcripts to limit the number of primers needed to cover the many involved gene segments.7With the recent availability of a panel of Vβ antibodies that covers more than 65% of all Vβ domains,8 flow cytometric analysis of the Vβ repertoire now promises to be a relevant alternative approach for molecular clonality studies.9,10This is especially true because reference values have been determined in healthy controls for the available individual Vβ antibodies.10
Here we present 2 different approaches of Vβ repertoire analysis: Vβ-Cβ RT-PCR and flow cytometry with Vβ antibodies. After validation on SB-defined clonal TCRαβ+ T-cell acute lymphoblastic leukemia (T-ALL) samples and a series of T-cell lines, the diagnostic applicability of the 2 methods was tested in a series of 47 mature T-cell proliferations. The feasibility and applications of both Vβ clonality assays are discussed.
Patients, materials, and methods
Patients, cell samples, and cell lines
Peripheral blood (PB) samples were collected from patients (n = 47) showing persistent mature TCRαβ+ T-cell proliferation, including T-cell chronic lymphocytic leukemia (T-CLL), T-cell large granular lymphocyte leukemia (T-LGL), T-cell non-Hodgkin lymphoma (T-NHL), T-cell prolymphocytic leukemia (T-PLL), and mycosis fungoides/Sézary syndrome (MF/SS). Mononuclear cells (MNCs) were isolated by Ficoll-Hypaque (density, 1.077 g/mL; Pharmacia, Uppsala, Sweden) density centrifugation and used for immunophenotyping and Vβ analysis and for DNA and RNA isolation.
TCRαβ+ or cytoplasmic TCRβ+(CyTCRβ+) T-cell lines (n = 12) and MNC fractions of PB or bone marrow (BM) samples from patients with TCRαβ+T-ALL (n = 16) were used as reference samples.
Immunophenotyping and analysis of Vβ domains of expressed TCRβ chains
MNCs were analyzed for cell membrane expression of T-cell markers and for expression of the HLA-DR antigen (L243), cytoplasmic TCRβ (βF1), TCRαβ (BMA031) and TCRγδ (11F2) with the following monoclonal antibodies (mAbs): CD2 (Leu-5b), CD3 (Leu-4), CD4 (Leu-3a), CD5 (Leu-1), CD7 (3A1), CD8 (Leu-2a), CD16 (Leu-11c), CD56 (Leu-19), and CD57 (Leu-7). The mAbs of the Leu series, as well as L243 and 11F2, were obtained from Becton Dickinson (San Jose, CA), the 3A1 hybridoma was obtained from ATCC (Rockville, MD), βF1 from T-cell Sciences (Needham, MA), and BMA031 from Immunotech (Marseilles, France). Immunofluorescence stainings were performed as described and evaluated on a FACScan flow cytometer using CellQuest software (Becton Dickinson).10
The samples were studied in more detail for Vβ domain expression using a panel of carefully selected Vβ mAbs (Table1).10 Data acquisition and analysis of the Vβ immunofluorescence stainings in T-ALL samples and T-cell lines were performed in double stainings using unconjugated Vβ mAbs, followed by incubation with fluorescein isothiocyanate (FITC)-conjugated goat antimouse immunoglobulin (CLB, Amsterdam, The Netherlands) and subsequently normal mouse serum to block free antigen binding sites of the second-step reagent, after which CD3-PE (Leu-4) was added.10 In T-cell lines with cytoplasmic TCRβ expression (βF1 reactivity), cells were permeabilized before Vβ staining. For the mature TCRαβ+ T-cell proliferations, the Vβ mAbs were used in combination with CD3-PerCP (Leu-4) and occasionally CD4-PE (Leu-3a) or CD8-PE (Leu-2a) to further define the immunophenotype of the T cells.10 The Vβ antibodies were from Immunotech (Marseilles, France), T-cell Sciences (Needham, MA), and T-cell Diagnostics (Cambridge, MA) (Table 1 for the origin of the Vβ antibodies). In addition, several samples were studied by means of 6 mixtures of clustered Vβ antibodies as a fast and easy method for the identification of single Vβ domain expression, as described by Van den Beemd et al.10 These 6 mixtures consisted of: MIX1, Vβ3 (CH92) and Vβ5.1 (IMMU157); MIX2, Vβ8.1/8.2 (56C5.2) Vβ12.2 (VER2.32), and Vβ17 (E17.5F3); MIX3, Vβ2 (MPB2/D5), Vβ16 (TAMAYA1.2), and Vβ23 (HUT78.7/AF23); MIX4, Vβ6.1 (CRI304.3), Vβ7.1 (Zoé), Vβ9.1 (FIN9), Vβ11.1 (C21), and Vβ14 (CAS1.1.3); MIX5, Vβ13.6 (JU74.3), Vβ18 (BA62.6), Vβ20 (ELL1.4), Vβ21.3 (IG125), and Vβ22 (IMMU546); and MIX6, Vβ5.2/5.3 (4H11), Vβ6.7 (OT145), and Vβ13.1/13.3 (BAM13).10
DNA and RNA isolation and cDNA synthesis
DNA was isolated from frozen MNCs and cell lines as described earlier.3 Total RNA was isolated from all samples using RNAzol (Tel-Test, Friendswood, TX). After oligo dT annealing for 3 minutes at 85°C, 2 μg total RNA was subsequently reverse transcribed in 40 μL volumes for 1 hour at 41°C using Superscript II RT enzyme (Life Technologies, Paisley, United Kingdom) in the presence of dNTPs and RNAguard (Amersham Pharmacia Biotech, Uppsala, Sweden).
