Increasingly, perturbations in cellular iron and ferritin are emerging as an important element in the pathogenesis of disease. These changes in ferritin are important not only in the classic diseases of iron acquisition, transport, and storage, such as primary hemochromatosis, but also in diseases characterized by inflammation, infection, injury, and repair. Among these are some of the most common diseases that afflict mankind: neurodegenerative diseases such as Parkinson disease1 and Alzheimer disease,2vascular diseases such as cardiac and neuronal ischemia-reperfusion injury,3,4 atherosclerosis itself,5 pulmonary inflammatory states,6 rheumatoid arthritis,7,8 and a variety of premalignant conditions and frank cancers.9 We are just beginning to learn the mechanisms and implications of alterations in ferritin and iron homeostasis from these natural “experiments.” However, what is increasingly apparent is that ferritin appears to be a key molecule that limits the extent, character, and location of the pro-oxidant stress that typifies inflammatory diseases, cancer, and conditions of altered oxygenation. The link between alteration in ferritin regulation and these diseases is forged through a diverse set of cellular stress pathways that alter ferritin subunit composition and/or content within cells. The 2 broad themes developed in this review are that understanding the signals and pathways that regulate ferritin may lead to insights into the pathophysiology of these diseases, and that attention to how ferritin responds to stress will in turn teach us more about the normal functions of this complex protein.
Ferritin structure and function
Ferritin is a ubiquitous and highly conserved iron-binding protein. In vertebrates, the cytosolic form consists of 2 subunits, termed H and L. Twenty-four ferritin subunits assemble to form the apoferritin shell. Each apoferritin molecule of 450 000 d can sequester up to approximately 4500 iron atoms.10 Depending on the tissue type and physiologic status of the cell, the ratio of H to L subunits in ferritin can vary widely, from predominantly L in such tissues as liver and spleen, to predominantly H in heart and kidney.11 The H-to-L ratio is not fixed, but is rather quite plastic: it is readily modified in many inflammatory and infectious conditions, and in response to xenobiotic stress, differentiation, and developmental transitions, as well as other stimuli. Ferritin H and L subunits are encoded by separate genes.12,13 Although a single functional H and L gene was thought to be expressed in all vertebrate species, a functional mitochondrial ferritin gene has recently been described.14Multiple pseudogenes are also present.15-17 Ferritin also has enzymatic properties, converting Fe(II) to Fe(III) as iron is internalized and sequestered in the ferritin mineral core. Use of recombinant ferritins has demonstrated that this function is an inherent feature of the H subunit of ferritin, which has a ferroxidase activity.18 The ferroxidase center is evolutionarily conserved,10 and ferroxidase activity is dramatically reduced following mutation of residues His65 and Glu62 in both human and mouse.18 19
Small quantities of ferritin are also present in human serum, and are elevated in conditions of iron overload and inflammation.20-22 Serum ferritin is iron-poor, resembles ferritin L immunologically, and may contain a novel “G” (glycosylated) subunit.23 Despite widespread use of serum ferritin as a clinical indicator of body iron stores, little is known of the source of this ferritin. However, the increase in serum ferritin in patients with mutations in ferritin L has led to the suggestion that serum ferritin and ferritin L derive from the same gene product24 (see below).
