Abstract
The molecular pathways of normal myeloid differentiation, as well as the mechanisms by which oncogenes disrupt this process, remain poorly understood. A major limitation in approaching this problem has been the lack of suitable cell lines that exhibit normal, terminal, and synchronous differentiation in the absence of endogenous oncoproteins and in response to physiologic cytokines, and whose differentiation can be arrested by ectopically expressed human oncoproteins. This report describes clonal, granulocyte-macrophage colony-stimulating factor-dependent myeloid cell lines that exhibit these properties. The cell lines were established by conditional immortalization of primary murine marrow progenitors with an estrogen-regulated E2a/Pbx1-estrogen receptor fusion protein. Clones were identified that proliferated as immortalized blasts in the presence of estrogen, and that exhibited granulocytic, monocytic, or bipotential (granulocytic and monocytic) differentiation on estrogen withdrawal. Differentiation was normal and terminal as evidenced by morphology, cell surface markers, gene expression, and functional assays. The differentiation of the cells could be arrested by heterologous oncoproteins including AML1/ETO, PML/RARα, PLZF/RARα, Nup98/HoxA9, and other Hox proteins. Furthermore, the study examined the effects of cooperating oncoproteins such as Ras or Bcr/Abl, which allowed for both factor-independent proliferation and differentiation, or Bcl-2, which permitted factor-independent survival but not proliferation. These myeloid cell lines provide tools for examining the biochemical and genetic pathways that accompany normal differentiation as well as a system in which to dissect how other leukemic oncoproteins interfere with these pathways.
Introduction
In normal hematopoietic progenitors, a program of specific gene expression orchestrates commitment and differentiation to mature cells of multiple different lineages. In acute leukemias, however, oncoproteins interfere with this genetic program, resulting in the unregulated proliferation of cells that no longer retain the capacity to differentiate normally.1 This is particularly evident in acute myeloid leukemias (AMLs), and many known myeloid oncoproteins can block the differentiation of normal progenitors cultured in vitro in granulocyte-macrophage colony-stimulating factor (GM-CSF) or interleukin-3 (IL-3). However, neither the genetic events that underlie normal myeloid differentiation nor the mechanism through which leukemic oncoproteins interfere with execution of this program are well understood.
Primary cells and myeloid cell lines offer useful, but limited, models to approach these questions. Although primary marrow progenitors demonstrate normal granulocytic and monocytic differentiation in IL-3 or GM-CSF, one is limited by the scarcity of cells, the difficulty in isolating homogenous populations, and the inability to verify expression of nontransforming oncoproteins. Useful myeloid cell lines that demonstrate inducible differentiation in response to changes in cytokines include FDCPmixA4 (GM-CSF + granulocyte colony-stimulating factor [G-CSF] + macrophage colony-stimulating factor [M-CSF]),2 32Dcl3 (G-CSF),3 M1-AML (IL-6),4 and FDB cells (GM-CSF),5 whereas those that respond to nonphysiologic stimuli include HL60 (high levels of retinoic acid [RA], 12-o-tetradecanoylphorbal 13-acetate [TPA], dimethyl sulfoxide [DMSO]),6,7 EML (GM-CSF and RA),8 MPRO (RA),9 NB4 (RA),10and U937 cells (RA, TPA, DMSO, or vitamin D3).11 Although these lines supply an unlimited number of clonal cells, most are limited by the fact that they contain undefined genetic changes such that their differentiation is often incomplete, asynchronous, or accompanied by cell death. Because they are already blocked to differentiation in response to either IL-3 or GM-CSF, it is unclear whether induction by other extrinsic factors proceeds through normal differentiation pathways. Furthermore, myeloid oncoproteins whose action it is to block differentiation induced by IL-3 or GM-CSF cannot be assayed in these prearrested cell lines.
Therefore, an optimal myeloid cell line model would (1) lack constitutive expression of interfering oncoproteins, (2) exhibit conditional and terminal differentiation in GM-CSF, and (3) be blocked in differentiation by common leukemic oncoproteins. Here we describe such a system, established by a conditional version of E2a/Pbx1, a fusion oncoprotein that results from the t(1;19) translocation of pediatric pre-B acute lymphocytic leukemia (ALL),12-14that blocks myeloid differentiation in murine marrow culture.15 E2a/Pbx1 was made conditional by inserting the hormone-binding domain (HBD) of the estrogen receptor (ER) between the E2a transactivation domains and the Pbx1 DNA-binding homeodomain.16 The E2a/Pbx1-ER fusion proteins were estrogen dependent at the level of transactivation and transformation, and, like wild-type E2a/Pbx1, were capable of arresting the differentiation of GM-CSF–dependent myeloid progenitors. Progenitor clones were established which, on removal of estrogen, differentiated to mature granulocytes, monocytes, or to both types of cells. Accompanying differentiation, the clones exhibited normal changes in cell surface markers, gene expression, and cellular function. Having established and characterized these conditionally immortalized myeloid progenitor clones, we further analyzed the effect, on differentiation, of other leukemic oncoproteins such as AML1/ETO, PML/RARα, and Nup98/HoxA9, as well as cooperating oncoproteins Ras, Bcr/Abl, and Bcl-2.
Materials and methods
Construction of estrogen-dependent versions of E2a/Pbx1
In human t(1;19) pre-B ALL, both E2a/Pbx1a (825 amino acids) and E2a/Pbx1b (742 amino acids) are expressed, differing only in residues C-terminal to the Pbx1 homeodomain. Sequences encoding residues 282 to 595 of the Gly400Val human ER, with flanking MluI restriction sites, were amplified by high-fidelity polymerase chain reaction (PCR; Pfu polymerase, Stratagene, La Jolla, CA). The Gly400Val mutant ER was used because the point mutation renders the receptor insensitive to the low levels of estrogen found in fetal bovine serum (FBS) as well as to the estrogenic effects of compounds such as phenol red.17 The estrogen-binding domain was inserted into 2 versions of E2aPbx1b (E2a/Pbx1Δ487-578 and E2a/Pbx1Δ487-623) that lack Pbx1 sequences N-terminal to the Pbx1 homeodomain (Figure1A) and that have unique MluI sites in place of the deleted residues,18 resulting in fusions designated EPΔ578ER and EPΔ623ER, respectively. EPΔ578ER and EPΔ623ER were subcloned into the murine stem cell virus (MSCV) retroviral vectors MSCVneo and MSCVpac.19
Estrogen-dependent forms of E2a/Pbx1 are produced by replacement of Pbx1 sequences with the ER HBD.
(A) EPΔ578ER and EPΔ623ER were created by internal fusion of the HBD (amino acids 282-595) of the Gly400Val mutant human ER. The HBD replaces Pbx1 sequences, upstream of the DNA-binding homeodomain, that are dispensable for the biochemical and transforming properties of E2a/Pbx1. (B) EPΔ578ER and EPΔ623ER demonstrate conditional transformation of NIH3T3 fibroblasts. NIH3T3 fibroblasts infected with empty vector (MSCV) or stably expressing E2a/Pbx1, EPΔ578ER, or EPΔ623ER, were assayed for density-dependent growth by culture in the presence or absence of 1 μM β-estradiol. (B, inset) Immunoblot (α-E2a antibody) analysis of E2a/Pbx1 and EPΔ623ER (and EPΔ578ER, data not shown) in NIH3T3 fibroblasts demonstrates equivalent protein expression in the presence and absence of estrogen. (C) Combined Hox-independent and Hox-dependent transcriptional activation was assayed in NIH3T3 fibroblasts by cotransfection of constructs encoding wild-type E2a/Pbx1, EPΔ578ER, EPΔ623ER (denoted Δ578ER, and Δ623ER, respectively) and the 6xTGATTGAT_luciferase reporter in the presence (solid bars) or absence (empty bars) of 1 μM β-estradiol. (C, inset) The concentration of β-estradiol required for maximal transactivation of EPΔ623ER in NIH3T3 fibroblasts was determined by measuring luciferase activity in transfected cells cultured over a range of β-estradiol concentrations. (D) Hox-independent transactivation was assayed on the 6xTGATTGAT_luciferase reporter in human pre-B Nalm-6 cells, which lack significant expression of endogenous Hox protein partners. (E) Hox partner-dependent cooperative transactivation was assayed in Nalm-6 cells on the 6xTGATTTAT_luciferase reporter by cotransfection of E2a/Pbx1 constructs with constructs encoding HoxC8 (E) or HoxA9 (F). Lanes are described in the legends accompanying each graph. In panels C-F, luciferase activity was first normalized to the internal Renilla luciferase transfection control, and the fold transactivation was determined in comparison to cells cotransfected with empty MSCV vector DNA and reporter plasmid. The results represent the average of duplicate or triplicate transfections performed 2 times on separate days.