Southern blot analysis
Southern blot (SB) analysis of the TCRB genes was performed as described.3 The 32P-labeled TCRBJ1, TCRBJ2, and TCRBC genomic DNA probes (DAKO, Carpinteria, CA) were used in subsequent hybridizations of EcoRI- andHindIII-digested DNA to determine the rearrangement status of the T-cell lines and patient samples.5
PCR amplification
Oligonucleotide primers used for amplification of Vβ-Cβ transcripts are given in Table 2. Most Vβ family-specific primers were adapted from those published by Gorski et al,7 but several primers were added to maximize recognition of Vβ gene segments within a given family and to minimize cross-annealing to other Vβ gene families at the 3′ primer ends. The Cβ primer was also adapted from Gorski et al.7 The quality of the studied cDNA samples was determined through RT-PCR analysis of the ubiquitously expressed ABL gene. PCR amplification of the TCRB genes of the cDNA samples was performed in multiple tubes (n = 31), each containing one of the Vβ family primers and the Cβ primer (Table 2). Reactions were performed in 20 μL volumes, containing one fortieth (1 μL) of the cDNA reaction mixture, 2.5 pmol Vβ family primer, 2.5 pmol Cβ primer, 0.2 mM dNTPs, 1.5 mM MgCl2, and 0.2 U AmpliTaqDNA polymerase in reaction buffer II (Applied Biosystems, Foster City, CA). PCR reaction conditions for the Perkin-Elmer 480 thermal cycler (Applied Biosystems) were initial denaturation of 3 minutes at 94°C, followed by 35 to 40 cycles of 1 minute at 94°C, 1 minute at 60°C, 2 minutes at 72°C, and a final extension step of 10 minutes at 72°C.
Heteroduplex analysis
After amplification, half (10 μL) the PCR mixtures were loaded on 1% agarose gels to evaluate PCR product formation with the various Vβ-Cβ primer combinations. PCR products were visualized by ethidium bromide using UV light. The other 10 μL PCR reaction mixture was subjected to heteroduplex analysis to discriminate between monoclonal and polyclonal PCR products.11 In short, heteroduplex analysis consisted of 5′ denaturation at 94°C immediately followed by 60′ renaturation at 4°C before electrophoresis on 6% nondenaturing polyacrylamide gels (polyacrylamide:bisacrylamide 29:1) in 0.5× TBE buffer.11Ethidium bromide-stained homoduplex or heteroduplex PCR products were visualized with UV light.
Sequencing
If heteroduplex analysis showed PCR products to be clonal, the homoduplexes were excised from the gel and eluted as described.12 Eluted products were either directly sequenced or reamplified in a second-step PCR reaction using the same primers as in the initial reaction. Sequencing was performed on the ABI377 fluorescence sequencer (Applied Biosystems) using the dye terminator cycle sequencing kit and AmpliTaq FS (Applied Biosystems).12 Assignment of Vβ, Dβ, Jβ, and Cβ gene segments and reading frames of the involved TCRB gene rearrangements was performed as described.13 14
Results
Molecular and flow cytometric Vβ analysis in T-cell lines
To study the feasibility of Vβ analysis in clonal T-cell populations, we used a panel of 12 T-cell lines (Table3). Although only 4 of these cell lines showed TCRαβ membrane expression, all had cytoplasmic expression of TCRβ chains. SB analysis revealed clonal TCRB gene rearrangements in all cell lines. In 6 of the T-cell lines, 2 clear Vβ-Cβ RT-PCR products were found, and in the other 6 only one was found (Table 3). The Vβ-Cβ products of the various cell lines contained V gene segments from many distinct Vβ families, with a slight predominance of Vβ2-Cβ products (n = 3). Remarkably, in most identified transcripts, Jβ2 region gene segments were used, which fits with the predominance of Vβ-Jβ2 gene rearrangements in immature T-cell malignancies.5 Further sequencing of the clonal products revealed that all except one of the cell lines showed a single in-frame Vβ-Cβ RT-PCR product (Table 3). In cell line KT-1, both an in-frame Vβ18-Cβ and an in-frame Vβ15-Cβ transcript were found, whereas in the other 5 cell lines with 2 clonal Vβ-Cβ products only one appeared in-frame.
