Abstract
Nonhuman primates are useful large animal model systems for the in vivo study of hematopoietic stem cell biology. To better understand the degree of similarity of the hematopoietic systems between humans and baboons, and to explore the relevance of such studies in nonhuman primates to humans, this study was designed to compare the global gene expression profile of bone marrow CD34+ cells isolated from these 2 species. Human complementary DNA (cDNA) filter arrays containing 25 920 human cDNAs were surveyed for this purpose. The expression pattern and relative gene abundance of the 2 RNA sources were similar, with a correlation coefficient of 0.87. A total of 15 970 of these cDNAs were expressed in human CD34+ cells, of which the majority (96%) varied less than 3-fold in their relative level of expression between human and baboon. Reverse transcriptase–polymerase chain reaction analysis of selected genes confirmed that expression was comparable between the 2 species. No species-restricted transcripts have been identified, further reinforcing the high degree of similarity between the 2 populations. A subset of 1554 cDNAs, which are expressed at levels 100-fold and greater than background, is described, which includes 959 expressed sequence tags and uncharacterized cDNAs, and 595 named genes, including many that are clearly involved in hematopoiesis. The cDNAs reported here represent a selection of some of the most highly abundant genes in hematopoietic cells and provide a starting point to develop a profile of the transcriptosome of CD34+cells.
Introduction
Nonhuman primates are important experimental models for hematopoietic stem cell transplantation and biology, because the behavior of hematopoietic stem and progenitor cells in primates closely resembles that in man.1-3 The use of nonhuman primates permits a degree of experimental freedom to perturb hematopoiesis not possible in man, which might end in a genetic analysis of hematopoiesis, not only under steady-state conditions, but also under conditions of stress. The baboon (Papio anubis) is particularly useful in this regard because it is closely related to humans and shows cross-reactivity with many of the reagents used to study human hematopoiesis. Recent studies have initiated a description of the overall pattern of gene expression in murine bone marrow stem cells,4,5 but by contrast, relatively little is known of the expression patterns of human bone marrow hematopoietic stem cells or the baboon marrow stem and progenitor cells. To support our studies in baboon hematopoiesis, and facilitate their extrapolation into human systems, we attempted to gain a better understanding of the similarity of gene expression between human and baboon bone marrow stem cells. Thus, it was necessary first to describe and compare the expression profiles of this tissue for each species. Human and baboon bone marrow cells that were positive for the CD34 antigen (CD34+ cells) were used for these comparisons, because they represent a marrow fraction enriched for both primitive hematopoietic stem and progenitor cells.6-8
Human complementary DNA (cDNA) filter arrays were used to establish the expression profiles for both species, because there is no comparable product available for baboon cDNA analysis and a high nucleotide sequence homology between these 2 species was anticipated.9,10 The cDNA filter arrays used (GeneFilters) contained 25 920 cDNAs from the UniGene dataset (http://www.ncbi.nlm.nih.gov/unigene/index.html),15including both known genes and uncharacterized expressed sequence tags (ESTs), permitting the survey of one fourth to one half of the estimated 50 000 to 100 000 genes in the genome. Here we report the results of these studies, which describe the transcriptosome of CD34+ cells, demonstrating very comparable gene expression patterns in CD34+ cells in these 2 species, and validating the utility of human cDNA arrays for baboon studies.
Materials and methods
Collection and selection of CD34+ marrow cells
Healthy adult baboons (P anubis) weighing 9 to 10 kg were used for these studies. The animals were housed under conditions approved by the Association for the Assessment and Accreditation of Laboratory Animal Care. Bone marrow aspirates were obtained from the humeri and iliac crest of adult baboons under ketamine and xylazine (1 mg/kg) anesthesia under guidelines established by the Animal Care Committee of the University of Illinois at Chicago. Human bone marrow aspirates from the iliac crest were obtained from normal human adult donors after informed consent was obtained, as approved by the Institutional Review Board of the University of Illinois at Chicago. Marrow mononuclear cells were isolated from the marrow as previously described.1 Briefly, the marrow was heparinized, diluted 1:15 in phosphate-buffered saline (PBS), and fractionated over 60% Percoll (Pharmacia LKB, Uppsala, Sweden) by centrifugation at 500g for 30 minutes at 20°C. The interphase mononuclear cells were resuspended in PBS containing 0.2% bovine serum albumin and human immune globulin (Sigma Chemical, St Louis, MO) and labeled with the biotin-conjugated mouse antihuman CD34+ antibodies; monoclonal antibody (MoAb) 12-811 for baboon, and Qband/1012 for human cells, washed, and relabeled with streptavidin-conjugated rat antimouse antibody-containing iron microbeads (Miltenyi Biotech, Auburn, CA). The CD34+ cells were then selected by passing the CD34+ cell-antibody-iron bead complex through a magnetic column. The purity of the CD34+ fraction was estimated by flow cytometry using a fluorescein isothiocyanate (FITC)-conjugated antihuman CD34+ antibody K6.11 for baboon cells and MoAb HPCA-2 for human cells.