The critical role of ferritin in cellular and organismal iron homeostasis is intimately linked to its primary and best-studied function—iron sequestration. Iron in heme is necessary for the transport, binding, and release of oxygen; the ready availability of iron for incorporation to heme is essential to organismal survival. Iron is also essential for the function of enzymes that participate in numerous critical cellular processes, including the cell cycle, the reductive conversion of ribonucleotides to deoxyribonucleotides, electron transport, and others. However, iron also donates electrons for the generation of the superoxide radical, and can participate in the generation of hydroxyl radicals via the Fenton reaction (Fe (II) + H2O2 → Fe (III) + OH− + OH.).25 Thus, iron status dramatically affects the generation of oxygen as well as ferryl and nitrogen radicals. The toxicity of iron in cellular systems is attributable in large part to its capacity to participate in the generation of such reactive species, which can directly damage DNA, lipids, and proteins, leading to profound cellular toxicity. At an organismal level, iron balance is maintained with exquisite care.26,27 Ferritin, by capturing and “buffering” the intracellular labile iron pool,28-31 plays a key role in maintaining iron homeostasis. It is not surprising, then, that homozygous murine knockouts of ferritin H are lethal.32
Recently, it has become evident that regulatory factors, in addition to those that regulate iron flux, have an important impact on cellular ferritin (Table 1). In fact, ferritin can be viewed not only as part of a group of iron regulatory proteins that include transferrin and the transferrin receptor, but also as a member of the protein family that orchestrates the cellular defense against stress and inflammation.33 This review focuses on the molecular mechanisms and biologic implications of ferritin regulation by cytokines, oxidants, oncogenes, growth factors, and other stimuli, as well as their relevance to the complex and still poorly understood events that perturb ferritin and iron homeostasis in a number of disease states.
Iron-mediated regulation of ferritin
Not only does ferritin sequester iron in a nontoxic form, but levels of “labile” iron regulate cellular ferritin levels, protecting cells from damage triggered by excess iron. Iron-mediated, largely posttranscriptional pathways of ferritin regulation have been identified through a series of elegant experiments over the last 15 years.34-42 The events that coordinate ferritin regulation are described briefly below and are illustrated in Figure1. The reader is also referred to detailed reviews of this subject.38,43 Although there is widespread agreement that these regulatory mechanisms are utilized in many cell types, additional regulatory pathways may be operant in erythroid cells, due to the specialized function of these cells in hemoglobin synthesis.44
Foundations of ferritin biology: IRE/IRP and iron-mediated regulation.
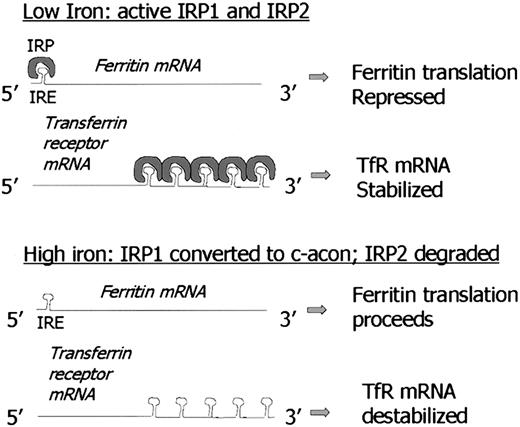
Note model of 5′ IRE repression of ferritin translation in low-iron conditions is similar to mechanism of iron-mediated regulation of erythroid ALA synthase (e-ALAS) gene. Similarly, destabilization of TfR mRNA under high-iron conditions by binding of IRP to 3′ IRE is similar to proposed regulation of divalent metal transporter-1 (DMT-1). C-acon indicates cytosolic cis-aconitase; IRP1, iron regulatory protein 1; IRP2, iron regulatory protein 2; IRE, iron responsive element.
Foundations of ferritin biology: IRE/IRP and iron-mediated regulation.
Note model of 5′ IRE repression of ferritin translation in low-iron conditions is similar to mechanism of iron-mediated regulation of erythroid ALA synthase (e-ALAS) gene. Similarly, destabilization of TfR mRNA under high-iron conditions by binding of IRP to 3′ IRE is similar to proposed regulation of divalent metal transporter-1 (DMT-1). C-acon indicates cytosolic cis-aconitase; IRP1, iron regulatory protein 1; IRP2, iron regulatory protein 2; IRE, iron responsive element.