Flow cytometric Vβ analysis using a panel of Vβ family antibodies (Table 1) showed reactivity with one of the Vβ mAbs from the panel in 10 of the 12 cell lines, with a specificity profile that completely correlated with the presence of the identified in-frame Vβ-Cβ transcript (Table 3). Cell line KT-1, having in-frame Vβ15-Cβ and Vβ18-Cβ transcripts, reacted with the Vβ18 antibody. However, double (membrane or cytoplasmic) Vβ expression could not be confirmed or excluded in this cell line because Vβ15-specific antibodies were not available. In 2 cell lines, no reactivity was observed with any of the Vβ mAbs of the panel, despite cytoplasmic TCRβ chain (βF1 mAb) detection. In the RPMI 8402 cell line, this could easily be explained by the lack of a Vβ24-specific antibody, but for the DND-41 cell line the appropriate Vβ18 antibody was present in the panel. The fact that DND-41 expresses a βδ TCR rather than an αβ TCR might, however, hamper proper detection of the recognized epitope of the Vβ18 domain expressed in this cell line.
Molecular and flow cytometric Vβ analysis in T-ALL samples
In addition to the 12 T-cell lines, we studied 16 TCRαβ+ T-ALL for their Vβ expression profile using molecular techniques and flow cytometric analysis (Table 3). Vβ-Cβ RT-PCR heteroduplex analysis revealed a single clonal in-frame Vβ-Cβ transcript in 6 of the 16 T-ALL samples. The remaining 10 samples had at least 2 clonal Vβ-Cβ transcripts; in 2 cases, even 3 and 4 clonal Vβ-Cβ transcripts were present, suggestive of subclone formation with a minor clone (less than 10%) not detectable by SB (Table 3) but readily detectable by RT-PCR. In contrast to the low frequency of bi-allelic in-frame Vβ-Cβ products in T-cell lines, 6 of 10 T-ALL samples were found to contain double in-frame Vβ-Cβ transcripts (Table 3). The identified Vβ-Cβ products in the T-ALL represented Vβ segments from many Vβ families, with a predominance of Vβ3-Cβ products (n = 5). Gene segments of the Jβ1 region were identified in 17 Vβ-Cβ products, whereas in 12 products, Jβ2 segments could be found.
Reactivity with one of the Vβ mAbs was seen in 8 of the 16 T-ALL samples. In all 8 samples, the Vβ antibody reactivity pattern was completely in line with the results of the molecular Vβ analysis, confirming the single (in-frame) transcript in most of these 8 samples. Although samples T077 and T140 with 2 in-frame Vβ-Cβ transcripts might have shown double Vβ protein expression, this did not occur, as evidenced by the single Vβ8 (T077) and Vβ3 (T140) membrane expression on these cells. Potential double Vβ expression in a few other cases (T012, T044, T145) remained undetected because of the lack of the relevant Vβ mAbs for both in-frame alleles in the panel. In 8 T-ALL samples, no reactivity of the entire T-ALL cell population with any of the Vβ antibodies from the panel was seen, despite the clear presence of a TCRαβ molecule on the membrane. In virtually all instances, this nonreactivity can most probably be attributed to expression of the Vβ-Cβ product for which no Vβ mAb is available (eg, Vβ4 in T015 or Vβ24 in T012) or might be caused by a family member not covered by the available Vβ family antibody (eg, members of the Vβ5, Vβ6, and Vβ7 families). The precise members of each Vβ gene family could not be recognized in our study because the position of our Vβ primers is close to the 3′ end of the segments, thereby resulting in PCR products that do not include sufficient sequence information for Vβ member identification. Sample T139 with in-frame Vβ3-Cβ and Vβ11-Cβ products is remarkable in that the cells should have been recognized with either the Vβ3 or the Vβ11 mAb, both of which belong to a single-member gene family. However, for reasons that remain unclear, this did not occur. One possible explanation might be that the actual epitope in either the Vβ3 or the Vβ11 chain was modified or influenced by the junctional region or by the involved Jβ segment.