Estrogen-dependent forms of E2a/Pbx1 are produced by replacement of Pbx1 sequences with the ER HBD.
(A) EPΔ578ER and EPΔ623ER were created by internal fusion of the HBD (amino acids 282-595) of the Gly400Val mutant human ER. The HBD replaces Pbx1 sequences, upstream of the DNA-binding homeodomain, that are dispensable for the biochemical and transforming properties of E2a/Pbx1. (B) EPΔ578ER and EPΔ623ER demonstrate conditional transformation of NIH3T3 fibroblasts. NIH3T3 fibroblasts infected with empty vector (MSCV) or stably expressing E2a/Pbx1, EPΔ578ER, or EPΔ623ER, were assayed for density-dependent growth by culture in the presence or absence of 1 μM β-estradiol. (B, inset) Immunoblot (α-E2a antibody) analysis of E2a/Pbx1 and EPΔ623ER (and EPΔ578ER, data not shown) in NIH3T3 fibroblasts demonstrates equivalent protein expression in the presence and absence of estrogen. (C) Combined Hox-independent and Hox-dependent transcriptional activation was assayed in NIH3T3 fibroblasts by cotransfection of constructs encoding wild-type E2a/Pbx1, EPΔ578ER, EPΔ623ER (denoted Δ578ER, and Δ623ER, respectively) and the 6xTGATTGAT_luciferase reporter in the presence (solid bars) or absence (empty bars) of 1 μM β-estradiol. (C, inset) The concentration of β-estradiol required for maximal transactivation of EPΔ623ER in NIH3T3 fibroblasts was determined by measuring luciferase activity in transfected cells cultured over a range of β-estradiol concentrations. (D) Hox-independent transactivation was assayed on the 6xTGATTGAT_luciferase reporter in human pre-B Nalm-6 cells, which lack significant expression of endogenous Hox protein partners. (E) Hox partner-dependent cooperative transactivation was assayed in Nalm-6 cells on the 6xTGATTTAT_luciferase reporter by cotransfection of E2a/Pbx1 constructs with constructs encoding HoxC8 (E) or HoxA9 (F). Lanes are described in the legends accompanying each graph. In panels C-F, luciferase activity was first normalized to the internal Renilla luciferase transfection control, and the fold transactivation was determined in comparison to cells cotransfected with empty MSCV vector DNA and reporter plasmid. The results represent the average of duplicate or triplicate transfections performed 2 times on separate days.
Cell culture
Cells were maintained in a 37°C humidified incubator with 5% CO2. NIH3T3 murine fibroblasts and 293T human embryonic kidney cells were grown in Dulbecco modified Eagle medium (DMEM) with 4.5 g/L glucose, 10% FBS (Gemini, Woodland, CA), and penicillin-streptomycin-glutamine (Gibco BRL, Rockville, MD). Nalm-6 pre-B cells and primary murine hematopoietic cells were grown in RPMI 1640 with 10% FBS and penicillin-streptomycin-glutamine. GM-CSF–dependent cells were maintained by the addition of 1:100 conditioned media (about 10 ng/mL GM-CSF) from a B16 melanoma cell line stably transfected with a murine GM-CSF construct. Beta-estradiol (E2758, Sigma, St Louis, MO), where applicable, was added to the medium at a final concentration of 1 μM from a 10 000 × stock in 100% ethanol.
Analysis of transcriptional activation
Luciferase reporter constructs have been previously described.18 Both the 6xTGATTGAT (PRS, Pbx Responsive Sequence) and 6xTGATTTAT are cloned upstream of a minimal Fos promoter driving luciferase expression in the pGL3 basic luciferase vector (Promega, Madison, WI). NIH3T3 fibroblasts were transfected (Superfect, Qiagen, Valencia, CA) in 6-well plates according to manufacturer's specifications. The transfection mixture included 1 μg expression plasmid, 1 μg of the 6xPRS luciferase reporter plasmid, and 0.1 μg of a Renilla luciferase plasmid DNA as internal transfection control. Nalm-6 cells were electroporated in a final volume of 200 μL at a concentration 5 × 106cells/mL. Each electroporation reaction contained 6 μg of each appropriate expression plasmid, 6 μg Hox-protein plasmid where applicable, 6 μg of the luciferase reporter construct, and 0.5 μg of the Renilla luciferase control plasmid DNA. In both cases, cells were harvested at 48 hours and luciferase activity measured using the Dual-Luciferase Reporter Assay System (Promega). Fold transactivation was calculated relative to control cells transfected with empty vector DNA.
Analysis of density-dependent growth
The NIH3T3 fibroblasts were infected with helper-free retrovirus encoding wild-type and inducible versions of E2a/Pbx1. Stably expressing cells, as well as cells transduced with empty vector virus, were selected 7 days in 1 mg/mL G418 in the absence of estrogen. Equivalent numbers of cells were plated in triplicate into 60-mm dishes in the presence and absence of 1 μM β-estradiol. Half-media changes were performed every 2 days and the total number of live, adherent cells was determined after 14 days. Fold density was calculated in comparison to the number of cells transduced with empty-vector retrovirus.
Immunoblot analysis of E2a/Pbx1 proteins
Protein from 5 × 104 cells was resolved by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membrane (Pall, Ann Arbor, MI). Membranes were blocked in tris-buffered saline-Tween (TBS-T) with 5% nonfat milk, and immunoblotting was performed using monoclonal mouse anti–human E2a (E12/E47) antibody (G193-86, Pharmingen, San Diego, CA). Primary antibody was detected with a horseradish peroxidase (HRP)–linked rabbit antimouse secondary antibody (NEB, Beverly, MA). Bound antibody was detected using SuperSignal West Pico Chemiluminescence (Pierce, Rockford, IL).
Retroviral infection of primary murine marrow progenitors
We isolated bone marrow mononuclear cells on a Ficoll-Paque gradient (Pharmacia, Piscataway, NJ) following harvest from the femurs and tibia of female Balb/c mice injected intraperitoneally with 5-fluorouracil (150 mg/kg) 5 days prior to harvest. Marrow progenitors were purified by negative selection of lineage-positive cells on a magnetic column with a murine progenitor antibody cocktail (Stemcell Technologies, Vancouver, BC, Canada). Progenitors were prestimulated for 48 hours in Iscoves modified Dulbecco medium (IMDM) containing 15% FBS, 50 ng/mL stem cell factor (SCF), 25 ng/mL IL-3, and 25 ng/mL IL-6.
Helper-free retrovirus was prepared by calcium phosphate transfection (Invitrogen, Carlsbad, CA) of 293T cells with MSCV retroviral constructs and an ecotropic packaging construct. Then, 25 000 marrow progenitors were infected with 1 mL retroviral supernatant by spinoculation (2500g, 2 hours, 22°C) in the presence of Lipofectamine (1:1000, Gibco BRL). Following spinoculation, the cells were cultured in RPMI media containing GM-CSF and 1 μM β-estradiol as described above. Immortalized myeloid progenitors were enriched by the serial passage of nonadherent cells over the course of approximately 3 weeks.
Single-cell clones were prepared by limiting dilution.E2a/Pbx1-mediated conditionalmyeloid (ECoM)–GM and ECoM-G clones were established from both populations immortalized by EPΔ578ER and EPΔ623ER, whereas the ECoM-M cell line was derived from a population immortalized by EPΔ623ER (see “Results”).
Wright-Giemsa staining, nitroblue tetrazolium reduction assay, nonspecific esterase assay, and phagocytosis assay
Wright-Giemsa staining (3 minutes Wright, 9 minutes 20% Giemsa) was performed on cytocentrifuge preparations of cells (Thermo Shandon, Pittsburgh, PA).
Activity of NADPH oxidase was assayed by nitroblue tetrazolium (NBT) reduction. The 2.5 × 105 cells in 200 μL growth media were added to 800 μL 0.125% NBT (Sigma) in the presence of 2 × 10−7 M TPA. The cells were incubated 25 minutes at 37°C, washed, cytocentrifuged onto a coverslip, counterstained with safranin O (5 minutes, 0.5% safranin O in 20% ETOH), and examined for the dark purple deposits indicative of a positive NBT reduction.
Nonspecific esterase (NSE) activity was assayed as previously described.20 Briefly, cytocentrifuge preparations of cells were fixed in cold phosphate-acetone-formaldehyde and stained with α-naphthyl acetate and fast blue BB salt in a Tris-maleate buffer. Cells were washed and counterstained with neutral red (1% for 5 minutes). All reagents were purchased from Sigma.