The content of cytoplasmic ferritin is regulated by the translation of ferritin H and L mRNAs in response to an intracellular pool of “chelatable” or “labile” iron.45,46 Thus, when iron levels are low, ferritin synthesis is decreased; conversely, when iron levels are high, ferritin synthesis increases. Although in certain circumstances there is an increase in ferritin mRNA in response to iron,47 the regulatory response of ferritin to iron is largely posttranscriptional,48 and is due to the recruitment of stored mRNA from monosomes to polysomes in the presence of iron.46 This process is mediated by interaction between RNA binding proteins and a region in the 5′ untranslated region of ferritin H and L mRNA termed the iron responsive element (IRE) that has a “stem-loop” secondary structure. There are 2 RNA binding proteins, iron regulatory proteins 1 and 2 (IRP1 and IRP2), that bind to this stem loop structure and inhibit mRNA translation. However, the proteins are regulated differently: IRP1 is an iron-sulfur cluster protein that exists in 2 forms. When iron is abundant, it exists as a cytosolic aconitase. When iron is scarce, it assumes an open configuration associated with the loss of iron atoms in the iron-sulfur cluster, and can bind the IRE stem loop, acting as a repressor of ferritin translation. In contrast, IRP2 is regulated by degradation: IRP2 protein is abundant in iron scarcity, but is degraded rapidly in iron excess through targeting of a unique 73 amino acid sequence.49 Although both IRP1 and IRP2 bind the IRE and exert an inhibitory effect on ferritin synthesis, there is evidence that IRP1 and IRP2 may have distinct tissue-specific roles.50-52 Thus, IRP2 knockout mice exhibit a pronounced misregulation of iron metabolism in the intestinal mucosa and central nervous system,53 suggesting that the function of IRP2 in these tissues cannot be complemented by IRP1. Further, relative ratios of IRP1/IRP2 differ in a tissue-specific fashion, with IRP1 being more abundant than IRP2 in liver, kidney, intestine, and brain, and less abundant in pituitary and a pro–B-lymphocytic cell line.51 Action of IRP proteins can be further modulated through the activation of signal transduction cascades. For example, activation of protein kinase C (PKC) by phorbol esters phosphorylates IRP1 and increases its binding to the IRE.54 Similarly, PKC can activate IRP2, but through phosphorylation of different serine residues.54 Recently, a protein distinct from IRP1 and IRP2 that binds to a 5′ stem loop structure of mitochondrial complex 1 has been identified.55
Perhaps the most interesting feature of the IRE-IRP interaction is the conservation of the IRE sequence in other genes that regulate iron homeostasis. For example, the 3′ untranslated region (UTR) of the transferrin receptor gene contains 5 tandem IRE sequences. IRE-IRP binding lengthens transferrin receptor mRNA half-life, ultimately leading to increased transferrin receptor display on the cell surface in situations of iron depletion. Thus, similar RNA-protein binding motifs can have strikingly different biologic effects when located in different positions on different genes. In the case of ferritin, IRP binding results in inhibition of translation, whereas in the transferrin receptor, IRP binding increases transferrin receptor mRNA half-life.56 In addition to ferritin H and L and transferrin receptor, inducible eALA synthase (the enzyme catalyzing the rate-limiting step in heme biosynthesis), mitochondrial aconitase, and DMT-1 (the recently discovered divalent metal transporter, also termed DCT-1 and Nramp2), have functional IRE sequences.57IRE sequences have been identified in a number of other genes including ferroportin1/IREG1/MTP127,58 59; however, whether they function in IRP binding has not yet been determined.