Vβ repertoire analysis for clonality assessment in mature T-cell proliferations
Although the results of the Vβ analysis in T-cell lines and T-ALL samples showed that the clonal cell population could easily be identified by Vβ mAb reactivity or by lack of reactivity with any of the Vβ mAbs in the panel, the relevant diagnostic application of Vβ gene analysis in daily practice concerns the analysis of postthymic mature lymphoproliferations of the T-cell lineage. For this reason we chose to study the applicability of Vβ repertoire analysis in a large series of 47 mature TCRαβ T-cell proliferations (including T-NHL, T-CLL, T-PLL, and T-LGL) that, in the past decade, had been sent to our laboratory with a strong suspicion of clonality (Table 4). All samples were indeed found to contain clonal TCRB rearrangements in SB analysis. Next, the samples were subjected to Vβ-Cβ RT-PCR, followed by heteroduplex analysis to confirm or exclude the clonal character of the obtained RT-PCR products. This revealed the presence of dominant clonal Vβ-Cβ transcripts in all 42 samples from which RNA could be isolated. In 26 cases only a single transcript was detectable, whereas in 14 cases double Vβ-Cβ products were seen (Table 4). Remarkably, in 2 samples (93-067 and 96-019), 3 dominant clonal products were apparent. SB analysis of case 96-019 already suggested more than 2 rearranged alleles, but in case 93-067 this was not anticipated from the SB pattern because no additional bands were apparent. Further sequencing in the latter case revealed that the 3 PCR products contained distinct junctional regions, excluding cross-annealing of primers as an explanation for the occurrence of multiple bands.
To compare the molecular and flow cytometric Vβ results in these T-cell proliferations, we sequenced the Vβ-Cβ RT-PCR products. Unfortunately, sequence analysis could not be performed in the 5 samples from which RNA was lacking. In-frame Vβ-Cβ transcripts were indeed identified in all 26 evaluable cases that showed only a single RT-PCR product (Table 4). Remarkably, in most (at least 9 of 16) sequenced cases with multiple Vβ-Cβ products, more than one (distinct) in-frame Vβ-Cβ transcript was found.
Flow cytometric analysis of the Vβ repertoire was performed in all 47 cases. Most of the studied samples (n = 31) appeared to have a restricted Vβ reactivity pattern, with predominance of a single Vβ mAb reactivity (exemplified in Figure 1). Although this restricted reactivity concerned many different Vβ specificities, Vβ2 (n = 5) and Vβ3 (n = 4) were observed more frequently than others. The latter may not be too surprising given the relatively high frequency of, especially, Vβ2+ TCRαβ cells in healthy controls (Table 1 contains median values).10 In the other 16 cases, the complete lack of reactivity of the suspect leukemic cell population with any of the individual Vβ mAbs or the 6 Vβ mixtures of the panel10was considered indirect evidence for the “clonal” character of these cells (Figure 2).
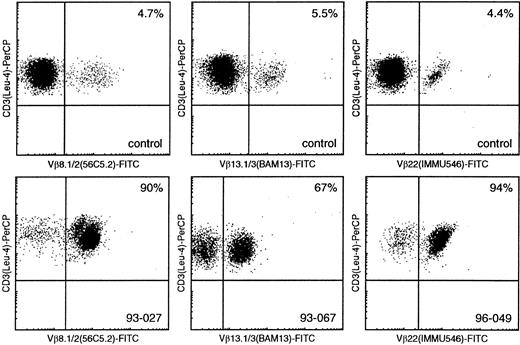
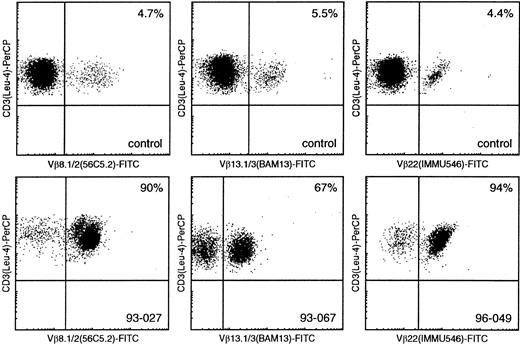
Flow cytometric Vβ analysis in patients with mature T-cell proliferations compared with healthy controls.
