Abstract
In acute myelogenous leukemia (AML) and adult T-cell leukemia, it has been demonstrated that the transcription factor LIL-STAT is constitutively activated. To identify and characterize this unknown LIL-STAT protein, electrophoretic mobility shift assay (EMSA) and oligoprecipitation assays were performed by using lipopolysaccharide/interleukin-1 (IL-1)–responsive element (LILRE) oligonucleotide probes. EMSA demonstrated a significant increase in LIL-STAT binding to the LILRE oligonucleotides after interferon γ (IFN-γ) and IL-6 stimulation of THP-1 cells. In unstimulated THP-1 and AML cells, LILRE oligonucleotide probes bound only to STAT1 α and β isoforms. The LILRE element showed a significant increase in binding of both α and β isoforms of STAT1 and STAT3 upon IFN-γ and IL-6 stimulation. Similar results were observed with human monocytes upon IL-6 or IFN-γ stimulation. These studies indicate that LIL-STAT consists of STAT1 and STAT3 proteins that bind to the LILRE DNA consensus site in a stimulus-dependent way.
Introduction
The signal transducers and activators of transcription (STAT) proteins are a family of transcription factors involved in cytokine and growth factor signaling and are critical for the proliferation and differentiation of hematopoietic cells. The DNA-binding target of STAT dimers upon receptor activation is the consensus interferon γ (IFN-γ) activation site (GAS), which was initially reported to bind the IFN-γ–induced STAT1. So far, 7 different STAT genes have been identified.1
Recently, a novel GAS-binding STAT factor was characterized with unique binding activity for a lipopolysaccharide (LPS)/interleukin-1β (IL-1β)–responsive element (LILRE) sequence motif of 8 nucleotides, TTCCTGAGA. LILRE is located within the enhancer region of the pro–IL-1β gene. Activation of this LIL-STAT factor was mediated by LPS, IL-1β, and IL-6 stimulation in different cell lines and could be shifted in electrophoretic mobility shift assay (EMSA) by antibodies directed against the N-terminus of STAT1, but not by those specific for the C-terminus of STAT1. In addition, no cross-competition existed with the other GAS-binding oligonucleotides.2 Spontaneous expression of LIL-STAT has been demonstrated in acute myeloid leukemia (AML) cells and adult T-cell leukemia.3,4 In addition, it was shown that the expression was dependent on the stage of differentiation. In primitive hematopoietic CD34+ cells, LIL-STAT expression was present, but it was absent in differentiated monocytes and granulocytes.3 However, it is unclear whether LIL-STAT is a novel STAT protein separate from the well-defined STAT1 to STAT6 proteins.
STAT activation can be detected by elevated DNA-binding activity, as measured in EMSAs using an oligonucleotide probe corresponding to specific GAS-binding sites. STAT1 homodimers bind to the GAS site of the FcγRI gene, but not STAT3 homodimers.5 In contrast, both STAT1 and STAT3 bind to the high-affinity synthetic derivative of the c-fos promoter m67 hSIE (mutated humansis-inducible element)6 and S1-S3 (consensus binding sequence for STAT1 or STAT3).7 8
In the present study, we demonstrate by EMSA and oligoprecipitation assays that binding of α and β isoforms of STAT1 and STAT3 to the LILRE DNA motif is stimulus dependent and differs between monocytic leukemic cells and monocytes.
Study design
Cell culture, AML cells, cytokines, and antibodies
Human monocytic leukemic cells, namely THP-1 and AML cells, were isolated as described before.3 Nuclear proteins were extracted from leukemic cells and monocytes9 after 10 minutes of incubation with 10 ng/mL recombinant human IFN-γ (Endogen, Woburn, MA) or 10 ng/mL IL-6.10
Peripheral blood cells or bone marrow cells were obtained from AML patients after informed consent. AML blasts were cultured at 37°C in RPMI 1640 medium (Flow, Rockville, MD) supplemented with 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.1% fetal calf serum for 4 hours before cytokine stimulation and subsequent nuclear extract preparation. Mononuclear cells were obtained from healthy blood donors after informed consent, and monocytes were isolated as described earlier.3
Polyclonal antibodies directed against the N-terminal domain (amino acids 1-194) of STAT1 (G16930) and monoclonal antibodies against the C-terminal domain of STAT1 (S21120) were purchased from Transduction Laboratories (Lexington, KY). Phospho-STAT1 (Y701; no. 06-657) and phosphotyrosine PY100 were obtained from Upstate Biotechnology (Lake Placid, NY). Polyclonal antibodies directed against the C-terminal domain of STAT1 (sc-346) and STAT3 antibodies (C-20: sc-482; F2: sc-8019) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Phospho-STAT3 (Y705) antibodies (no. 9131) were purchased from Cell Signaling Technology (Beverly, MA). Antibodies were used according to recommendations from the manufacturer.