Phagocytosis was assayed using fluorescein isothiocyanate (FITC)–labeled heat-killed Escherichia coliBioparticles (Molecular Probes, Eugene, OR). The 107BioParticles and 106 cells were incubated for 1 hour at 37°C with shaking. Following cytocentrifugation onto Superfrost Plus slides (Fisher, Pittsburgh, PA), the nuclei were counterstained with bisBenzimide (Hoechst 33258, Sigma), and the F-actin counterstained with TRITC-phalloidin (Sigma). Images were captured with a DeltaVision deconvolution microscope system and the data sets were deconvolved and analyzed using SoftWorx software (both from Applied Precision, Issaquah, WA).
Flow cytometric analysis
The FITC-labeled monoclonal antibodies GR-1(Ly6G) and Mac-1(CD11b) were purchased from Pharmingen and F4/80 from Serotec (Kidlington, Oxford, United Kingdom). Then, 106 cells were labeled 30 minutes at 4°C in phosphate-buffered saline (PBS)/1% FBS/0.1% NaN3, washed, and resuspended in the same buffer with 2 μg/mL propidium iodide. Flow cytometry data were acquired with the program CELLQuest on either a FACScan (Becton Dickinson, San Jose CA) or FACSCalibur bench-top flow cytometer attached to a Macintosh G3 computer. Live cells were gated for analysis by forward and side scatter signals and lack of propidium iodide staining.
Northern blot analysis
Cells were maintained in 15-cm dishes in the presence or absence of 1 μM β-estradiol. Cytoplasmic RNA was purified (RNEasy, Qiagen), and a portion subjected to polyA selection (Oligotex, Qiagen). Then, 15 μg cytoplasmic RNA or 5 μg polyA-selected RNA was resolved by formaldehyde-1% agarose gel electrophoresis and the RNA transferred to a positively charged nylon membrane (GeneScreen Plus, NEN, Boston, MA). Membranes were dried for 2 hours at 80°C in a vacuum oven prior to hybridization. The 32PdCTP-labeled DNA probes were prepared from 100 ng DNA subjected to random-hexamer oligolabeling (Pharmacia). Hybridization in Ultrahyb (Ambion, Austin, TX) and washing were carried out at 42°C according to the manufacturer's protocols. In general, probes for transcription factors were hybridized to blots containing polyadenylated messenger RNA (mRNA), and probes for all other gene products were hybridized to blots containing total RNA.
Cell-cycle analysis by DNA content
Washed cells were resuspended in PBS, fixed by the addition of ice-cold 100% ethanol to a final concentration of 70%, and stored overnight at 4°C. Cells were incubated with RNase A (0.1 mg/mL, 15 minutes, 37°C), followed by propidium iodide staining (50 μg/mL, 30 minutes, 4°C). DNA content (FL2-Area) was measured using the Doublet Discrimination Module (DDM) set on FL2, and MulticycleAV, Version 3.11 (Phoenix Flow Systems, San Diego, CA) was used to calculate cell-cycle phases.
Retroviral infection of ECoM-G cells with heterologous oncoproteins
Helper-free retrovirus was generated from wild-type E2a/Pbx1, Hoxa7, Hoxa9, Nup98/HoxA9, Hoxb8, AML1/ETO, PML/RARα, and PLZF/RARα constructs cloned into the multiple cloning site of the MSCVneo and MSCVpac retroviral vectors. ECoM-G cells (250 000) from 3 different clones were transduced by spinoculation (as described above) and selected 5 days in G418 (1 mg/mL) or puromycin (1 μg/mL). The cells were washed and plated in media without estrogen. Only those cells that were capable of continued and indefinite proliferation in the absence of estrogen were expanded to generate RNA for Northern analysis.
Results
Estrogen-dependent forms of E2a/Pbx1 are produced by replacement of Pbx1 sequences with the ER HBD
Fusion of the ER HBD to the N- or C-terminus of full-length E2a/Pbx1 produced proteins that were estrogen-dependent at the level of transcription, but were unstable (expressed at only 10%-30% the level of wild type), and failed to exhibit estrogen-dependent transformation (Xinyu Fu, unpublished observations, August 1997). Therefore, we replaced Pbx1 sequences N-terminal to the homeodomain, which are dispensable for the biochemical and transforming properties of E2a/Pbx1,18 with the ER HBD (as described in “Materials and methods”), creating EPΔ578ER and EPΔ623ER proteins (Figure1A). As shown below, EPΔ578ER and EPΔ623ER are stable, are constitutively expressed, and demonstrate estrogen-dependent biochemical and oncogenic functions.
Wild-type E2a/Pbx1 exhibits 2 forms of transactivation that can be separately assessed on different luciferase reporter constructs.18 Transactivation through TGATTGAT motifs is accomplished by both a Hox-partner–dependent mechanism and a Hox-independent mechanism likely mediated by E2a/Pbx1 dimerization, whereas transactivation through TGATTTAT motifs is strictly Hox-dependent. Estrogen responsiveness of EPΔ578ER and EPΔ623ER was first assayed in NIH3T3 fibroblasts, which express endogenous Hox proteins, using the 6xTGATTGAT_luciferase reporter (Figure1C). Hox-independent activation through TGATTGAT motifs was next assessed in human Nalm-6 pre-B cells, which lack significant levels of Hox proteins21 (Figure 1D). Hox-dependent cooperative transactivation was assayed by transfection of E2a/Pbx1 and Hoxc8 (Figure 1E) or Hoxa9 (Figure 1F) constructs into Nalm-6 cells along with the 6xTGATTTAT_luciferase reporter. Collectively, the strikingly similar patterns of transactivation in these experiments demonstrated that activation by wild-type E2a/Pbx1 is unaffected by estrogen, and that EPΔ578ER and EPΔ623ER can participate in both Hox-dependent and Hox-independent transcriptional activation in a strictly estrogen-dependent manner. The weaker transactivation of EPΔ578ER, as compared to EPΔ623ER, parallels the weaker cooperativity of the parental EPΔ578 protein with Hox partners in both DNA-binding and transactivation assays.18 Estrogen (β-estradiol at 1 μM) was sufficient for inducing maximal transactivation (Figure 1C, inset) and estrogen-induced luciferase activity could be detected within 45 minutes, with maximal activity attained at 8 hours (data not shown).
E2a/Pbx1 induces proliferation and focus-formation in monolayer cultures of NIH3T3 fibroblasts.22 Stably expressing NIH3T3 cells were established to assess the transforming function of EPΔ578ER and EPΔ623ER proteins in relation to wild-type E2a/Pbx1. In density-dependent proliferation assays, wild-type E2a/Pbx1 induced an approximate 3.5-fold increase in total cell number (Figure 1B). In the absence of estrogen, neither EPΔ578ER nor EPΔ623ER stimulated cell proliferation, whereas the addition of estrogen induced a 1.2-fold and 4.5-fold increase in cell number, respectively. This is consistent with the relative transforming strength of EPΔ578 and EPΔ623 proteins.18 Upon removal of estrogen, cells conditionally transformed by EPΔ578ER or EPΔ623ER reverted to the growth densities exhibited by parental NIH3T3 fibroblasts, demonstrating that the transformation induced by E2a/Pbx1 is fully reversible (data not shown). Failure to induce proliferation in the absence of estrogen was not due to diminished stability or expression because the wild-type E2a/Pbx1, EPΔ578ER (data not shown), and EPΔ623ER proteins were of equal abundance in the presence and absence of estrogen (Figure 1B, inset). The doublet corresponds to 2 versions of the fusion protein that arise from alternative splicing within the E2AmRNA.18
EPΔ578ER and EPΔ623ER conditionally immortalize factor-dependent primary murine myeloid progenitors
Expression of E2a/Pbx1 in primary murine marrow immortalizes GM-CSF–dependent myeloid progenitors,15 whereas uninfected cells differentiate into short-lived granulocytes and adherent monocytes. Both EPΔ578ER (12 of 12 cultures) and EPΔ623ER (6 of 6) immortalized progenitors as efficiently as E2a/Pbx1, but only in the presence of 1 μM estrogen. Wild-type E2a/Pbx1-, EPΔ578ER-, and EPΔ623ER-immortalized progenitors were phenotypically identical (EPΔ578ER shown in Figure 2A). All cell lines were dependent on exogenous GM-CSF, which could be substituted with IL-3. The cells were unresponsive to G-CSF, or to M-CSF, undergoing apoptotic cell death within 24 hours when cultured in these, or in the absence of cytokines (data not shown).
EPΔ578ER-immortalized progenitors differentiate to granulocytes and monocytes upon the removal of estrogen.
Wright-Giemsa–stained cytocentrifuge preparations were prepared from samples of EPΔ578ER progenitors cultured in the presence (A) or absence (B) of estrogen for 5 days. (C) Cell surface staining with FITC-labeled GR-1 (Ly6G), Mac-1 (CD11b), and F4/80 of EPΔ578ER progenitors cultured in the presence or absence of estrogen for 5 days was determined by flow cytometry. Profiles of unstained cells or cells stained with B220 antibody were performed to ensure that observed changes were not due to increased autofluorescence or to nonspecific antibody binding.