Using Vβ8.1/8.2, Vβ13.1/13.3, and Vβ22 (FITC-labeled) double immunofluorescence stainings with CD3-PerCP, only small percentages of CD3+/Vβ+ cells can be identified in healthy controls (upper panel), whereas similar double stainings in samples 93-027, 93-067, and 96-049 enable identification of large T-cell populations with single Vβ8.1/8.2, Vβ13.1/13.3, and Vβ22 expression, respectively (lower panel).
Flow cytometric Vβ analysis in patients with mature T-cell proliferations compared with healthy controls.
Using Vβ8.1/8.2, Vβ13.1/13.3, and Vβ22 (FITC-labeled) double immunofluorescence stainings with CD3-PerCP, only small percentages of CD3+/Vβ+ cells can be identified in healthy controls (upper panel), whereas similar double stainings in samples 93-027, 93-067, and 96-049 enable identification of large T-cell populations with single Vβ8.1/8.2, Vβ13.1/13.3, and Vβ22 expression, respectively (lower panel).
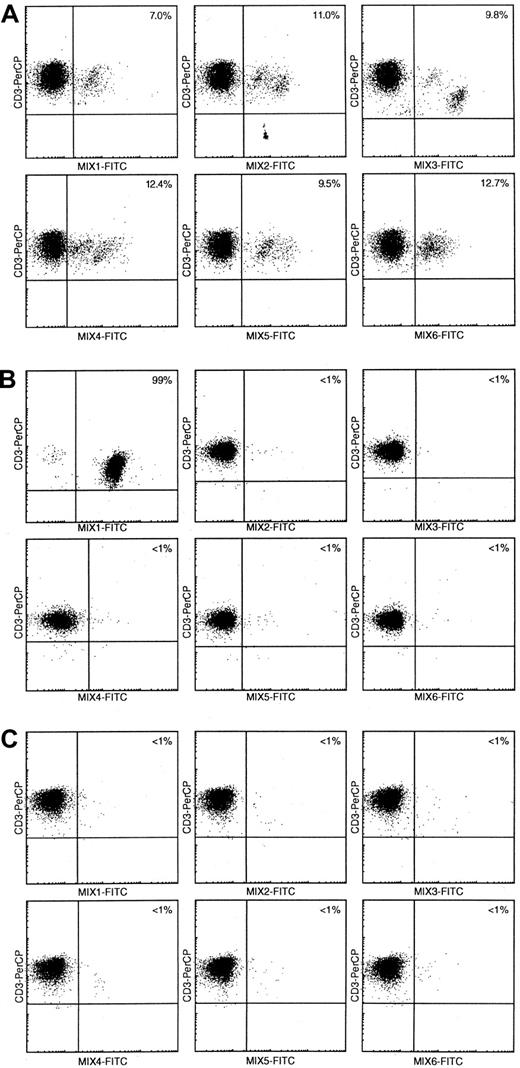
Flow cytometric Vβ repertoire analysis using Vβ mAb mixtures in mature T-cell proliferation patients.
(A) In healthy controls, 60% to 65% of CD3+ cells are recognized in double immunofluorescence stainings with 6 different Vβ mAbs mixtures in combination with CD3-PerCP.10 (B) Using comparable stainings in T-CLL patient 98-086, 99% of CD3+ cells are recognized by Vβ mAbs mix 1, whereas all other 5 mixes only recognize less than 1% of all CD3+cells. This indicates the presence of a large, presumably clonal CD3+ T-cell population with single Vβ expression. (C) Double immunofluorescence stainings with the 6 Vβ mAbs mixtures resulted in less than 5% CD3+/Vβ+ cells in T-CLL patient 98-002, suggesting the presence of a large, presumably clonal CD3+ T-cell population with single Vβ expression not recognized by any of the Vβ mAbs in the current panel.
Flow cytometric Vβ repertoire analysis using Vβ mAb mixtures in mature T-cell proliferation patients.
(A) In healthy controls, 60% to 65% of CD3+ cells are recognized in double immunofluorescence stainings with 6 different Vβ mAbs mixtures in combination with CD3-PerCP.10 (B) Using comparable stainings in T-CLL patient 98-086, 99% of CD3+ cells are recognized by Vβ mAbs mix 1, whereas all other 5 mixes only recognize less than 1% of all CD3+cells. This indicates the presence of a large, presumably clonal CD3+ T-cell population with single Vβ expression. (C) Double immunofluorescence stainings with the 6 Vβ mAbs mixtures resulted in less than 5% CD3+/Vβ+ cells in T-CLL patient 98-002, suggesting the presence of a large, presumably clonal CD3+ T-cell population with single Vβ expression not recognized by any of the Vβ mAbs in the current panel.