Oligonucleotides and EMSA
Oligonucleotides containing GAS element (underlined nucleotide sequences) were synthesized for the FcγRI (Fab constant transmembrane region of IgG receptor): 5′-AGCTGTATTCCCAGAAAAGGAAC-3′ (sense strand); m67hSIE (sis-inducible element): 5′-CTAGCATTTCCCGTAAATCATC-3′ (sense strand); and LILRE: 5′-AGCTTATAAGAGGTTTCACTTCCTGAGAGTCGA- 3′ (sense strand) (Life Technologies, Glasgow, Scotland). Double-stranded oligonucleotides were radiolabeled with α32P-dATP by filling in the 5′-protruding ends using a random primer DNA-labeling protocol (Life Technologies). EMSAs were performed as described previously.3
Oligoprecipitation assay
5′-Biotin–labeled oligonucleotides were synthesized for S1-S3 (STAT1-STAT3 consensus binding site)7: 5′-biotin-AGCTTAGGTTTCCGGGAAAGCAC-3′; and LILRE: 5′-biotin-AGCTTATAAGAGCTTTCACTTCCTGAGAGTCGA-3′3 4and purified by polyacrylamide gel electrophoresis (PAGE; Gibco Life Technologies). Four micrograms of double-stranded biotin-labeled oligonucleotides was incubated with 200 μL streptavidin-sepharose HP beads (Amersham Pharmacia Biotech, Uppsala, Sweden) for 1 hour at 4°C in a “head-over-head” rotor. After 2 wash steps in buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT], 1 mM orthovanadate, 1× protease inhibitor cocktail [Roche Boehringer, Mannheim, Germany]), the oligobead complex was dissolved in buffer A to a final concentration of 40 ng/μL. Thirty-five microliters of oligobead solution was incubated with 50 to 100 μg nuclear protein in EMSA buffer containing 2.5 μg poly [dI-C] (Roche Boehringer Mannheim), 1 mM sodium orthovanadate, 1 mM DTT, and 1 × protease inhibitor cocktail for 16 hours at 4°C in a head-over-head rotor. Protein concentrations were determined according to a Bradford assay (BioRad Laboratories, Hercules, CA). Competitive oligoprecipitation assays were performed by the addition of unlabeled LILRE (5′-AGCTTATAAGAGCTTTCACTTCCTGAGAGTCGA-3′) and TRE double-stranded oligonucleotides (TPA responsive element in c-jun promoter) (5′-GATCCGGGTGACATCATGG- 3′). Finally, oligonucleotide-bound proteins were washed 2 times in buffer A and dissolved in sodium dodecyl sulfate (SDS) sample buffer.