RNA and DNA preparation
Total RNA was extracted from 1 to 5 × 106 human and baboon CD34+ cells using an Ultraspec RNA Isolation kit (Biotecx Laboratories, Houston, TX) according to the manufacturer's protocol. The quantity of total RNA was determined by A260absorbance, and quality was verified by analysis on 1% agarose gels using standard techniques. Genomic DNA was prepared from the HL60 human cell line (American Type Culture Collection, Rockville, MD) and baboon peripheral blood cells using Trizol reagent (Life Technologies, Grand Island, NY) according to the manufacturer's specification.
Uniformly labeled cDNA probes were prepared from 3 μg total RNA by priming with 2 μg oligo-dT, followed by elongation with 300 U Superscript II reverse transcriptase (Life Technologies) in the presence of 100 μCi of 33P dCTP (Amersham Pharmacia Biotech, Piscataway, NJ). The labeled probe was purified from unincorportated nucleotides and other small molecules with Probequant G-50 (Amersham Pharmacia Biotech).
Hybridization of cDNA probes to GeneFilters
Five releases (GF200-204) of human GeneFilters (Research Genetics, Huntsville, AL) were prehybridized for 2 hours at 42°C in MicroHyb solution (Research Genetics), with the addition of 1 μg/mL each of polyA (Research Genetics) and human Cot1 DNA (Life Technologies). The blots were then hybridized overnight in the same MicroHyb solution with the addition of 2 × 106 cpm/mL heat-denatured probe. The blots were washed twice at 50°C with 2 × standard sodium citrate (SSC), 1% sodium dodecyl sulfate (SDS) for 20 minutes, and once at room temperature in 0.5 × SSC, 1% SDS with gentle agitation for 15 minutes, prior to imaging. For re-use of membranes, the filters were stripped in 0.5% SDS for 1 hour at room temperature with gentle agitation as recommended by the manufacturer and re-exposed to confirm complete stripping.
Exposure, imaging, and analysis of filter membranes
The hybridized filters were imaged using a phosphor imaging screen (Molecular Dynamics, Sunnyvale, CA), exposed for 3 to 4 days, imaged using a Storm phosphor imaging system (Molecular Dynamics) at 50-μ resolution, and analyzed using PathwaysII from Research Genetics following the manufacturer's guidelines. Using this program, individual cDNA spots were identified and fit to a grid, and their intensity measurements were recorded as raw intensities. The background for a particular experiment, provided as a reference, was calculated by averaging the measured intensities between the 2 grids of the filter. This background information was used to assign levels of expression of the genes. Data from poor hybridizations, such as those that had unacceptably high background or nonuniform control spots intensities across the membrane, were not considered for further analysis and discarded. To compare expression of a cDNA spot between 2 probes that were sequentially hybridized to the same filter, the intensities were normalized using the algorithm provided by the PathwaysII software, using either control spots or all data points as reference. The data were exported as Excel files for further analysis. Because PathwaysII uses an older, somewhat outdated version of UniGene (build versions 18, 19, 39, and 42) and substantial changes have been made in the UniGene database since then, we updated our cDNAs list using UniGene build version 118 as a reference (current as of April 2000). To accomplish this, both the UniGene and GeneFilter dataset were reformatted to Microsoft Access database. The GenBank accession numbers of the GeneFilter dataset were then matched against the UniGene database to update the cluster identification, gene name, and gene description.