Ferritin genes and mutations
Before one can approach the regulation of ferritin by signals other than cellular iron, it is essential to have an understanding of the overall gene structure of ferritin. This topic, as well as ferritin protein structure, has been extensively reviewed, and the reader is referred to excellent and detailed reviews of the protein, its crystal structure, and the implications for iron catalysis and storage.10,60 61
The structure of ferritin genes and proteins are highly conserved, likely due to the critical role of ferritin in the maintenance of iron homeostasis in species ranging from plants to humans. In vertebrates, the structure of cytoplasmic ferritin genes in all species studied thus far shows 3 introns and 4 exons, with the intron-exon boundaries occurring at similar locations.62 Sequences that code for a stem loop structure—the IRE—in the 5′ untranslated region of the ferritin mRNA are particularly conserved among species. A recently reported mitochondrial ferritin gene is the exception to this rule: it is intronless, and contains a domain with only weak homology to the classical IRE.14
The importance of the IRE in ferritin regulation is highlighted by the discovery of an autosomal dominant disorder resulting in hyperferritinaemia and cataracts, which can be attributed to point mutations in the IRE of ferritin L mRNA, leading to the constitutive activation of ferritin L translation and high serum ferritin in the absence of iron excess (Table 2). A recently discovered autosomal dominant mutation in the IRE of ferritin H leads to increased affinity of the IRE for IRP, reduced ferritin H protein, and iron overload.63 A dominantly inherited mutation in the C-terminal domain of ferritin L with an associated decrease in serum ferritin and abnormal deposition of ferritin and iron in the brain has also been described: this mutation has been suggested to underlie a new syndrome termed “neuroferritinopathy.”64
In nonvertebrate animals and plants, structural analysis and functional studies in many species have either failed to reveal structures compatible with posttranscriptional regulation by iron, or have revealed sequences that are not functional IRP binding sites. Instead, transcriptional regulation of ferritin is most frequently (although not universally) identified as a primary mode of response to exogenous iron as well as stress-related signals.65-72
Ferritin regulation by cytokines and inflammation
The cytokine tumor necrosis factor alpha (TNFα; cachectin) is synthesized by stimulated macrophages and other cell types. Specific binding of TNFα to cell surface receptors triggers apoptotic pathways in susceptible cells; its pleiotrophic effects also mediate, in concert with other inflammatory cytokines, many of the events in inflammation and septic shock (for review see Beutler and Cerami,73Tsuji and Torti,74 and Baud and Karin75). TNFα is also an important contributor to the syndrome of cancer cachexia and other chronic inflammatory conditions,76diseases in which ferritin levels are frequently altered.5,8 77
TNFα and interleukin 1α (IL-1α), another proinflammatory cytokine, transcriptionally induce the H chain of ferritin, suggesting that pathways related to inflammation and stress can impact on ferritin regulation33,78 (Figure 2). In mesenchymal cells (normal human skeletal myoblasts and myocytes, adipocytes, human and mouse fibroblasts), these TNFα and IL-1α effects were striking, selective for ferritin H mRNA, and resulted in the accumulation of a population of H-rich ferritin proteins, substantially altering the cellular subunit composition and content of ferritin.33 This observation provided a plausible physiologic explanation for the differential regulation of ferritin subunits in inflammation. Since cytokines markedly affect aspects of iron homeostasis in malignancy and inflammatory diseases,79 the observation that ferritin H could be selectively transcriptionally regulated provided a molecular model to explain the linkage between inflammation and the modulation of subunit composition and content of ferritin, largely inexplicable based on posttranscriptional regulation alone.
Mouse and human ferritin H genes.
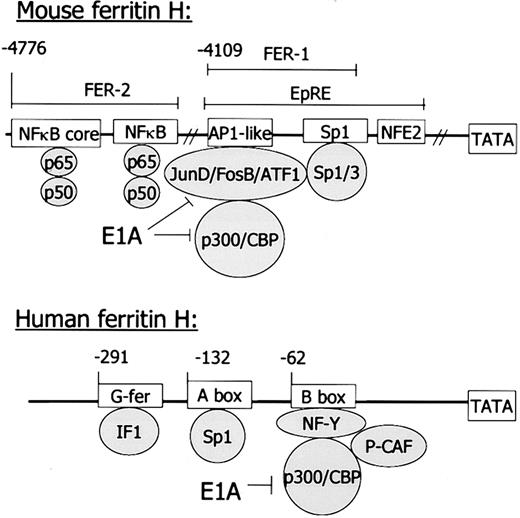
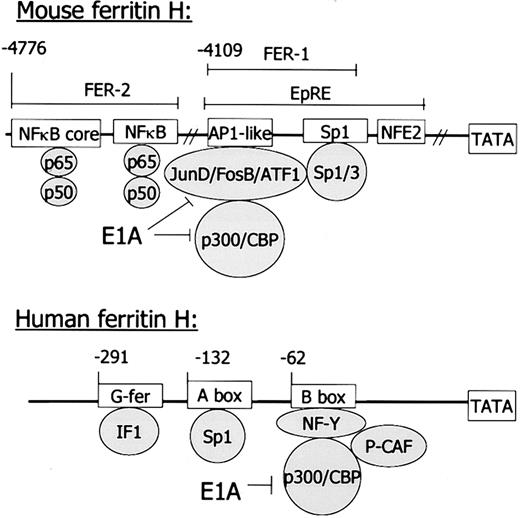
In the murine gene, regulatory regions reside between 4 kb and 5 kb distal to the transcriptional start site; in the human gene, regulatory regions are just 5′ of the TATA box. However, similar sequence motifs and binding proteins suggest similar functional regulation of these genes. IF1 has not been definitively identified as binding to the G-fer region of the human ferritin H gene.115 See text for further explanation.