Western blot analysis
Oligoprecipitates were boiled for 5 minutes before separation on 10% SDS-PAGE. Subsequent transfer of proteins to a membrane and antibody incubation have been described earlier.10
Results and discussion
Binding of STAT1 and STAT3 to different GAS elements
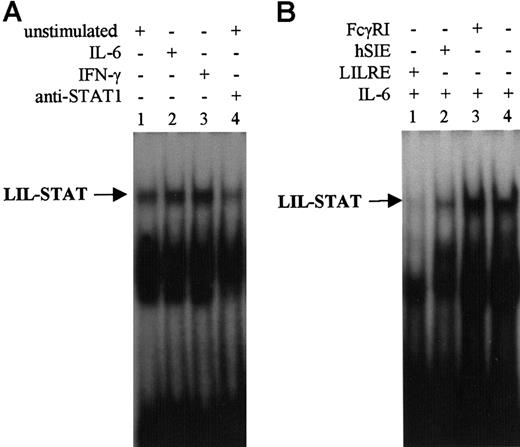
Nuclear extracts were isolated from THP-1 and B1 cells after stimulation with IL-6 and IFN-γ. The binding of STAT proteins was monitored by EMSA analysis using LILRE as oligonucleotide probe. IL-6 and IFN-γ stimulation of THP-1 cells increased STAT binding to the radiolabeled LILRE probe (Figure 1A). LIL-STAT binding was completely inhibited by addition of excess LILRE oligonucleotides and partially inhibited by hSIE oligonucleotides (Figure 1B). In contrast, LIL-STAT binding was not inhibited by addition of unlabeled FcγRI oligonucleotides (Figure 1B). By incubating nuclear extracts of unstimulated THP-1 cells with polyclonal anti-STAT1N antibodies, it was shown that the LIL-STAT binding decreased (Figure 1A) as well as upon IL-6 and IFN-γ stimulation (data not shown). To identify STAT oligonucleotide binding proteins as detected in EMSAs, we subsequently performed oligoprecipitations.
EMSA using LILRE as radiolabeled probe.
(A) EMSA of nuclear extracts isolated from unstimulated (lanes 1 and 4), IL-6–stimulated (lane 2), and IFN-γ–stimulated (lane 3) THP-1 cells. The sample in lane 4 has been incubated with STAT1N (G16930) antibodies. (B) EMSA of nuclear extracts isolated from IL-6–stimulated (lanes 1-4) B1 cells. To assess specificity of LILRE oligobinding, a 100-fold molar excess of unlabeled LILRE, hSIE, and FcγRI oligonucleotides (lanes 1, 2, and 3) was added to compete for radiolabeled LILRE binding. Furthermore, NF-κB (p52) antibodies were included as control for nonspecific antibody binding (lane 4). Figures represent the results of 3 independent experiments (n = 3). MW indicates molecular weight.
EMSA using LILRE as radiolabeled probe.
(A) EMSA of nuclear extracts isolated from unstimulated (lanes 1 and 4), IL-6–stimulated (lane 2), and IFN-γ–stimulated (lane 3) THP-1 cells. The sample in lane 4 has been incubated with STAT1N (G16930) antibodies. (B) EMSA of nuclear extracts isolated from IL-6–stimulated (lanes 1-4) B1 cells. To assess specificity of LILRE oligobinding, a 100-fold molar excess of unlabeled LILRE, hSIE, and FcγRI oligonucleotides (lanes 1, 2, and 3) was added to compete for radiolabeled LILRE binding. Furthermore, NF-κB (p52) antibodies were included as control for nonspecific antibody binding (lane 4). Figures represent the results of 3 independent experiments (n = 3). MW indicates molecular weight.
Identification of STAT1 and STAT3 binding to the LILRE GAS site in myeloid leukemia cells and monocytes
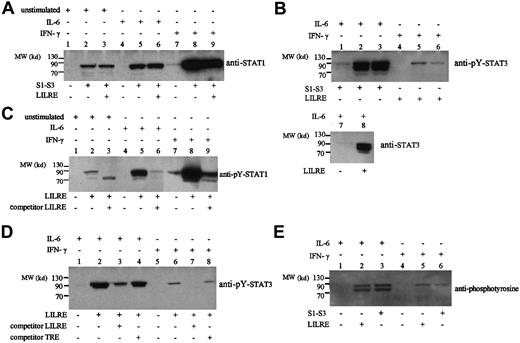
Biotinylated double-stranded LILRE and STAT1-STAT3 (S1-S3) oligonucleotides coupled to streptavidin-coated sepharose beads were incubated with nuclear extracts from unstimulated, IL-6–stimulated, and IFN-γ–stimulated THP-1 cells, AML cells, and monocytes. The oligoprecipitated proteins were separated on SDS-PAGE and analyzed by Western blot using antibodies against STAT1 and STAT3. As shown for unstimulated THP-1 cells, there was binding of STAT1 to LILRE and S1-S3 oligonucleotides (Figure2A). In the case of IL-6 and IFN-γ stimulation, there was a strong increase of STAT1 binding to both GAS oligonucleotide elements (Figure 2A). Increased binding of tyrosine-phosphorylated STAT1 was observed after stimulation with IL-6 and IFN-γ (Figure 2B). The specificity of STAT1 binding to LILRE was assayed by competitive oligoprecipitation, in which addition of excess amounts of double-stranded LILRE oligonucleotides abrogated STAT1 binding (Figure 2B). The identification of full-length STAT1 protein bound to the LILRE oligonucleotide by antibodies directed against STAT1 contradicts earlier results.2 3 In those EMSA studies, the binding to the LILRE oligonucleotide was diminished after incubation with polyclonal STAT1N antibodies, but not in the presence of polyclonal STAT1C antibodies (G16930). Based on these experiments, STAT1 was excluded as a LILRE STAT binder. A possible explanation for this discrepancy might be that the conformation of STAT1 is changed when binding to the LILRE oligonucleotide. The binding of STAT1 to the LILRE sequence might change the epitope that is recognized by the polyclonal STAT1C antibodies.