Polymerase chain reaction analysis
For reverse transcriptase–polymerase chain reaction (RT-PCR), first-strand cDNA was generated from approximately 1 μg RNA that had been DNase-treated with RNase free DNase I (Life Technologies). The RNA was then used to make first-strand cDNA in a 20-μL reaction volume with (+RT) or without (−RT) reverse transcriptase using Superscript II Reverse Transcriptase kit from Life Technologies according to the manufacturer's recommended protocol followed by RNase H treatment. If not stated otherwise, 1/20 volume of the +/−RT reaction mix was used for the PCR reaction in presence of 1 × PCR buffer (PerkinElmer Cetus, Boston, MA), 1.5 mM MgCl2, 200 μM dNTPs, 1 μM each of forward and reverse primers, and 1 U Amplitaq polymerase (PerkinElmer Cetus) in a 20-μL reaction volume using the following cycles: initial denaturation at 95°C for 5 minutes, followed by each cycle at 95°C for 30 seconds, annealing at 58°C /65°C depending on the primer pair for 30 seconds, amplification at 72°C for 30 seconds, and the final amplification was for 5 minutes at 72°C. PCR analysis of genomic DNA was similarly performed, using 200 ng genomic DNA instead of first-strand cDNA.
Comparison of expression levels by semiquantitative RT-PCR
To compare the expression of individual genes, RT-PCR was performed using primer pairs designed based on the sequence of the cDNA clones that was included on the GeneFilter. The PCR was done from 25 to 40 cycles with increments of 5 cycles, except for β2-microglobulin, which was done at 18, 22, 25, and 30 cycles. The PCR products were analyzed on a 3% agarose gel stained with ethidium bromide, and the amount of DNA was quantitated as band intensities using GelDoc software from Biorad (Hercules, CA). The level of expression of each gene was normalized against the level of β2-microglobulin expression between these 2 species. The relative expression between human and baboon cDNA was estimated by measuring the ratio of intensity of DNA product, comparing only those measurements that fell within the linear range of PCR amplification cycles; multiple determinations, when performed, were averaged. The sequences of forward (F) and reverse (R) primers are: transmembrane 4 superfamily member 4 (TM4SF4), F-AAGCGATTTGCGATGTTCACCTC, R-GAGGCTCTCGGCACTTGTTCC; protein tyrosine kinase 9 (PTK9), F-GATTCCTTTGTTTTACCCCTGTTGGAG, R-TTGCTGC ATACAACATTTTTTGAC; cytochrome P450, subfamily I (dioxin-inducible), polypeptide 1 (glaucoma 3, primary infantile) (CYP1B1), F-GTAATGGTGTCCCAGTATAA GTAATGAG-3′, R-TCATGAATGCTTTTAGTGTGTGC-3′; colony-stimulating factor 3 receptor (granulocyte) (CSF3R), F-CTGAAGTTATAGGAAACAAGC ACAAAAGGC, R-GCCCATGACTAAAAACTACCCCAGC; β2-microglobulin (B2M), F-CCTGAA TTGCTATGTGTCTGGG, R-TGATGCTGCTTACATGTCTCGA; R82595, F-GCTCGTAGCAACATTTTCG- TAATAGCC, R-GGACCCATCGTGGTT ACCGTG; AA676327, F-ATATTTCGGTAACTTTTGACCCTAAG, R-CAGGGGCAA TTTTGAGGTATG; R85439, F-GGCAGGGCTCTAAATGGAAGTAGTTG, R-CTCAG AAGTGTTTTGTAGCAAGGCTGC; AA487912, F-AAACAGTGACTTATCCCGCTAC CC, R-GGGTGGGTTTACTCTTAGAATCGC; N25920, F-CAGATGGAGGGTTTATG AGTGAGGCTGG, R-GCTTGTTCTTTGGG- GATTGTGGTGC;R05886, F-TAGGCG TGAGAAGCATATAGAGGC, R-AGTGAATAAGCAAGAAATCAGGGTG;N74363, F-ACAAAGGGCTGTTTACTGAGAGACCTGAGC, R-GGCATAACTCACACCCATT TGTTT- ACCTGC; N55359, F-GGCAGAATCTACTGGGCATCTTGTAATC, R-AGTTTTGGTGGTCCAGGGAAGGTAC.
Results
Correlation of gene expression between human and baboon CD34+ cells
The CD34+ cell populations were isolated from bone marrow aspirates by immunomagnetic cell sorting using antibodies to the human CD34 antigen that cross-react with baboon CD34+cells. The human marrow cell population was 90% pure, as determined by FACS analysis with antihuman CD34+ antibody. Using the same method, the baboon CD34+ cells measured 77% purity. This measurement in baboon cells is an underestimate of the true degree of purity due to the relative nonspecificity of the antihuman CD34+ antibody K6.1 (used for quantitation by flow cytocytometry) with baboon cells, resulting in a weaker fluorescence signal and lower estimates of purity than can be measured in comparable human cells, but it is within the range that is normally observed with this method.