Mouse and human ferritin H genes.
In the murine gene, regulatory regions reside between 4 kb and 5 kb distal to the transcriptional start site; in the human gene, regulatory regions are just 5′ of the TATA box. However, similar sequence motifs and binding proteins suggest similar functional regulation of these genes. IF1 has not been definitively identified as binding to the G-fer region of the human ferritin H gene.115 See text for further explanation.
The regulatory elements in the murine gene that respond to cytokines have been mapped in detail, and the DNA binding proteins that mediate this response have been identified (Figure 2). Ferritin H is regulated by TNFα through a cis-acting region (FER2) located 4.8 kb upstream of the transcriptional start site that binds the transcription factor NFκB.80 Two of the multiple NFκB subunits are specifically involved in this binding, p50 and p65. They bind to an NFκB full consensus sequence and to an adjacent NFκB core sequence. Both sequences are necessary for maximal ferritin H induction by TNFα.
Cytokines also have transcriptional effects on ferritin in other cell types. Ferritin induction in macrophages may be particularly important, given their central role in iron homeostasis as scavengers of old and damaged red blood cells, a critical and quantitatively important element in whole body iron turnover. In the U937 macrophage cell line, the proinflammatory cytokines TNFα and interferonγ increased mRNA for ferritin H, but not ferritin L. IL-1β had no effect on ferritin mRNA levels. TNFα uniquely resulted in a greater proportion of iron incorporated into ferritin.81 In A549 cells (a human cell line with properties of type 2 alveolar pneumocytes), TNFα and IL-1β in the presence or absence of exogenous iron, induced predominately H ferritins and increased iron uptake. In contrast, treatment with iron alone (transferrin-bound or non–transferrin-bound iron) induced the synthesis of L ferritin.82Overexpression of myotonic dystrophy protein kinase in C2C12 myocytes led to an increase in ferritin H mRNA, presumably by inducing IL-1.83
Cytokines also regulate ferritin posttranscriptionally. Early experimental models involving induction of inflammation in rats with turpentine demonstrated increased ferritin synthesis in the liver84; an increase in ferritin synthesis but not ferritin mRNA was also seen in liver slices from turpentine-treated rats.85 A subsequent study revealed an increase in ferritin mRNA in the livers of turpentine-treated rats; in this series of experiments, concomitant nitric oxide (NO)–mediated induction of IRP activity prevented a coordinate increase in ferritin protein (see below).86 The response of ferritin in cultured hepatocytes treated with defined cytokines has also been investigated. In the HepG2 hepatic cell line, induction of ferritin synthesis was observed with a number of cytokines: IL-1β, IL-6, TNFα. In each case, desferrioxamine inhibited the increase in ferritin, suggesting a posttranscriptional induction in liver cells.87 The mechanism of this posttranscriptional regulation of ferritin by IL-1β has been evaluated in detail88; it is not mediated by protein binding to the IRE, but rather to a distinct G+C–rich region of the mRNA distinct from the IRE.89 A similar sequence is present on genes for other acute phase proteins. IL-1β also appears to affect ferritin accumulation posttranscriptionally in human astrocytoma cells and as a consequence reduces the labile iron pool.90