LILRE/S1-S3 oligoprecipitations of nuclear extracts from THP-1 cells followed by immunoblot analysis.
(A) Oligoprecipitation by LILRE (lanes 2, 5, and 8) and S1-S3 (lanes 3, 6, and 9) oligonucleotides of nuclear extracts isolated from unstimulated (lanes 1-3), IL-6–stimulated (lanes 4-6), and IFN-γ–stimulated (lanes 7-9) THP-1 cells. Subsequent immunoblot analysis was performed on isolated proteins by using STAT1 antibodies (G16930). Lanes 1, 4, and 7 show control precipitations using only streptavidin-coated sepharose beads. (B) LILRE/S1-S3 oligoprecipitation of nuclear extracts isolated from IL-6–stimulated (lanes 1-3) and IFN-γ–stimulated (lanes 4-6) THP-1 cells, followed by immunoblot analysis using phosphotyrosine-STAT3 (Tyr705) antibodies (no. 9131). Lanes 1 and 4 show control precipitations using beads without coupled oligonucleotides. Additional LILRE oligoprecipitation assays were performed on nuclear extracts isolated from IL-6–stimulated THP-1 cells, followed by immunoblot analysis using STAT3 (F2) antibodies. Lane 7 shows a control precipitation using streptavidin sepharose beads without biotinylated LILRE oligonucleotides. (C) Oligoprecipitation by LILRE (lanes 2, 5, 8) and LILRE/competitive LILRE oligonucleotides (lanes 3, 6, 9) of nuclear extracts from unstimulated (lanes 1-3), IL-6–stimulated (lanes 4-6), and IFN-γ–stimulated (lanes 7-9) THP-1 cells. LILRE-bound proteins were analyzed by Western blot using phosphotyrosine (pY) STAT1 (Y701) antibodies. (D) Competitive oligoprecipitation assay on nuclear extracts from IL-6–stimulated (lanes 1-4) and IFN-γ–stimulated (lanes 5-8) THP-1 cells, followed by immunoblot analysis using pY-STAT3 antibodies. Precipitations were performed using biotin-labeled LILRE oligonucleotides bound to streptavidin sepharose beads (lanes 2-4; 6-8). Excess amounts (12 μg) of double-stranded LILRE oligonucleotides (lanes 3, 7) and TRE oligonucleotides (lanes 4, 8) were added to LILRE-coupled streptavidin beads. Lanes 1 and 5 show control precipitations using uncoupled streptavidin sepharose beads. (E) LILRE/S1-S3 oligoprecipitation of nuclear extracts isolated from IL-6–stimulated (lanes 1-3) and IFN-γ–stimulated (lanes 4-6) THP-1 cells, followed by immunoblot analysis using phosphotyrosine antibodies (PY100). Lanes 1 and 4 show control precipitations using uncoupled streptavidin sepharose beads. Figures represent the results of 3 independent experiments (n = 3). MW indicates molecular weight.
LILRE/S1-S3 oligoprecipitations of nuclear extracts from THP-1 cells followed by immunoblot analysis.