Radioactively labeled RNA-based probes prepared from each cellular population were hybridized to 5 nylon filter membrane arrays (GeneFilters releases 200-204, containing a total of 25 920 cDNAs) and phosphoimaged, and the resultant image was analyzed to determine the relative hybridization signal intensity for each cDNA with each probe. Each cDNA on the array is derived from a single clone from the IMAGE consortium (http://image.llnl.gov) representing the 3′-end of a unique UniGene cluster. All data were obtained by sequential hybridization to a single filter set, to provide the most accurate comparisons between probes and avoid variability in cDNA spotting. Duplicate experiments were performed when possible, but were limited by the lifetime of the filters, which in general could be successfully rehybridized no more than 3 to 4 times. It was not possible to use pooled baboon marrow donors because of the limited availability of animals, and thus we opted not to use pooled human donors either, recognizing that our method is not sensitive enough to detect small differences between individual donors.
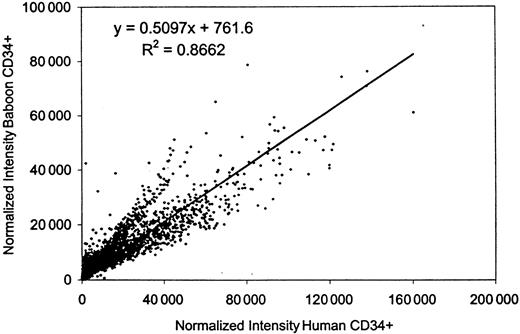
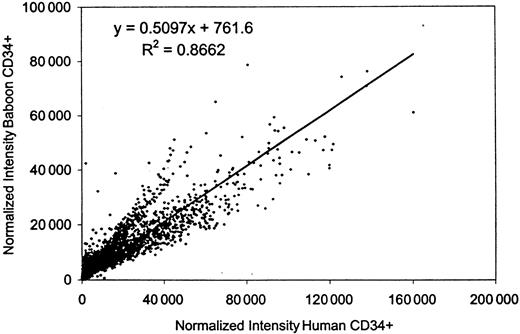
Normalized signal intensities for individual cDNA spots from these hybridizations were compared by scatter analysis, and revealed that the gene expression patterns in human and baboon cells were similar, with an overall correlation of 0.87. The composite data for all hybridizations are summarized on a scatter plot (Figure1). The measured raw intensity of the hybridization signal relative to the filter background is used as an indicator of the relative abundance of the cDNA. For these experiments, a cut-off level of raw intensity (nonnormalized) of 3-fold over background was used to indicate that a gene is definitively expressed in human cells. By this criterion, human CD34+ cells displayed positive expression for approximately 15 970 (62%) of the 25 920 cDNAs present on these filters. This gene list excludes many housekeeping genes, which are measured on the GeneFilters as hybridization controls but are not included for normalization by Pathways II software. (For information on all the spotted cDNA for each filter including the housekeeping genes, refer to the Research Genetics ftp website, ftp://ftp.resgen.com/pub/genefilters/.) The baboon-derived probes showed a consistently higher hybridization background, approximately 3-fold higher, than the human-derived probes, so it was not possible to apply the same cut-off level for this species (baboon). However, 13 447 cDNAs (84%) gave a signal with the baboon probe that varied less than 2-fold from the human level of expression, whereas almost all of the genes (15 407 or 96.5%) were expressed within 3-fold of each other. Much of the measured differences in expression level is likely to be due to experimental variation; we have found that about 3% of cDNAs will vary more than 3-fold on repeat hybridization with these probes (data not shown). Other measured differences between the human and baboon RNAs probably reflect true differences in expression, but in either case, the variation is not great. Thus, we conclude that the human and baboon CD34+ cells express virtually the same spectrum of genes, with similar though not identical levels of expression.
Correlation of gene expression between human and baboon CD34+ cells.
The normalized intensities of all the data points (25 920) from 5 releases of GeneFilters (GF200-GF204) hybridized to the baboon-derived CD34+ probe were compared to those resulting from the human-derived CD34+ probe by scatter analysis, using Microsoft Excel software.
Correlation of gene expression between human and baboon CD34+ cells.