Cytokines may also affect ferritin translation indirectly through their ability to induce iNOS (nitric oxide synthase) and hence increase NO.50,91,92 NO in turn causes the activation of both IRP1 and IRP2 (although effects on IRP2 appear to exhibit some cell type specificity), effects that may be particularly important in inflammatory conditions. Mechanisms hypothesized to underlie NO-mediated induction of IRP binding activity include cluster disassembly (IRP1), intracellular iron chelation (IRP1 and IRP2), or increased de novo synthesis (IRP2).93
Secretion of ferritin is stimulated by cytokines. In primary cultured human hepatocytes, IL-1α and IL-6 induced a transient secretion of ferritin at 24 hours followed by a decline to baseline, whereas TNF treatment resulted in a sustained increase in ferritin secretion, reaching a level 10 times that found in untreated cells.94Similar effects on secreted ferritin were shown in HepG2 liver cells. In this study, both TNFα and IL-1β induced the secretion of ferritin, and the combination was at least additive. Iron in the form of either iron-dextran or ferric nitrilotriacetate (FeNTA) also induced the secretion of ferritin to about the same level as the combination of cytokines. Secretion was inhibited by brefeldin A, an inhibitor of Golgi function, and by an inhibitor of transcription.95
Cytokines play a pivotal role in the cellular response to infection, and ferritin plays a prominent role in the cytokine response. Lipopolysaccharide (LPS; endotoxin), a component of the outer membrane of gram-negative bacteria, elicits a variety of reactions that involve ferritin. Although stimulation of a number of inflammatory cytokines is associated with LPS, the cellular reaction is complex, and in animals can involve vascular leak syndrome, the coagulation cascade, activation of complement, and prostaglandin synthesis. LPS administered endotracheally to rats induced ferritin protein but not mRNA.96 Similarly, tail vein injection of rats with LPS increased immunoreactive ferritin in the spleen.97 Viral infection with mengo picornavirus has been reported to lead to an increase in ferritin.98 Cyclopentenone prostaglandins (A-type prostaglandins A1, A2, J2, etc), which are involved in inflammatory and febrile responses as well as viral replication, induced L chain ferritin, heme oxygenase, and HSP 70 in human monocytes.99
Ferritin is also involved in the inflammatory processes of atherosclerosis. In a study to determine the genes regulated in atherogenesis, cDNA libraries were constructed from atherosclerotic aorta and screened for genes differentially expressed in normal and atherosclerotic plaques.5 Ferritin H and L mRNAs were markedly induced in the aortas of rabbits fed an atherogenic diet for 6 weeks. In situ hybridization revealed that both H and L ferritin were induced in endothelial cells and in macrophages. Cells in culture were then used to model elements of the atherosclerotic process. In the THP-1 monocytic cell line and in aortic smooth muscle, ferritin was up-regulated by IL-1 and TNFα, but not TGF, platelet-derived growth factor (PDGF), or oxidized low-density lipoprotein (LDL).5
Regulation of ferritin by hormones, growth factors, and second messengers
Transcription of the human ferritin H gene is induced in response to both hormones and second messengers, including cAMP. Thecis-acting elements mediating these responses have mapped to a relatively small region in the proximal promoter of the human ferritin H gene (Figure 2).