(A) Oligoprecipitation by LILRE (lanes 2, 5, and 8) and S1-S3 (lanes 3, 6, and 9) oligonucleotides of nuclear extracts isolated from unstimulated (lanes 1-3), IL-6–stimulated (lanes 4-6), and IFN-γ–stimulated (lanes 7-9) THP-1 cells. Subsequent immunoblot analysis was performed on isolated proteins by using STAT1 antibodies (G16930). Lanes 1, 4, and 7 show control precipitations using only streptavidin-coated sepharose beads. (B) LILRE/S1-S3 oligoprecipitation of nuclear extracts isolated from IL-6–stimulated (lanes 1-3) and IFN-γ–stimulated (lanes 4-6) THP-1 cells, followed by immunoblot analysis using phosphotyrosine-STAT3 (Tyr705) antibodies (no. 9131). Lanes 1 and 4 show control precipitations using beads without coupled oligonucleotides. Additional LILRE oligoprecipitation assays were performed on nuclear extracts isolated from IL-6–stimulated THP-1 cells, followed by immunoblot analysis using STAT3 (F2) antibodies. Lane 7 shows a control precipitation using streptavidin sepharose beads without biotinylated LILRE oligonucleotides. (C) Oligoprecipitation by LILRE (lanes 2, 5, 8) and LILRE/competitive LILRE oligonucleotides (lanes 3, 6, 9) of nuclear extracts from unstimulated (lanes 1-3), IL-6–stimulated (lanes 4-6), and IFN-γ–stimulated (lanes 7-9) THP-1 cells. LILRE-bound proteins were analyzed by Western blot using phosphotyrosine (pY) STAT1 (Y701) antibodies. (D) Competitive oligoprecipitation assay on nuclear extracts from IL-6–stimulated (lanes 1-4) and IFN-γ–stimulated (lanes 5-8) THP-1 cells, followed by immunoblot analysis using pY-STAT3 antibodies. Precipitations were performed using biotin-labeled LILRE oligonucleotides bound to streptavidin sepharose beads (lanes 2-4; 6-8). Excess amounts (12 μg) of double-stranded LILRE oligonucleotides (lanes 3, 7) and TRE oligonucleotides (lanes 4, 8) were added to LILRE-coupled streptavidin beads. Lanes 1 and 5 show control precipitations using uncoupled streptavidin sepharose beads. (E) LILRE/S1-S3 oligoprecipitation of nuclear extracts isolated from IL-6–stimulated (lanes 1-3) and IFN-γ–stimulated (lanes 4-6) THP-1 cells, followed by immunoblot analysis using phosphotyrosine antibodies (PY100). Lanes 1 and 4 show control precipitations using uncoupled streptavidin sepharose beads. Figures represent the results of 3 independent experiments (n = 3). MW indicates molecular weight.
Furthermore, the observation that LILRE protein binding could be inhibited by excess of hSIE oligonucleotides (Figure 1B), a well-known STAT1-STAT3 binding sequence, suggests possible STAT3 binding to the LILRE site. To test this assumption, we performed additional oligoprecipitations and analyzed them by Western blot by using antibodies against phosphotyrosine (pY) STAT3 (Figure 2C). No phospho-STAT3 protein binding was detected for the LILRE or S1-S3 oligoprecipitations using unstimulated THP-1 nuclear extracts (data not shown). However, after IL-6 stimulation, 2 proteins of approximately 90 and 84 kd could be demonstrated. IL-6 stimulation strongly induced binding of both α and β STAT3 isoforms to the LILRE and S1-S3 oligonucleotide-coupled beads (Figure 2C). A minor small band of about 70 kd was detected as well. This may be a proteolytic product of STAT3 α or β that is still capable of DNA binding. STAT3 α and β were precipitated by LILRE on nuclear extracts from IL-6–stimulated THP-1 cells using STAT3 (F2) antibodies (Figure 2C, lane 2), and minor pY-STAT3 α binding was observed in IFN-γ–stimulated THP-1 cells (Figure 2C, lanes 5-6). To further assess the specificity of LILRE oligoprecipitation, we completely abolished the pY-STAT1 and pY-STAT3 binding by unbiotinylated double-stranded LILRE oligonucleotides (Figure 2B,D). In contrast, the addition of nonspecific TRE oligonucleotides did not affect the pY-STAT3 binding (Figure 2D). Similarly, the binding of pY-STAT1 to LILRE was inhibited by unlabeled LILRE (Figure 2B), but no inhibition was obtained in the presence of TRE oligonucleotides (data not shown).