The normalized intensities of all the data points (25 920) from 5 releases of GeneFilters (GF200-GF204) hybridized to the baboon-derived CD34+ probe were compared to those resulting from the human-derived CD34+ probe by scatter analysis, using Microsoft Excel software.
cDNAs highly expressed in both human and baboon
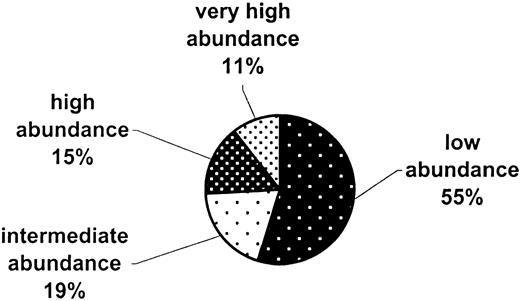
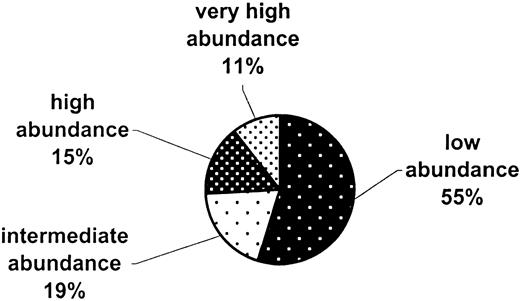
The 15 407 cDNAs that are commonly expressed in human and baboon CD34+ cells were arbitrarily placed into several groups (Figure 2) based on their spot intensities relative to background in the human data set: very high abundance (100-fold and over), 1619 cDNAs; high abundance (25-fold to < 100-fold), 2376 cDNAs; intermediate abundance (10-fold to < 25-fold), 2976 cDNAs; low abundance (3-fold to < 10-fold), 8436 cDNAs.
Abundance categories of the common genes in human and baboon CD34+ cells.
A total of 15 407 cDNAs whose expression varies less than 3-fold between human and baboon CD34+ RNAs were arbitrarily grouped into 4 relative expression categories, from low to very high abundance. The categories, based on the signal intensity of the human RNA relative to filter background, are as follows: no expression (< 3-fold), low abundance (3-fold to < 10-fold), intermediate (10-fold to < 25-fold), high 25-fold to < 100-fold), and very high abundance (100-fold and higher).
Abundance categories of the common genes in human and baboon CD34+ cells.
A total of 15 407 cDNAs whose expression varies less than 3-fold between human and baboon CD34+ RNAs were arbitrarily grouped into 4 relative expression categories, from low to very high abundance. The categories, based on the signal intensity of the human RNA relative to filter background, are as follows: no expression (< 3-fold), low abundance (3-fold to < 10-fold), intermediate (10-fold to < 25-fold), high 25-fold to < 100-fold), and very high abundance (100-fold and higher).
The very highly abundant genes identified by Pathways II analysis were then updated to the most current UniGene release (version 118, April 2000), and examined in detail. A total of 1554 UniGene clusters remained after updating. This list included 595 named genes and 959 ESTs and uncharacterized cDNAs. This list of highly abundant genes and ESTs is available as an appendix to the online version of this article and is also available on our hematopoietic stem cell website (http://westsun.hema.uic.edu/html/expression.html). The named genes represent a wide variety of functional categories such as growth factors and cytokines, receptors and cell surface molecules, intracellular signaling molecules, cell cycle proteins, and so forth. A sample of these genes, sorted by functional category, is presented in Table 1. Note that this list includes many of the genes (typed in bold) that would be expected to be present in CD34+ cells, such as receptors for interleukin 3 and colony-stimulating factor 3. Interestingly, many expected hematopoietic genes are not in this category, because their level of expression is relatively low; for example, the CD34 antigen is expressed at a relatively low level, only 6-fold above background (for human).
A large fraction, over 61% of these highly expressed cDNAs, are ESTs and uncharacterized cDNAs. Although many of these genes are uncharacterized, the UniGene database provides some information about their similarity to known proteins. Furthermore, many of the named genes represent full-length cDNAs that have not been fully studied or are only partially characterized, though some function is suggested by homology to known proteins. A partial list of some of these interesting ESTs and partially characterized named genes is presented in Table2. Further characterization of the ESTs in this database represents a potential wealth of new information about the CD34+ transcriptosome.