Comparison of the data from molecular and flow cytometric analyses revealed complete concordance between the identified in-frame Vβ-Cβ transcript and the expressed Vβ domain of the TCRβ chain in 26 cases (Table 4). Remarkably, in case 95-121, we identified Vβ6.7- and Vβ13.1/13.3-positive cells, which is in line with the presence of the corresponding in-frame Vβ-Cβ transcripts in this sample and the presence of 2 clones as found by SB analysis and immunophenotyping. In addition, in sample 91-030, 2 populations (Vβ5.2/5.3 and Vβ22) were found, but here no clear evidence was detected for further immunophenotypical heterogeneity or for clonal Vβ22-Cβ transcripts, suggesting that the Vβ22-positive population concerned either a small subclone or a polyclonal Vβ22+ cell population. The latter is more likely because a monoclonal subclone of 10% Vβ22+ T-cells should have been identified in PCR heteroduplex analysis; in contrast, only polyclonal (heteroduplex) Vβ22-Cβ PCR products were observed in sample 91-030.
In 16 cases no single Vβ reactivity of the clonal cell population was observed, despite the presence of clonal Vβ-Cβ RT-PCR products in 15 of these samples (one was not studied through RT-PCR). Close examination revealed that in all 15 cases, the clonal transcripts contained gene segments derived from multimember Vβ families (Vβ5, Vβ6, Vβ7, and Vβ13) known to be incompletely covered by the respective mAbs in the current panel. The position of the various primers for these Vβ families of gene segments, however, did not allow for a detailed analysis into the exact gene member that was used. Nevertheless, even in these cases without Vβ mAb reactivity, the molecular and flow cytometric results were not discordant.
Vβ oligoclonality in mature T-cell proliferations
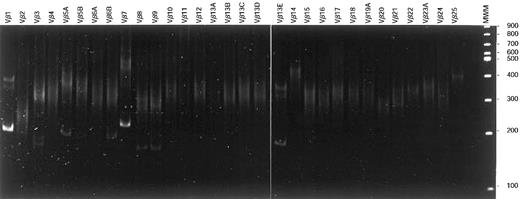
Although in all analyzed mature T-cell proliferations 1 or 2 dominant Vβ-Cβ RT-PCR products could be identified, in several samples a whole array of additional Vβ-Cβ products of variable, but mostly weak, intensity were found, next to the dominant clonal Vβ-Cβ product(s), as exemplified in Figure3. Close examination of these oligoclonal samples disclosed that virtually all were diagnosed as T-LGL leukemias, whereas only a few concerned patients with T-CLL or Sézary syndrome. Despite their oligoclonal character, in all cases a dominant clonal cell population was observed, as evidenced by the flow cytometric data.
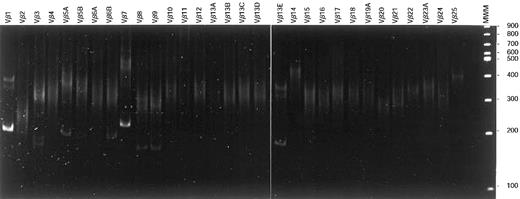
Vβ-Cβ RT-PCR heteroduplex analysis.
After reverse transcription of total RNA of T-LGL patient 86-041, cDNA was PCR amplified using Vβ family primers in combination with a Cβ primer. On heteroduplex analysis, 2 major clonal products (Vβ1-Cβ and Vβ7-Cβ) belonging to the dominant clone were observed in addition to several weaker clonal Vβ-Cβ products. These latter products represented rearranged alleles of small subclones that were not identified with flow cytometric Vβ analysis.
Vβ-Cβ RT-PCR heteroduplex analysis.
After reverse transcription of total RNA of T-LGL patient 86-041, cDNA was PCR amplified using Vβ family primers in combination with a Cβ primer. On heteroduplex analysis, 2 major clonal products (Vβ1-Cβ and Vβ7-Cβ) belonging to the dominant clone were observed in addition to several weaker clonal Vβ-Cβ products. These latter products represented rearranged alleles of small subclones that were not identified with flow cytometric Vβ analysis.