To exclude the binding of tyrosine-phosphorylated proteins other than STAT1-STAT3, we performed additional oligoprecipitation experiments and analyzed them by immunoblot using phosphotyrosine antibodies (PY100). Again, tyrosine-phosphorylated proteins were identified by LILRE and S1-S3 oligoprecipitations in nuclear extracts isolated from IL-6–stimulated and IFN-γ–stimulated THP-1 cells. In the IL-6 sample, 2 tyrosine-phosphorylated proteins of approximately 85 and 90 kd were precipitated by LILRE and S1-S3 oligonucleotides, resembling the molecular weights of STAT3 α and β isoforms. In the IFN-γ sample, a protein band of about 90 kd was detected, which correlates with the molecular weight of STAT1 (Figure 2E). Importantly, no other tyrosine-phosphorylated proteins of different molecular weights could be identified by LILRE and S1-S3 oligoprecipitation.
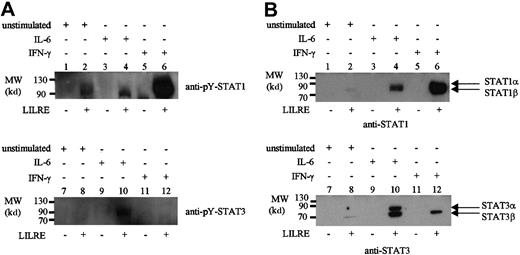
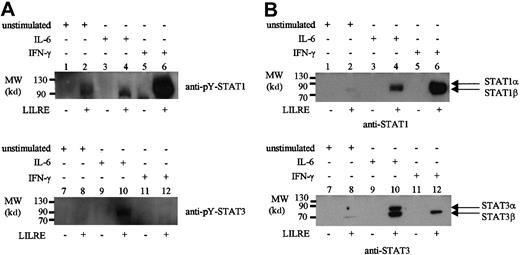
Finally, oligoprecipitations were performed to identify STAT binding to LILRE in AML cells, which showed a relatively high LILRE binding activity on EMSA (data not shown). As a result, pY-STAT1 was precipitated in the unstimulated AML cells with high LILRE binding (Figure 3A). Upon IFN-γ stimulation, the STAT1 LILRE binding was increased, whereas IL-6 stimulation showed the same amount of STAT1 binding as the unstimulated AML sample (Figure3A). The binding of STAT1 to LILRE-coupled sepharose beads is LILRE oligonucleotide-mediated because no STAT1 binding was observed in the control precipitations using only streptavidin sepharose beads (Figure3A, lanes 1, 3, and 5). pY-STAT3 was bound to the LILRE oligonucleotides in IL-6–stimulated AML cells, and was not precipitated in the unstimulated condition or after IFN-γ stimulation (Figure 3A, lanes 8 and 12).
LILRE oligoprecipitation of nuclear extracts isolated from AML cells and monocytes.
Lanes 1, 3, 5, 7, 9, and 11 represent control precipitations using streptavidin sepharose beads with no bound LILRE oligonucleotides. Lanes 2, 4, 6, 8, 10, and 12 show precipitations using LILRE-coupled streptavidin sepharose beads. (A) Nuclear extracts isolated from unstimulated (lanes 1, 2, 7, and 8), IL-6–stimulated (lanes 3, 4, 9, and 10), and IFN-γ–stimulated (lanes 5, 6, 11, and 12) AML cells followed by immunoblotting using pY-STAT1 and pY-STAT3 antibodies. Each precipitation was performed using 50 μg nuclear protein. (B) Nuclear proteins isolated from unstimulated (lanes 1, 2, 7, and 8), IL-6–stimulated (lanes 3, 4, 9, and 10), and IFN-γ–stimulated (lanes 5, 6, 11, and 12) monocytes. Each precipitation was performed using 100 μg nuclear protein. Figures are representative of 3 independent experiments (n = 3). MW indicates molecular weight.