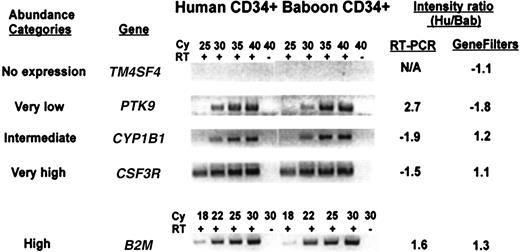
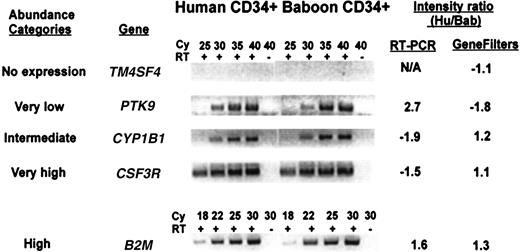
Several known genes from each abundance category were selected to verify their relative level of expression in both species by semiquantitative RT-PCR. Representative examples are shown in Figure3. Each gene tested was found to be expressed at comparable levels in both species, although the abundance category was not always accurate, especially in the lower abundance genes. For example, PTK9 is expressed at a level 5-fold above background in human cells, but its signal appears stronger than CYPB1, measured at 20-fold above background. The measurement of the absolute level of expression of a cDNA using filter hybridization is related to many factors, including the amount of DNA placed on the filter (which cannot be accurately controlled), and the efficiency of hybridization. Thus, the assignment of a gene to a relative abundance category can only be regarded as approximate and may require additional confirmation.
Comparison of the expression level between human and baboon CD34+ cells for genes selected from different abundance categories, by semiquantitative RT-PCR.
Five known genes representative of each of the abundance categories described in Figure 2 were analyzed by RT-PCR using primers from the 3′-untranslated region of the gene. The PCR reactions were done with (+) or without (−) addition of reverse transcriptase (RT) for the indicated cycle number (Cy). The genes tested are: TM4SF4, transmembrane 4 superfamily member 4; PTK9, protein tyrosine kinase 9; CYP1B1, cytochrome P450, subfamily 1 (dioxine-inducible), polypeptide 1 (glaucoma 3, primary infantile);CSF3R, colony-stimulating factor 3 receptor; B2M, β2-microglobulin. The intensity measured with GeneFilters is compared to that measured by RT-PCR.
Comparison of the expression level between human and baboon CD34+ cells for genes selected from different abundance categories, by semiquantitative RT-PCR.
Five known genes representative of each of the abundance categories described in Figure 2 were analyzed by RT-PCR using primers from the 3′-untranslated region of the gene. The PCR reactions were done with (+) or without (−) addition of reverse transcriptase (RT) for the indicated cycle number (Cy). The genes tested are: TM4SF4, transmembrane 4 superfamily member 4; PTK9, protein tyrosine kinase 9; CYP1B1, cytochrome P450, subfamily 1 (dioxine-inducible), polypeptide 1 (glaucoma 3, primary infantile);CSF3R, colony-stimulating factor 3 receptor; B2M, β2-microglobulin. The intensity measured with GeneFilters is compared to that measured by RT-PCR.
Species-specific transcripts
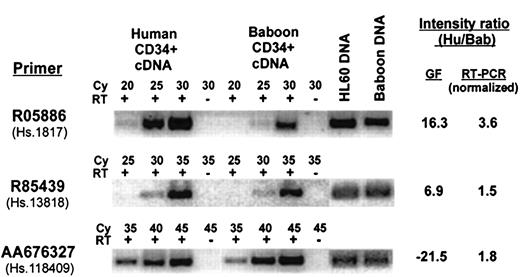
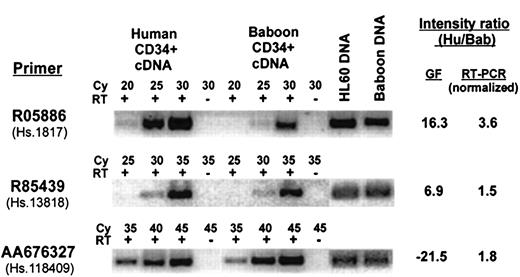
Although there were a number of cDNAs that did not appear to be highly correlated (ie, their expression varied more than 3-fold between species), there were a few genes whose measured intensity suggested that they were preferentially expressed in only one species. To identify these genes, the GeneFilters dataset was searched for cDNAs that were unexpressed in one species (defined as a raw intensity of < 3-fold background) and were clearly expressed in the other species (> 3-fold background) with a normalized intensity ratio of more than 3-fold between species. There were only 14 cDNAs that fit these criteria, 6 baboon and 8 human, which include 6 known genes and 8 ESTs. PCR primer pairs for all 14 cDNAs were designed to match the sequence of the human clones that were present on the filter membrane; the pairs were tested for their ability to amplify both genomic DNA and reverse-transcribed RNA from both species. Six primer pairs (4 human and 2 baboon) were successfully validated on both species in this manner, and these were further analyzed by semiquatitative RT-PCR, using an additional normalization factor for PCR efficiency on genomic DNA from both species. The ratio of expression for each gene, as measured by semiquantitative RT-PCR, compared to that measured on GeneFilters, is summarized in Table 3, and representative examples are shown in Figure4. The use of normalization factors, one as a control for PCR efficiency of human-specific primers against baboon, and another for RT reaction, adds complexity and probably some inaccuracy in quantitative comparison of gene expression between the 2 species, so the measured levels can only be regarded as