Discussion
Vβ repertoire analysis for identification of clonal cell populations
Clonality assessment of mature T-cell proliferations has long been performed by means of SB analysis. Although SB analysis is highly reliable owing to an almost complete lack of false-positive and false-negative results, it is time consuming and labor intensive, and it requires relatively large amounts of high-quality DNA. The latter is generally unavailable in paraffin-embedded tissues or small tissue biopsies. For this reason, PCR analysis of TCRG gene rearrangements has been used as an alternative strategy.15-17 However, the relatively restricted diversity of TCRG gene rearrangements and the resultant high-background amplification of similar rearrangements in normal T cells limits the potential of this approach. Because theTCRB recombination diversity is essentially larger than that of TCRG genes, analysis of the Vβ repertoire ofTCRB genes has been put forward as a diagnostic strategy for clonality studies in suspect T-cell proliferations. This approach can also be used to study the actual TCRB repertoire in other disease states with high T-cell activity, such as autoimmune diseases,18-22 immunodeficiencies,23-25 and alloreactivity in patients who have undergone transplantation.26-28 Until recently, this type of analysis mainly concerned PCR-based assays.18,21,22,26,29,30 However, a much faster and more quantitative analysis of the TCRβ repertoire is now possible through the use of mAbs directed against the Vβ domains of TCRαβ molecules, which cover 65% to 70% of Vβ domains in blood T lymphocytes of children and adults.10 31-34
We performed parallel molecular and flow cytometric Vβ analyses in a series of SB-defined TCRαβ T-ALL samples and T-cell lines to validate both approaches for the detection of clonality. Indeed, clonal Vβ-Cβ transcripts could easily be detected in all T-ALL samples and T-cell lines. Moreover, the corresponding Vβ mAb reactivity patterns also indicated the presence of large, single Vβ domain (“clonal”) cell populations in these cases, indicating that both methods are suitable for detecting aberrant T-cell proliferations. However, patients with T-ALL generally do not have diagnostic dilemmas for which Vβ analysis is required because thymus derived T-ALL is relatively easy to diagnose using mAbs against the nuclear enzyme TdT and various CD antigens. Given that the main diagnostic problems generally concern suspect mature (postthymic) T-cell proliferations, we studied a series of 47 such mature T-cell proliferations, proven to be clonal based on SB analysis. RT-PCR heteroduplex analysis of Vβ-Cβ transcripts in all 42 analyzed cases confirmed the (mono)clonal character of the studied cell samples. Flow cytometric Vβ repertoire analysis of the suspect mature T-cell proliferations identified single Vβ domain expression in 31 (66%) of 47 cases. Together with the single Vβ reactivity in 10 of 12 T-cell lines and in 8 of 16 T-ALL samples, this means that 49 (65%) of 75 cases were picked up through flow cytometric Vβ repertoire analysis. In 16 of the 47 cases showing extensive T-cell proliferation, the combined Vβ antibody reactivity covered less than 20% of all CD3+ T cells (instead of approximately 65%), which is indirect proof for the presence of a large T-cell population with single, though unidentified, Vβ domain expression (Table 4).
Flow cytometric Vβ repertoire analysis can thus be used as a (quantitative) screening method for the detection of large, aberrant T-cell populations with single Vβ domain expression. To define single Vβ expression, the mean normal Vβ values should be used, also taking into account the differences in the use of particular Vβ domains between CD4+ and CD8+ T-cell populations and observations of a more restricted Vβ usage of especially CD8 T lymphocytes in the elderly.10,35-39 Taken together, this means that we consider T-cell expansions aberrant if they concern more than the mean Vβ value plus 3 SD (generally more than 20%) of the peripheral blood T cells or if the suspect population exceeds 2.0 × 109/L. The arbitrary cut-off of 2.0 × 109/L is considered an important criterion for diagnosing T-cell LGL leukemia,40 though even lower absolute counts might sometimes occur. Use of a 6-tube test kit with Vβ mAbs mixtures, each covering maximally 10% to 15% of the Vβ repertoire (Figure 2), further enables the screening for Vβ repertoire restrictions.10 Meanwhile, an attractive commercial 8-tube kit (IO Test Beta Mark) has become available (Immunotech/Beckman Coulter, Marseilles, France); in each of the 8 tubes, 3 distinctly labeled Vβ antibodies are present that not only allow detection of single Vβ expression but also direct identification of the involved Vβ domain or family.