LILRE oligoprecipitation of nuclear extracts isolated from AML cells and monocytes.
Lanes 1, 3, 5, 7, 9, and 11 represent control precipitations using streptavidin sepharose beads with no bound LILRE oligonucleotides. Lanes 2, 4, 6, 8, 10, and 12 show precipitations using LILRE-coupled streptavidin sepharose beads. (A) Nuclear extracts isolated from unstimulated (lanes 1, 2, 7, and 8), IL-6–stimulated (lanes 3, 4, 9, and 10), and IFN-γ–stimulated (lanes 5, 6, 11, and 12) AML cells followed by immunoblotting using pY-STAT1 and pY-STAT3 antibodies. Each precipitation was performed using 50 μg nuclear protein. (B) Nuclear proteins isolated from unstimulated (lanes 1, 2, 7, and 8), IL-6–stimulated (lanes 3, 4, 9, and 10), and IFN-γ–stimulated (lanes 5, 6, 11, and 12) monocytes. Each precipitation was performed using 100 μg nuclear protein. Figures are representative of 3 independent experiments (n = 3). MW indicates molecular weight.
In our previous study, we detected LILRE binding activity in CD34+ cells, which was absent upon differentiation into monocytes and granulocytes.3 Therefore, nuclear extracts isolated from unstimulated, IL-6–stimulated, and IFN-γ–stimulated monocytes were tested by LILRE oligoprecipitation. In contrast to THP-1 cells, unstimulated monocytes did not show LILRE binding of STAT1 α and STAT3 α, but only a very small amount of the β subunit. IL-6 stimulation clearly showed binding of both STAT1 and STAT3 α and β subunits (Figure 3B). In the case of IFN-γ stimulation, STAT1 bound to the LILRE sequence but in concert with the STAT3 β subunit (Figure 3B). These results are in contrast to previous results demonstrating no LILRE binding activity in IFN-γ–stimulated monocytes as detected by EMSA.3 The discrepancy might be explained by the fact that LILRE oligoprecipitations were performed in the presence of much more nuclear protein than used in the EMSA.
Based upon our studies, we have identified STAT1 and STAT3 as being the STAT variants that bind to the LILRE DNA element, depending on the stimulus. Unfortunately, because of the low resolution of the LILRE EMSA, we cannot discriminate between STAT1 and STAT3 homodimers or determine whether STAT1-STAT3 heterodimers are involved. However, based upon the relative abundance of STAT3 bound to the LILRE oligonucleotide upon IL-6 stimulation, and of STAT1 upon IFN-γ stimulation, we conclude that the LILRE complex most likely consists of STAT3 or STAT1 homodimers in monocytic leukemia cells and monocytes, depending on the stimulus. Additionally, the involvement of other proteins that interact with STAT1 and STAT3 bound to the LILRE oligonucleotide is possible. Recent reviews have already described the interactions of STATs with other proteins that are also stimulus- and cell-type specific.11,12 The binding of STAT1 and STAT3 to the LILRE site present in the IL-1β gene promoter might modulate IL-1β gene expression. Indeed, IL-6 and IFN-γ have been identified as positive modulators of IL-1β production in macrophage cell lines.13,14 The finding of STAT1-STAT3 protein association with the LILRE binding site in the IL-1β gene promoter is in line with previous reports showing constitutive activation of STAT1-STAT3 and IL-1β gene expression in AML and adult T-cell leukemia cell lines.3,4 15-19 Whether constitutive LILRE binding in AML or adult T-cell leukemia cells affects IL-1β gene expression remains to be elucidated.
We thank Dr Lyndsay Drayer for critically reading the manuscript.
Supported by the Bekales Foundation (Brussels, Belgium) and the Foundation for the Development of Research and Diagnostics of Hematopoietic Malignancies at Groningen.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Edo Vellenga, Division of Hematology, Department of Internal Medicine, Academic Hospital Groningen, Hanzeplein 1, 9713 GZ, PO Box 30001, Groningen, The Netherlands; e-mail:e.vellenga@int.azg.nl.