estimates. Nonetheless, most of the genes, except for 2 designated by Unigene Cluster ID Hs.1817 and Hs.215595, showed little if any differential between the 2 species and fall within 3-fold of each other, well within the arbitrary cut-off that was set for Table 1. Only Hs.1817 and Hs.215595 were confirmed to be expressed at somewhat higher levels in human than baboon (3.6-fold and 5.4-fold, respectively), although the differences were small and not as great as was measured on the filters. The results showing differential expression of Hs.1817 are included in Figure 4. Thus, none of the 6 genes tested showed expression restricted to one species, though some appear to be differentially expressed. This result suggests that the experimental variation in the GeneFilter hybridization system is greater than the actual variation between the 2 species. Additional work will be required to determine if there are any bona fide species-specific genes within either species.
Comparison of the expression level between human and baboon CD34+ cells for apparent species-specific genes selected from Table 3.
Representative analysis by semiquantitative RT-PCR for 3 transcripts from Table 3 with apparent species-specific expression as measured on GeneFilters (GF), using primers designed from the 3′-untranslated region of the gene. The PCR reactions were done with (+) or without (−) addition of reverse transcriptase (RT) for the indicated cycle number (Cy). The intensity measured with GeneFilters is compared to that measured by RT-PCR, normalized to genomic DNA. Intensity ratio measurement is shown as positive when expression in humans is higher than baboons, and negative when the reverse is true.
Comparison of the expression level between human and baboon CD34+ cells for apparent species-specific genes selected from Table 3.
Representative analysis by semiquantitative RT-PCR for 3 transcripts from Table 3 with apparent species-specific expression as measured on GeneFilters (GF), using primers designed from the 3′-untranslated region of the gene. The PCR reactions were done with (+) or without (−) addition of reverse transcriptase (RT) for the indicated cycle number (Cy). The intensity measured with GeneFilters is compared to that measured by RT-PCR, normalized to genomic DNA. Intensity ratio measurement is shown as positive when expression in humans is higher than baboons, and negative when the reverse is true.
Discussion
By its ability to simultaneously detect and quantitate the expression level of thousands of genes at one time, cDNA array technology is greatly improving our understanding of the complex patterns of gene expression in eukaryotic cells. Here, we use this technology to profile the gene expression patterns of CD34+marrow cells in human and baboon cell populations. Although there were a number of technical limitations in this study due to the comparison of 2 species, in general we found that baboon-derived probes are suitable for use on human cDNA arrays.
Expression studies on cDNA arrays require a fairly large number of cells to isolate an appropriate amount of RNA for probe preparation. Because of this constraint, it was necessary to purify the CD34+ cells by immunomagnetic columns rather than FACS, which would require prolonged sorting. The stress imposed by the prolonged sorting time required to prepare this number of cells can dramatically reduce cell viability and yield of CD34+ cells and may alter their gene expression profile. Because of the weak cross-reactivity of antihuman CD34+ antibody against baboon CD34+ antigen, it is difficult to accurately determine the level of purity of baboon CD34+ cell population. Thus, the purity of baboon CD34+ may be an underrepresentation. At any rate, despite the heterogeneity of the cell populations examined and the limited number of subjects studied, we determined that bone marrow cells derived from the 2 closely related species have similar patterns of gene expression. Although we expected to find many molecular similarities between human and baboon CD34+cells, the results suggest that the transcriptosomes are nearly identical, supporting experimental studies over the years that have demonstrated similar biologic activity. Our inability to identify any species-specific transcripts further supports the similarity of the 2 populations, though further studies will be required to address this issue.