The results in this study suggest that detection of a T-cell population with restricted Vβ usage in principle implies clonality of the involved proliferation. The fact that detection of single Vβ expression is not necessarily equal to clonality is illustrated by, for example, case 91-030 showing a small Vβ22+ population of approximately 10% that was not found to be clonal in molecular analysis. Although this expanded Vβ22+ population was smaller than 20%, we think that in doubtful cases it generally remains important to confirm the presumed clonality by amplification of the involved Vβ-Cβ product. In case of large Vβ unreactive T-cell populations, the clonal character should be established by PCR using a complete set of Vβ family primers or by other molecular approaches, such as PCR analysis of TCRG genes or SB analysis ofTCRB genes. The detection limits of both Southern blot analysis (5%-10%) and PCR-based assays (1%-5%) are superior over flow cytometric analysis of the Vβ repertoire, which has a sensitivity of 20% for single Vβ domain expression because of the background of normal Vβ usage. Nevertheless, this sensitivity of the latter method might be improved if additional immunophenotypic markers are included to detect Vβ usage in combination with a specific T-cell phenotype such as CD4, CD8, CD56, and CD57.1
Finally, a major advantage of flow cytometric Vβ analysis over PCR-based assays is that once established and confirmed, the Vβ-restricted T-cell population can easily and quantitatively be monitored in combination with other markers during and after therapy, using age-dependent reference values for comparison.10 41
Oligoclonality of T-LGL proliferations
During analysis of the mature T-cell proliferations, we observed that especially in many T-LGL samples, multiple weak additional products were seen next to 1 or 2 Vβ-Cβ transcripts belonging to the immunophenotypically dominant clone. Limited sequencing of these clonal products did not show similar Vβ gene segments or junctional region sequences. This observation provides further evidence for the hypothesis raised earlier, which is that T-LGL derive from polyclonal or oligoclonal proliferations of antigen-activated cytotoxic T-cells and that, in some situations, transformation or dysregulation of growth or apoptosis results in T-LGL leukemia showing a more restricted and dominant Vβ usage (expressed as percentage MNCs) and a raised absolute cell count of abnormal cells.42-45 Furthermore, the generally indolent course of this type of T-cell proliferation and the relatively lower white blood cell counts (compared to T-CLL and T-PLL proliferations) are also in line with a pretransformation state of oligoclonal proliferations of activated T-cells.
Monoreactivity in T-ALL and mature T-cell populations
Remarkably, in many T-ALL (6 of 10) and mature T-cell proliferations (at least 9 of 16) with bi-allelic TCRB gene rearrangements and, to a lesser extent, in T-cell lines (1 of 6 cases), double in-frame transcripts were observed. In theory this could lead to double Vβ expression in particular samples. However, in virtually all cases that could be evaluated, the clonal T-cell population reacted with only one of the Vβ mAbs; monoreactivity cannot be proven formally in a few samples because of the lack of appropriate Vβ antibodies in the panel. In case 95-121, the 2 identified in-frame Vβ-Cβ transcripts appeared to be derived from 2 distinct T-cell populations, given the results of flow cytometric analysis. Taken together, the data strongly suggest that in cases with double in-frame Vβ-Cβ products, monospecificity of the TCR is guaranteed by regulation at the level of translation of TCRβ chains or by preferential pairing of one TCRβ chain with the involved TCRα chain.
We conclude from our data that flow cytometric Vβ repertoire analysis is a fast and relatively cheap alternative tool that can be used as a powerful screening method in patients with suspect T-cell proliferations. Formally, however, clonality as deduced from single Vβ domain expression (more than 20% of cells) or from the lack of reactivity of a large (more than 50% to 60%) TCRαβ+T-cell population would still need proof by molecular assays. Nevertheless, flow cytometric analysis has the additional advantages of looking within T-cell subsets and precise quantitation of the Vβ+ cells. An additional interesting application is the flow cytometric monitoring of Vβ+ leukemic cells during and after therapy, once a single Vβ expression has been determined.
We thank Prof dr R. Benner for support and Ms J. Boon for secretarial assistance. We also thank the following clinicians and scientists for submitting samples from patients with suspect mature T-cell proliferations: H. J. Adriaansen, P. B. Berendes, J. W. Gratama, E. Harthoorn-Lasthuizen, C. van der Heul, J. van Helden, J. C. Kluin-Nelemans, P. J. Lugtenburg, C. Lynas, L. Marcelis, E. Moreau, P. Sonneveld, W. Slieker, J. W. Smit, H. Storm, P. Vandenberghe, M. B. van ‘t Veer, and G. Verhoef.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Anton W. Langerak, Department of Immunology, University Hospital Rotterdam, Erasmus University Rotterdam, 3000 DR Rotterdam, The Netherlands; e-mail:langerak@immu.fgg.eur.nl.