EPΔ578ER-immortalized progenitors differentiate to granulocytes and monocytes upon the removal of estrogen.
Wright-Giemsa–stained cytocentrifuge preparations were prepared from samples of EPΔ578ER progenitors cultured in the presence (A) or absence (B) of estrogen for 5 days. (C) Cell surface staining with FITC-labeled GR-1 (Ly6G), Mac-1 (CD11b), and F4/80 of EPΔ578ER progenitors cultured in the presence or absence of estrogen for 5 days was determined by flow cytometry. Profiles of unstained cells or cells stained with B220 antibody were performed to ensure that observed changes were not due to increased autofluorescence or to nonspecific antibody binding.
Removal of estrogen from polyclonal populations of EPΔ578ER- or EPΔ623ER-immortalized myeloblasts evoked synchronous morphologic differentiation to mature neutrophils (about 80%), characterized by segmented nuclei and lightly staining cytoplasm, and to mature macrophages (about 20%), characterized by their larger size and oval nuclei (Figure 2B). Granulocytes were fully differentiated within 5 to 6 days and survived for an additional 24 to 48 hours, whereas monocytes were adherent and stopped proliferating after 7 to 9 days.
The expression of cell surface markers on one population of EPΔ578ER-immortalized cells was characterized by flow cytometry. In the presence of estrogen, cells did not stain with GR-1 (Ly6G) or F4/80 and stained weakly with Mac-1 (CD11b), whereas removal of estrogen resulted in dramatic up-regulation of these myeloid differentiation markers (Figure 2C), paralleling the morphologic differentiation to granulocytes and monocytes. Functional NADPH oxidase activity, as evidenced by dark blue intracellular deposits following the NBT reduction assay, was observed in fewer than 1% of progenitors in the presence of estrogen and in more than 99% of cells cultured 7 days in the absence of estrogen (data not shown).
Conditionally immortalized myeloid progenitors initiate a transcriptional program of differentiation following withdrawal of estrogen
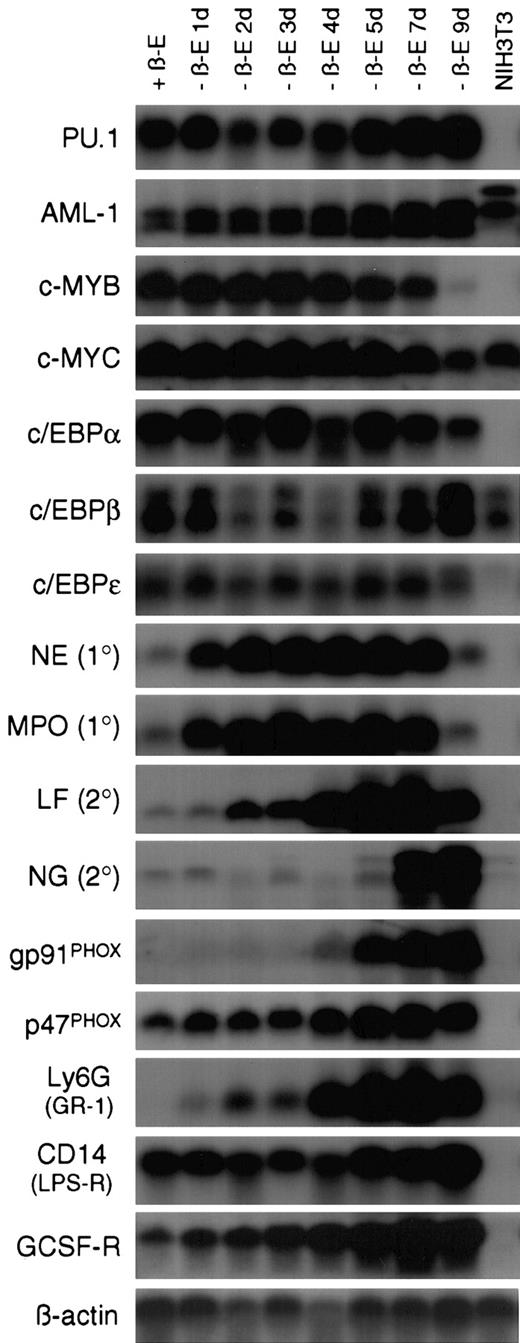
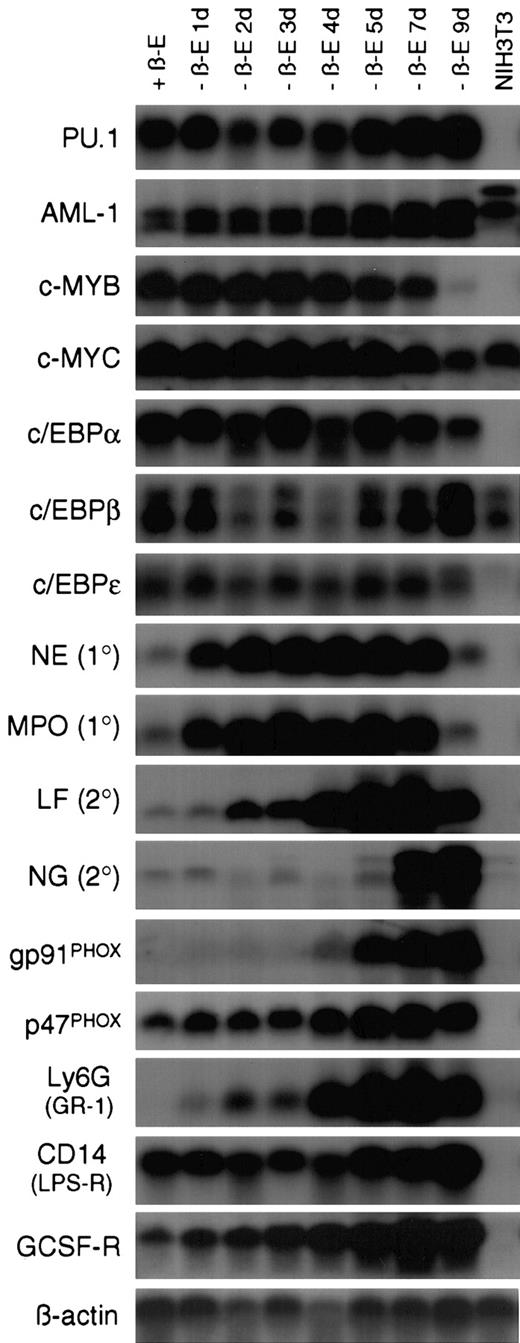
In some models of myeloid differentiation, phenotypic changes are not necessarily accompanied by normal expression of terminal differentiation markers.23 To address this concern, Northern blot analysis was performed on a 9-day time course of RNA samples collected from EPΔ578ER-immortalized cells differentiated in the absence of estrogen (Figure 3). The expression of genes encoding master transcriptional regulators, primary and secondary granule proteins, components of the NADPH oxidase complex, and other markers of myeloid differentiation was examined (reviewed by Tenen et al,24 Yamanaka et al,25Zhang et al,26 and Ward et al27).
Withdrawal of estrogen from EPΔ578ER progenitors initiates a transcriptional program of myeloid differentiation.
Northern blot analysis was performed on RNA samples prepared from EPΔ578ER progenitors maintained in estrogen (+β-E) as well as cultured out of estrogen (−β-E) for 1 to 9 days. RNA from NIH3T3 fibroblasts was used as a nonmyeloid control. In general, transcription factor probes were hybridized to blots prepared from polyA+-RNA, whereas other probes were hybridized to blots prepared from total RNA. Names of the probes are indicated to the left of each blot. Equivalent loading was verified by hybridization of a β-actin probe.
Withdrawal of estrogen from EPΔ578ER progenitors initiates a transcriptional program of myeloid differentiation.
Northern blot analysis was performed on RNA samples prepared from EPΔ578ER progenitors maintained in estrogen (+β-E) as well as cultured out of estrogen (−β-E) for 1 to 9 days. RNA from NIH3T3 fibroblasts was used as a nonmyeloid control. In general, transcription factor probes were hybridized to blots prepared from polyA+-RNA, whereas other probes were hybridized to blots prepared from total RNA. Names of the probes are indicated to the left of each blot. Equivalent loading was verified by hybridization of a β-actin probe.