The probe derived from the 3′ end of baboon RNA recognized human cDNAs fairly well under appropriate hybridization conditions. The concentration of Cot1 and oligo-dT, which are used for blocking nonspecific hybridization, were found to be crucial for this purpose. This is not unexpected because the genomes of the 2 species are highly conserved, and both have Alu sequences.13 14 In general, higher background resulting from the baboon probe may be a reflection that the Alu content is not identical, and might benefit from a readjustment of the hybridization conditions, especially Cot1 and oligo-dT concentration. Nonetheless, the hybridization signal obtained with the baboon probe was strong and resulted in a similar pattern to the one obtained with human probe. This suggests that human cDNA arrays are accurate substrates for baboon experiments, thereby facilitating translation of experimental results with this animal model to human relevance.
The studies were performed using a cDNA filter array system and radioactive probes. Although there may be limitations to the use of filters rather than solid cDNA supports, GeneFilters were especially attractive for these studies because they contain over 25 000 different cDNA clones, which cover an estimated 50% of the human genome, including a large proportion of uncharacterized cDNAs (ESTs). The use of UniGene clones for these arrays facilitates data comparison with other investigators because UniGene is a publicly available database that cross-references with GenBank, partial sequence information is available for each entry, and a representative clone can be obtained from commercial or public repositories.
The use of GeneFilters dictated an experimental design that differs from those using cDNA arrays on solid supports. Because 2 probes cannot be simultaneously hybridized and compared in a single experiment, reproducibility is maximized when the same membrane is reused for sequential hybridization to compare probes from different RNA sources. Due to limited membrane lifetime, it is not possible to repeat multiple experiments or compare expression patterns among different subjects, so the sampling error may be greater than for other methods for cDNA analysis. Thus, the results presented here should be regarded as a starting point for further confirmation and analysis.
The most reliable data obtained on these filters is the comparison of relative signal strength for a single gene between 2 probes. An absolute determination of the relative expression between different genes on one filter is less reliable, because the signal strength is dependent on many factors, such as the length of the clone and the hybridization efficiency of the probe, and the relative inaccuracies of spotting small amounts of DNA. Cross-comparisons of cDNA on different filters is less reliable. Here, we used the intensity of the hybridization signal relative to background as a means of comparison between filters, to estimate the relative level of expression of all of the genes on this dataset, recognizing that this is only an approximate, though generally reliable, measurement.
The gene list resulting from this study represents a selection of some of the most highly abundant genes in hematopoietic cells and provides a starting point to develop a profile of the predominant cDNAs that define CD34+ cells. Interestingly, a significant fraction of the genes identified on these filters is not unique to hematopoietic cells, but is present in other tissues. This reinforces the concept that a tissue is defined not only by the expression of tissue-specific genes, but also by the overall pattern and relative abundance of the sequences that are more widely expressed. Perhaps the most interesting result of this study is the fact that many of the cDNAs expressed at high level in these cells have not yet been identified or characterized. The gene and EST list presented here, and their relative expression levels, represent a potential wealth of new information about bone marrow stem cells and hematopoietic progenitor cells.
This study allows us to begin to compile a comprehensive description of the CD34+ transcriptosome with reference to the UniGenes represented in GeneFilters. Although by no means complete, our list of over 15 000 cDNAs comprises an estimated 25% to 50% of the genes expressed in CD34+ cells and also provides an approximation of their relative abundance. These data will be provided on our website when it is more fully characterized and catalogued. It will be interesting to compare these results to those obtained for the murine hematopoietic system4 when the availability of additional cDNA sequence makes such comparisons feasible. We believe that this gene set will prove to be useful for the production of customized cDNA arrays for bone marrow studies. It is expected that further study of more highly refined subsets of human and baboon marrow cells using cDNA arrays will reveal additional information on the genetic program of hematopoiesis in primates.
We are grateful to John Brandt for his assistance in procuring the baboon cells.
Supported by Public Health Service grants R01-CA-72593 (C.A.W.) and P01-75 606 (C.A.W. and R.H.); National Science Foundation grant MCB9980088 (J.L.), Leukemia Society of America (R.H.), the W. M. Keck Foundation Fellowship (N.M.), and a seed grant from the University of Illinois Office of the Vice Chancellor for Research (C.A.W.).
I.G. and T.S. contributed equally to this work.
Reprint:Carol A. Westbrook, Department of Medicine, Section of Hematology and Oncology, 900 S Ashland Ave, M/C 734, Chicago, IL 60607; e-mail: cwcw@uic.edu.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.