In both the presence and absence of estrogen, progenitors demonstrated stable expression of the Ets-family member PU.1, as well as members of the CCAAT/enhancer-binding protein (c/EBPα, β, and ε) family of transcription factors, all of which have been shown to be essential for normal myeloid development.28,29 c-Myb and c-Myc showed characteristic down-regulation late in differentiation, whereas AML1 showed marked (about 5-fold) up-regulation following the removal of estrogen. Based on this pattern, it is unlikely that E2a/Pbx1 establishes differentiation arrest by suppressing PU.1 orc/EBP gene expression, or by maintaining high levels of c-Myb, which has been shown to prevent differentiation.30It is unclear whether the up-regulation of AML1 following the removal of estrogen represents the normal up-regulation of AML1 during differentiation31 or whether E2a/Pbx1 might be suppressing its expression.
The sequential expression of primary and secondary granule genes also accompanied differentiation. Primary granule genes neutrophil elastase (NE) and myeloperoxidase (MPO) were rapidly up-regulated following estrogen withdrawal. This rapid up-regulation is important in the context of elucidating differentiation arrest by E2a/Pbx1, because it might allow for the identification, within the NE and MPO promoters, of transcriptional activators that are not produced, or transcriptional repressors that are not eliminated, in the presence of active E2a/Pbx1. Such genetic defects will provide molecular handles to identify the direct E2a/Pbx1 target genes that mediate differentiation arrest. Secondary granule genes lactoferrin (LF) and neutrophil gelatinase (NG) were not expressed in the myeloblasts and were activated at day 2 and day 7 of differentiation, respectively. Examination of 2 of the subunits of the NADPH oxidase complex revealed that p47PHOXwas constitutively expressed, whereas gp91PHOX was not expressed until day 4 of differentiation. The regulated expression of gp91PHOX is consistent with the timing of robust functional NADPH oxidase activity as assayed by NBT reduction. The cells also demonstrated transcriptional up-regulation of genes encoding cell surface receptors Ly6G (GR-1), CD14 (lipopolysaccharide receptor), and the G-CSF receptor (G-CSF-R). Ly6G expression paralleled the pattern of GR-1 staining seen by flow cytometric analysis (Figure4) and the low level of G-CSF-R expression in progenitors is consistent with their G-CSF unresponsiveness.
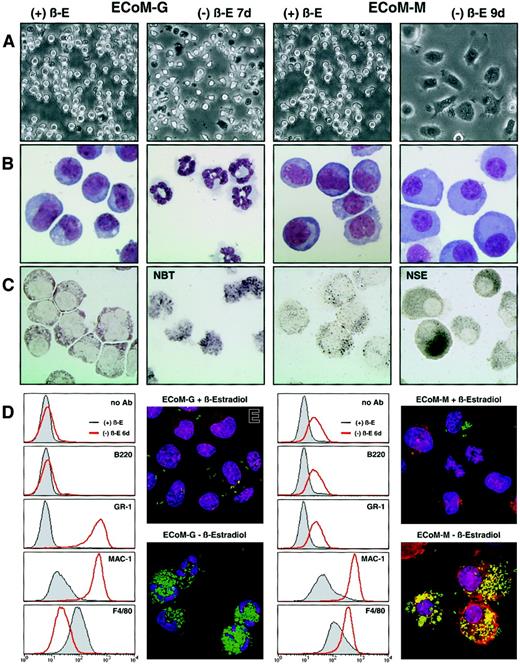
The EPΔ578ER and EPΔ623ER proteins immortalize progenitors that exhibit strict granulocytic or monocytic differentiation.
Single-cell progenitor's clones were generated that exhibited strict granulocytic or monocytic differentiation upon removal of estrogen, and they were designated ECoM-G (E2a/Pbx1-mediatedConditional Myeloid Differentiation-Granulocytic) and ECoM-M (Monocytic), respectively. (A) Light micrographs and (B) Wright-Giemsa stains were prepared from ECoM-G and ECoM-M cells in the presence (+β-E) and absence (−β-E) of estrogen as indicated above the photographs. (C) NBT reduction and NSE staining. The ECoM-G cells were functionally characterized based on NADPH oxidase activity (NBT reduction), which is up-regulated during myeloid maturation. The ECoM-M cells were analyzed for NSE activity, which is specifically expressed in mature macrophages. (D) Flow cytometric analysis of the cell surface staining with FITC-labeled Ly6G (GR-1), CD11b (Mac-1), and F4/80 on the ECoM-G and ECoM-M cells grown in the presence or absence of estrogen for 7 days. Note that the observed shift in GR-1 staining in the ECoM-M clone is due to increased autofluorescence. (E) Phagocytosis assay. The ability of the ECoM cells to internalize FITC-labeledE coli was evaluated in the presence and absence of estrogen. Hoechst and rhodamine-phalloidin were used to stain the nuclei, and polymerized actin, respectively.
The EPΔ578ER and EPΔ623ER proteins immortalize progenitors that exhibit strict granulocytic or monocytic differentiation.
Single-cell progenitor's clones were generated that exhibited strict granulocytic or monocytic differentiation upon removal of estrogen, and they were designated ECoM-G (E2a/Pbx1-mediatedConditional Myeloid Differentiation-Granulocytic) and ECoM-M (Monocytic), respectively. (A) Light micrographs and (B) Wright-Giemsa stains were prepared from ECoM-G and ECoM-M cells in the presence (+β-E) and absence (−β-E) of estrogen as indicated above the photographs. (C) NBT reduction and NSE staining. The ECoM-G cells were functionally characterized based on NADPH oxidase activity (NBT reduction), which is up-regulated during myeloid maturation. The ECoM-M cells were analyzed for NSE activity, which is specifically expressed in mature macrophages. (D) Flow cytometric analysis of the cell surface staining with FITC-labeled Ly6G (GR-1), CD11b (Mac-1), and F4/80 on the ECoM-G and ECoM-M cells grown in the presence or absence of estrogen for 7 days. Note that the observed shift in GR-1 staining in the ECoM-M clone is due to increased autofluorescence. (E) Phagocytosis assay. The ability of the ECoM cells to internalize FITC-labeledE coli was evaluated in the presence and absence of estrogen. Hoechst and rhodamine-phalloidin were used to stain the nuclei, and polymerized actin, respectively.
E2a/Pbx1 arrests myeloid differentiation in a DNA binding–dependent manner, possibly in cooperation with a Hox partner, or in larger complexes including other Pbx, Hox, and Meis proteins.32Although many Hox genes, especially of the A paralog, are implicated in myeloid differentiation,33 no expression ofHoxa5, Hoxa7, or Hoxa9 was detected in progenitors, though each transcript was clearly observed in NIH3T3 fibroblasts (data not shown). Furthermore, expression of Meis1, Meis2, and Meis3 was not detected in the myeloid progenitors, although their transcripts were also easily detected in NIH3T3 fibroblasts (data not shown).
Clones of EPΔ578ER and EPΔ623ER-immortalized progenitors execute strict granulocytic or monocytic differentiation in GM-CSF
Conditional immortalization by EPΔ578ER and EPΔ623ER allowed us to examine whether all progenitors, seemingly identical by morphologic criteria, were committed to identical patterns of differentiation. Populations of progenitors were cloned, clonality was verified by retroviral integration analysis using Southern blotting (data not shown), and phenotypes were examined by light microscopy (Figure 4A) and Wright-Giemsa staining (Figure 4B) 5 days after the removal of estrogen. Three types of clones exhibiting E2a/Pbx1-mediated conditional myeloid differentiation were identified: bipotential clones that differentiated to both granulocytes and monocytes (ECoM-GM), clones exhibiting restricted granulocytic differentiation (ECoM-G), and one clone exhibiting restricted monocytic differentiation (ECoM-M).
Within 4 days, ECoM-G cells differentiated homogeneously to granulocytes, whereas ECoM-M cells required 7 days for quantitative monocytic differentiation. Both ECoM-G and ECoM-M cells acquired functional NADPH oxidase activity (Figure 4C and data not shown, respectively), and the ECoM-M cells up-regulated NSE, a characteristic marker of normal macrophage development (Figure 4C). The ECoM-G cells showed a dramatic increase in GR-1 and Mac-1 staining 7 days after estrogen withdrawal, whereas the intensity of F4/80 staining was significantly reduced (Figure 4D). In contrast, the ECoM-M cells did not express GR-1 (the observed shift is due to increased autofluorescence), but were positive for, and showed increasing staining of, both Mac-1 and F4/80 during monocytic differentiation. Both ECoM-G and ECoM-M cells became functionally phagocytic, as demonstrated by the ability to engulf FITC-labeled E coliBioParticles (Figure 4E).
Phenotypic changes following the removal of estrogen were accompanied by reduced proliferation and G1 cell-cycle arrest as evidenced by DNA content analysis. Although both ECoM-G and ECoM-M progenitors in estrogen had a high S-phase fraction (G148%/G2 8%/S 44% and G1 35%/G213%/S 52%, respectively), the majority of the cells had accumulated in G1 following 7 days of differentiation in the absence of estrogen (G1 96%/G2 2%/S 2% and G1 83%/G2 11%/S 6%, respectively).
ECoM-G and ECoM-M cells exhibit lineage-specific and differentiation-specific gene expression
The parallel models of granulocyte and monocyte differentiation permitted the examination of lineage-specific gene expression (Figure5). ECoM-G and ECoM-M cells recapitulated well-established patterns of myeloid gene expression including the down-regulation of c-Myb and c-Myc, and the up-regulation of Egr-1, macrosialin, c-Fos, neutrophil collagenase (NC), c-Fms, and the macrophage scavenger receptor SRA-1. Zinc-finger protein and transcriptional repressor Gfi-1 was down-regulated in both ECoM-G and ECoM-M clones, whereas expression of family member Gfi-1B was down-regulated only in the ECoM-M clone (data not shown), similar to a previous report in which its down-regulation accompanied the cell-cycle arrest and up-regulation of p21 during IL-6–induced monocytic differentiation of M1-AML cells.34 Ear-2 was stably expressed throughout differentiation of both ECoM-G and ECoM-M cells in contrast to a previous report in which it was shown to be down-regulated during the G-CSF–induced granulocytic differentiation in 32Dcl3 cells and hypothesized to bind and inhibit AML1 activity.35 Similarly, Ets family member Fli-1 was also stably expressed. Expression of Fli-1 has been reported in human T-cell, B-cell, and myeloid leukemia cell lines36 and was shown to be critical for hematopoiesis in a Fli-1–deficient mouse.37 Expression of the aldoketo reductase mAKRa, whose function has not yet been elucidated, was restricted to the ECoM-G clone and was stable throughout differentiation in contrast to a previous report that showed decreased expression following the all-trans-RA (atRA)–induced granulocytic differentiation of EML-C1 and MPRO cells.38 Expression of Ets-2 was restricted to the ECoM-G clone, which was unexpected given previous reports of Ets-2 expression in normal and transformed macrophages.39
ECoM-G and ECoM-M clones exhibit lineage- and differentiation-specific patterns of gene expression.
Northern blot analysis of myeloid genes was performed on total RNA samples from ECoM-G and ECoM-M clones cultured in the presence (E2) or in the absence of estrogen for the indicated number of days. RNA from NIH3T3 fibroblasts was used as a nonmyeloid control. Names of the probes are indicated to the left of each blot. Equivalent loading was verified by hybridization of a β-actin probe.
ECoM-G and ECoM-M clones exhibit lineage- and differentiation-specific patterns of gene expression.
Northern blot analysis of myeloid genes was performed on total RNA samples from ECoM-G and ECoM-M clones cultured in the presence (E2) or in the absence of estrogen for the indicated number of days. RNA from NIH3T3 fibroblasts was used as a nonmyeloid control. Names of the probes are indicated to the left of each blot. Equivalent loading was verified by hybridization of a β-actin probe.
Overall, the differences in gene expression between ECoM-G and ECoM-M cells demonstrate that E2a/Pbx1 prevents differentiation but not lineage definition, and that these cell lines will be useful models to identify differences between granulopoiesis and monopoiesis.
Heterologous oncoproteins arrest the differentiation of ECoM-G cells when E2a/Pbx1 is inactivated
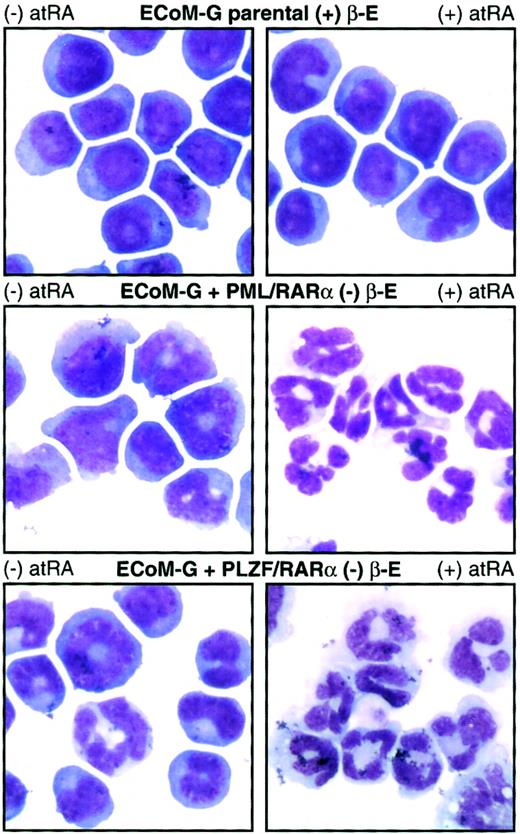
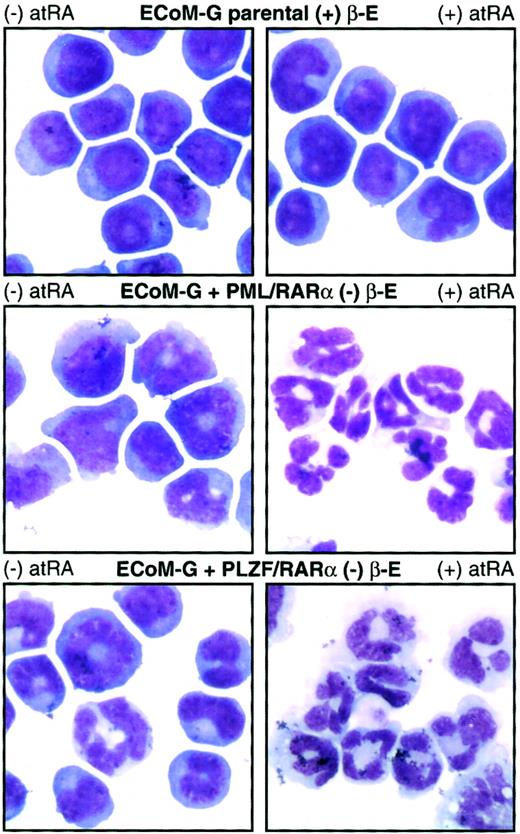
Although the ECoM cells are ideal for studying the differentiation block established by E2a/Pbx1, we wished to assay their utility in studying myeloid differentiation arrest by other oncoproteins. We introduced heterologous oncoproteins into ECoM-G clones by retroviral infection, verified expression by immunoblot analysis, and assayed for the ability to re-establish differentiation arrest following estrogen withdrawal. AML1/ETO, PML/RARα, PLZF/RARα, Hoxa9, Hoxb8, and wild-type E2a/Pbx1 prevented the granulocytic differentiation of specific clones, permitting their continued indefinite proliferation in the presence of GM-CSF. The resultant pattern of gene expression was consistent with differentiation arrest by the new oncoprotein (Figure6). Hoxa9, for example, permitted up-regulation of NE, gp91PHOX, Ly6G, and the G-CSF-R to levels similar to those of primary marrow immortalized by Hoxa9 alone.40 Similarly, whereas AML1/ETO permitted up-regulation of gp91PHOX and Ly6G, the expression of NE and G-CSF-R, whose activation requires AML1, remained low. This pattern is consistent with the dominant-negative function of AML1/ETO on AML1-responsive promoters.41 42 ECoM-G clones arrested in differentiation by PML/RARα or PLZF/RARα showed RA sensitivity not seen in the parental cells, providing mechanistic evidence of the function of the second oncoprotein. Although parental cells were completely unresponsive to treatment with 10 μM atRA, for 4 days, both the PML/RARα and PLZF/RARα derivatives stopped proliferating and underwent quantitative granulocytic differentiation (Figure 7, Table1). The ECoM-G cell lines can thus be used for analysis of the biochemical and genetic mechanisms by which oncoproteins prevent stage-specific myeloid differentiation.
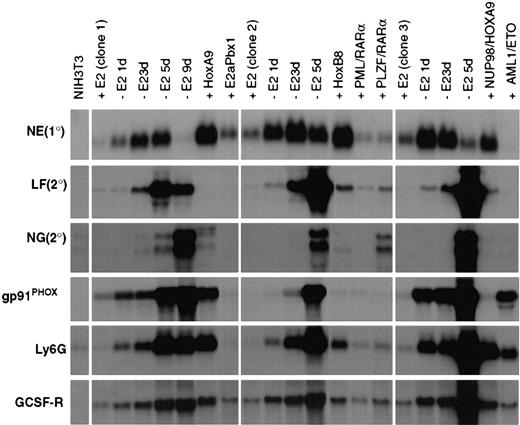
Gene expression in ECoM-G clones, whose differentiation arrest is re-established by heterologous oncoproteins, reveals profiles unique to the second oncoprotein.
Northern blot analysis of gene expression was performed on RNA samples from 3 ECoM-G clones cultured in the presence (+E2) and absence (−E2) of estrogen for the indicated number of days. RNA was also harvested from clones whose differentiation arrest in the absence of estrogen was re-established by Hoxa9, wild-type E2a/Pbx1, Hoxb8, PML/RARα, PLZF/RARα, Nup98/HoxA9, and AML1/ETO. These stable populations demonstrated continued and indefinite proliferation in the absence of estrogen and were cultured in the absence of estrogen for at least 4 to 5 weeks prior to the harvest of RNA. The heterologous oncoproteins were all cloned into MSCV-based vectors and introduced into the ECoM-G cells by retroviral infection. RNA from NIH3T3 fibroblasts was used as a nonmyeloid control.
Gene expression in ECoM-G clones, whose differentiation arrest is re-established by heterologous oncoproteins, reveals profiles unique to the second oncoprotein.
Northern blot analysis of gene expression was performed on RNA samples from 3 ECoM-G clones cultured in the presence (+E2) and absence (−E2) of estrogen for the indicated number of days. RNA was also harvested from clones whose differentiation arrest in the absence of estrogen was re-established by Hoxa9, wild-type E2a/Pbx1, Hoxb8, PML/RARα, PLZF/RARα, Nup98/HoxA9, and AML1/ETO. These stable populations demonstrated continued and indefinite proliferation in the absence of estrogen and were cultured in the absence of estrogen for at least 4 to 5 weeks prior to the harvest of RNA. The heterologous oncoproteins were all cloned into MSCV-based vectors and introduced into the ECoM-G cells by retroviral infection. RNA from NIH3T3 fibroblasts was used as a nonmyeloid control.
PML/RARα and PLZF/RARα arrest ECoM-G differentiation in the absence of estrogen and endow the cells with RA sensitivity.
Derivatives of one ECoM-G clone were established by retroviral infection with constructs encoding PML/RARα and PLZF/RARα followed by selection of immortalized cells in the absence of estrogen. Parental ECoM-G cells (maintained in estrogen) as well as PML/RARα-immortalized and PLZF/RARα-immortalized cells (in the absence of estrogen) were treated for 4 days with 10 μM atRA. Table 1presents data from Wright-Giemsa stains of cytocentrifuge preparations, as well as the percent of mitotic progenitors and mature granulocytes compiled from a minimum of 200 cells.
PML/RARα and PLZF/RARα arrest ECoM-G differentiation in the absence of estrogen and endow the cells with RA sensitivity.
Derivatives of one ECoM-G clone were established by retroviral infection with constructs encoding PML/RARα and PLZF/RARα followed by selection of immortalized cells in the absence of estrogen. Parental ECoM-G cells (maintained in estrogen) as well as PML/RARα-immortalized and PLZF/RARα-immortalized cells (in the absence of estrogen) were treated for 4 days with 10 μM atRA. Table 1presents data from Wright-Giemsa stains of cytocentrifuge preparations, as well as the percent of mitotic progenitors and mature granulocytes compiled from a minimum of 200 cells.
Interestingly, certain ECoM-G clones seemed more receptive than others to differentiation arrest, whereas the ECoM-M clone was entirely refractory to all oncoproteins tested. We observed that Hoxa9, Hoxb8, and AML1/ETO efficiently established differentiation arrest in multiple ECoM-G clones, whereas PML/RARα, PLZF/RARα, and Nup98/HoxA9 (see below) were more restricted in their target clones, arresting differentiation in only 1 of the 3 clones examined. This suggests that, regardless of the phenotypic similarity of the ECoM-G clones, differences exist in the expression of factors that cooperate with these oncoproteins to establish a new differentiation block.
Nup98/HoxA9 and Hoxa7 arrest the differentiation of ECoM-G cells when E2a/Pbx1 is inactivated
The ECoM-G cells provide a model in which to examine suspected oncoproteins and their effects on granulocytic differentiation. Both Nup98/HoxA9 (Figure 6) and Hoxa7 (data not shown) were capable of arresting ECoM-G differentiation following estrogen withdrawal—the first report to demonstrate that either protein can block in vitro myeloid differentiation. Despite their equivalent levels of expression, Nup98/HoxA9 was significantly less efficient than either Hoxa7 or Hoxa9 in arresting differentiation precipitated by estrogen withdrawal. This suggested that the mechanism of differentiation arrest by Nup98/HoxA9 was fundamentally different from that of Hoxa9 or Hoxa7. The small fraction (< 1%) of ECoM-G cells that continued to proliferate expressed a smaller version of the protein by Western analysis, indicating that some form of rearrangement had occurred. Nup98/HoxA9, identified as the product of the t(7;11) translocation in human myeloid leukemia,43,44 has also been shown to cause transformation of NIH3T3 fibroblasts45and to cause AML in mice.46 The pattern of gene expression in Nup98/HoxA9-immortalized ECoM-G cells was similar to that in Hoxa9-immortalized cells but for the lack of gp91PHOXexpression. Hoxa7 was initially shown to be coactivated with Meis1 by proviral integration in BXH-2 mice,47 and more recently shown to be coactivated with Meis1 in approximately 50% of human AML.48
Factor-independent ECoM cells can differentiate in the absence of GM-CSF
The ECoM-G and ECoM-M clones were rendered factor-independent following infection with retrovirus encoding H-RasL61 or Bcr/Ablp190. Removal of estrogen from these cells permitted differentiation as assessed by Wright-Giemsa staining (Table 2). Although the parental ECoM-G cells in the presence of GM-CSF underwent strict granulocytic differentiation (98%), ECoM-G_Ras and ECoM-G_Bcr/Abl cells demonstrated predominant monocytic differentiation (99% and 65%, respectively). In contrast, the differentiation of ECoM-M cells was not morphologically affected by expression of Ras or Bcr/Abl, though mature monocytes expressing Ras were no longer adherent. These morphologic results were supported by flow cytometric analysis. Differentiation of ECoM-G cells expressing Ras or Bcr/Abl was accompanied by increased F4/80 and decreased GR-1 staining, whereas expression of Ras or Bcr/Abl in ECoM-M cells did not significantly alter GR-1, MAC-1, or F4/80 staining (data not shown).
E2a/Pbx1 does not induce proliferation of myeloid progenitors
Although E2a/Pbx1 stimulates fibroblast proliferation, its ability to induce myeloid proliferation has not yet been investigated. If E2a/Pbx1 stimulated myeloid proliferation, then expression of BCL-2 should be sufficient to permit proliferation in the absence of GM-CSF or of other exogenous cytokines. Although Bcl-2α prevented apoptosis of ECoM-G and ECoM-M cells following factor withdrawal, neither cell line proliferated, indicating that E2a/Pbx1 does not stimulate myeloid progenitor proliferation (data not shown). This suggests that oncoproteins inducing proliferation, as well as protection from apoptosis, are required to complement E2a/Pbx1 function in leukemogenesis.
Discussion
In this paper we describe estrogen-dependent versions of E2a/Pbx1 that conditionally arrest the differentiation of clonal granulocytic, monocytic, or bipotential GM-CSF–dependent murine marrow progenitors. These ECoM clones provide models in which to (1) dissect the genetic circuitry of normal granulocytic and monocytic development, (2) identify E2a/Pbx1 target genes that contribute to differentiation arrest, (3) identify the mechanism by which other human myeloid oncoproteins interfere with the differentiation process, and (4) investigate the cooperative effects of Bcl-2, Ras, and Bcr/Abl on differentiation in vitro and on leukemogenesis in vivo.
Studies of normal myeloid differentiation49 have often relied on limited numbers of highly purified progenitors and sensitive techniques such as reverse transcription–PCR to evaluate gene expression. The ECoM cell lines are an inexhaustible source of clonal myeloid progenitors that, based on transcriptional and functional criteria, recapitulate normal myeloid development as it proceeds in GM-CSF and in the absence of endogenous oncoproteins or physiologically irrelevant inducers of differentiation. Combining the advantages of primary progenitors with synchronous differentiation and the ability to confirm expression of normal and mutant oncoproteins, the ECoM cells are a system in which to dissect the critical transcriptional events that accompany maturation and dictate lineage commitment.
The normal myeloid maturation following inactivation of E2a/Pbx1 demonstrates that expression of E2a/Pbx1 alone is sufficient to block differentiation. The reversibility of transactivation, transformation, and differentiation arrest suggests that E2a/Pbx1 does not establish an irreversible pattern of gene expression and, that by clinically targeting E2a/Pbx1, one might be able to reverse the in vivo differentiation block of t(1;19) pre-B ALL cells. We have not examined the in vivo behavior of the ECoM-G or ECoM-M clones though the cells would presumably differentiate normally in the absence of estrogen, as they do in vitro. Even if exogenous estrogen were supplied to maintain the activity of EPΔ578ER or EPΔ623ER, it is unlikely that the clones would be overtly leukemogenic given the observation that mice reconstituted with marrow retrovirally infected with wild-type E2a/Pbx1 still required a latency of 3 to 8 months to develop leukemia, denoting the necessity of a cooperating genetic event.50
The EPΔ578ER and EPΔ623ER proteins will be useful for identifying genetic targets of E2a/Pbx1. Previous studies have sought targets in NIH3T3 fibroblasts51,52 though the relevance of these targets is questionable based on their lack of expression in t(1;19)+ cell lines. Identification of targets in human non-t(1;19) pre-B cell lines53 has also been problematic because E2a/Pbx1 is toxic when expressed at a level comparable to that in t(1;19)+ leukemias.54 High-level expression of EPΔ623ER is possible in Nalm-6 cells maintained in the absence of estrogen (D.B.S., unpublished observation, November 1998) and direct E2a/Pbx1 targets could be identified by costimulation with cycloheximide and estrogen. In ECoM cell lines, genes up-regulated on the removal of estrogen provide a pool of targets whose transcriptional up-regulation is prevented by E2a/Pbx1. Determining how E2a/Pbx1 prevents expression of these targets provides a rational approach that will permit identification of the direct targets through which E2a/Pbx1 causes differentiation arrest. Dissecting the mechanisms of differentiation arrest is the first step in the development of molecularly targeted therapies.
The ECoM cells are also useful for identifying the mechanisms through which diverse oncoproteins arrest myeloid differentiation. To date, we have shown that the differentiation of ECoM-G clones can be arrested by Hoxa7, Hoxa9, Hoxb8, AML1/ETO, PML/RARα and PLZF/RARα, and that these oncoproteins establish unique blocks in differentiation as evidenced by their distinct patterns of gene expression in the immortalized cells. Although differentiation arrest by Hoxa9,55 Hoxb8,56 AML1/ETO,57PML/RARα,58 and PLZF/RARα58 has been demonstrated, this is the first report of in vitro myeloid differentiation arrest established by Hoxa7 or Nup98/HoxA9. Furthermore, expression of PML/RARα or PLZF/RARα, though tolerated in U937 and THP1 monocytic cells, is toxic to most granulocytic cell lines, such that this is a surprising demonstration of high-level, stable expression of these proteins in a granulocytic precursor (ECoM-G). The fact that E2a/Pbx1 and Hox proteins bind DNA cooperatively raised the concern that Hox partners could restore differentiation arrest by activating EPΔ578ER or EPΔ623ER in the absence of estrogen. However, this possibility was refuted by the following lines of evidence: (1) in the absence of estrogen, neither Hoxc8 nor Hoxa9 activates transcription with E2a/Pbx1 proteins (Figure 1), (2) a Pbx1-interaction mutant of HOXA9 retains its ability to rearrest differentiation in the ECoM-G cells,40(3) oncoproteins with disparate properties such as AML1/ETO or PML/RARα rearrest ECoM-G differentiation, and (4) those ECoM-G cells in which differentiation has been rearrested by PML/RARα or PLZF/RARα are sensitive to atRA, whereas parental cells are refractory.
The observation that ECoM-G cells rearrested in differentiation by PLZF/RARα were sensitive to the differentiating effects of atRA was at odds with certain reports in both human59 and murine cells,60 which have shown that PLZF/RARα-induced acute promyelocytic leukemias (APLs) are insensitive to treatment with atRA. However, similar to our findings, cultures of primary murine marrow immortalized by PML/RARα and PLZF/RARα were similarly sensitive to the effects of atRA,58 leukemic cells from a PLZF/RARα transgenic mouse showed atRA-induced granulocytic differentiation in the presence of spleen cell–conditioned media,61 and in certain clinical cases, PLZF/RARα-expressing APLs responded to atRA62 or combined atRA/GM-CSF therapy.63Coupled with the observation that PLZF/RARα can act as a transcriptional activator at high concentrations of atRA (10−6 to 10−5 M, as was used in our experiments),64 this suggests that in certain cellular contexts, and possibly in combination with certain cytokines, the mechanism by which PLZF/RARα prevents differentiation may be responsive to treatment with high concentrations of atRA.
The ECoM-G and ECoM-M cells expressing Bcl-2 did not proliferate in the absence of factor, indicating that E2a/Pbx1 alone does not induce myeloid proliferation, and is therefore unlikely to induce pre-B-cell proliferation. Although E2a/Pbx1 transforms NIH3T3 fibroblasts, we suspect that this is through a fibroblast-specific mechanism that does not occur in myeloid or pre-B lineages, and that oncoproteins that stimulate cell division will be necessary to complement the E2a/Pbx1-differentiation block in human pre-B cells. A fibroblast-specific proliferative mechanism would also explain our inability to promote the outgrowth of primary murine pre-B cells by coexpression of E2a/Pbx1 and Bcl-2 (D.B.S., unpublished observation, August 2000), and suggests that oncoproteins that induce both proliferation and protection from apoptosis may cooperate with E2a/Pbx1 in human pre-B-cell leukemia. This is consistent with a number of observations. First, the expression of E2a/Pbx1 is toxic in human pre-B-cell lines though coexpression of Bcl-2 protects against apoptosis.54 Second, T-cell leukemias in E2a/Pbx1 transgenic mice develop, with shortened latency, in cooperation with activated Pim165 or activated Notch expression.66 Third, the factor-independent proliferation of ECoM cells by expression of Ras or Bcr-Abl oncoproteins was consistent with a limited number of observations of RAS activation in ALL67 and of coexpression of E2a/Pbx1 and Bcr/Abl in pre-B ALL.68
Neither Ras nor Bcr/Abl, which mimic the downstream proliferative effects of GM-CSF signaling,69,70 prevented differentiation of the ECoM progenitors, indicating that the myeloid differentiation program was intrinsic to the cells and did not require continued GM-CSF signaling. Two recently reviewed models, the stochastic71 and the deterministic,72 have been proposed to describe the role of hematopoietic cytokines during differentiation. In the stochastic model, cytokines act as survival and proliferative factors, whereas lineage commitment decisions are intrinsic to the cell. The deterministic model suggests that the cytokine environment directs the pattern of gene expression, which determines the fate of the cell along a particular lineage. The differentiation of the ECoM cells in the absence of factor supports the stochastic model, in regard to E2a/Pbx1-immortalized myeloid progenitors, though the initial culture in GM-CSF may still have been required at earlier stages of development to “determine” the subsequent “stochastic” events. The observation in ECoM-G cells that Ras expression can bias predominant granulocytic differentiation toward the monocytic lineage has also been previously demonstrated in FDCP-mix73 as well as HL6074 and K562 cells.75
Although the ECoM cells are similar to murine cell lines derived by Hogg and coworkers using a c-Myb-ER fusion protein, clonal c-Myb-ER cell lines were not derived, nor were the cells assayed for their ability to score differentiation arrest by other oncoproteins.76 It should be noted that the expression of endogenous c-Myb in the ECoM cell lines is down-regulated late in differentiation (about 5 days after the removal of estrogen), indicating that the ECoM cells might represent an earlier stage of development whose differentiation can be disrupted by a broader spectrum of oncoproteins.
The EPΔ578ER and EPΔ623ER proteins provide an efficient and reproducible means of conditionally immortalizing cells along the myeloid lineage and potentially along other hematopoietic and nonhematopoietic lineages. Furthermore, the EPΔ578ER and EPΔ623ER proteins will allow for the conditional immortalization of marrow progenitors from mice harboring conditional knockout genes. Cloning of progenitors followed by infection with retrovirus encoding Cre-recombinase would establish a matched pair of clones and a powerful model to study in vitro differentiation in the absence of the gene in question.
We would like to thank Dan Tenen, Mary Dinauer, Renate Pilz, Martin Haas, Julie Leckstrom-Himes, Nancy Berliner, Arati Khanna-Gupta, Simon Williams, Yoshiaki Ito, Leighton Grimes, Beth Broome, and Rick Van Etten for supplying the probes for Northern analysis as well as BCL-2 and BCR/ABL expression constructs. Many thanks to Dennis Young (flow cytometry) and to James Feramisco, Stephen McMullen, and Carolan Buckmaster (deconvolution microscopy). A special thanks to Xinyu Fu and to Martina Pasillas for her excellent technical support. Much appreciation to Christopher Glass, Bruce Torbett, Katherine Calvo, Richard Lin, and Gernot Walter for critical reading of this manuscript.
Supported by Public Health Service grant CA56876. D.B.S. is supported by Department of Defense grant F49620-99-C-0054. M.P.K. is a scholar of the Leukemia and Lymphoma Society.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
David B. Sykes, Department of Molecular Pathology, University of California San Diego School of Medicine, 9500 Gilman Dr, La Jolla, CA 92093-0612; e-mail: dsykes@ucsd.edu.