There were 2 groups that identified ferritin H as a gene differentially expressed in response to thyrotropin in rodent cells.100,101 Subsequent work revealed that dibutyryl-cAMP recapitulated the effect of thyrotropin on ferritin H transcripts, albeit with different kinetics.102 Short fragments of the rat 5′ flanking region (up to 400 bp) but not longer fragments were responsive to dibutyryl-cAMP and thyrotropin in murine 3T3 cells and FRTL5 thyroid cells.103,104 Nuclear run-on assays confirmed the transcriptional effect of thyrotropin on ferritin H.105 The cAMP-dependent induction of ferritin was inhibited by ras in a rat thyroid cell line.106
Collectively, these experiments demonstrated that thyrotropin increased ferritin H transcription, probably by elevating cAMP. cAMP-mediated induction of ferritin H transcription was further defined in human HeLa cells.107 The human cAMP-responsive region (the B-box) binds a protein complex termed B-box binding factors (Bbf), comprised of the transcription factor NFY, the coactivator p300, and the histone acetylase p300/CBP associated factor (PCAF).108,109 The adenoviral oncogene E1A reduces the formation of this complex. Overexpression of p300 in HeLa cells reverses the E1A-mediated inhibition of the ferritin promoter driven by Bbf.110 Okadaic acid, a phosphatase inhibitor, stimulates H ferritin transcription in HeLa cells by increasing the interaction between the p300 coactivator molecule and other components of Bbf.111 In cells with low expression of human ferritin H, overexpression of the histone acetylase PCAF activates transcription from the B-box of ferritin H.112 The B-box may also mediate the increase in ferritin H mRNA that occurs during spontaneous differentiation of Caco-2 colon carcinoma cells108 and vascular smooth muscle.113 Other important regulatory elements in the human ferritin H gene include a region called the A-box at position −132, which contains an SP-1 consensus sequence.114
Since evaluation of the rodent cAMP regulatory region showed that longer promoter fragments exhibited a reduced rather than enhanced response to thyrotropin, these experiments also suggested the presence of negative cis-acting elements that may counteract the effect of cAMP and thyrotropin. Additional evidence for a negative regulator(s) of ferritin H transcription was obtained by Barresi et al.115 This group demonstrated that there is a stretch of 10 G's, which they termed “G-fer” between −272 and −291 of the human ferritin H gene. A 3-bp substitution mutation in this region increased promoter activity in HeLa cells, suggesting an inhibitory effect of this sequence on ferritin transcription. Inhibitory factor 1 (IF-1), which binds ubiquitously to G-rich sequences, was suggested to bind to this region.
Thryoid hormone may also regulate ferritin posttranscriptionally: T3 modulates the activity of IRP1, affecting its ability to bind to the ferritin IRE, possibly through induction of signal transduction cascades that result in phosphorylation of IRP1.116 T3 and TRH also induce the phosphorylation of IRP2.51
Similarities between the murine and human ferritin H gene highlight the conservation not only of ferritin function, but of ferritin regulation across species. The murine ferritin H gene contains similar elements to those described above; however, they are located almost 5 kb 5′ to the corresponding regulatory elements identified in the human ferritin H gene (see Figure 2). The murine ferritin H gene contains a basal enhancer FER1 that also binds p300 and is inhibited by E1A.117 Contained within FER1 is a region of dyad symmetry that binds SP1, like the A-box of the human ferritin H gene. However, to date the FER1 region has not been shown to respond to cAMP.
In addition to thyroid hormone, insulin and IGF-1 have also been implicated in regulation of ferritin at the mRNA level. Insulin and IGF-1 both induced mRNA for H and L ferritin in C6 glioma cells.118 There was no additive effect on ferritin induction when both hormones were combined at the optimal concentration of each, suggesting that insulin might be acting through the IGF-1 receptor.118 In contrast to the equal induction of ferritin H and L by insulin and IGF-1, in pancreatic cells high glucose caused selective induction of ferritin H mRNA, with a 4-fold to 8-fold increase in ferritin H mRNA, a 75% to 90% decrease in ferritin L, and an overall 3-fold increase in ferritin as assayed by immunostaining.119
Hematopoietic differentiation and ferritin
Among the most carefully studied areas of ferritin biology have been the changes in ferritin and other proteins of iron metabolism that occur during hematopoietic differentiation. This emphasis is appropriate for many reasons: the availability of iron during erythropoiesis is a critical aspect of mammalian homeostasis; the unique role of macrophages and monocytes in iron handling is essential to the understanding of iron recycling; and finally, the experimental systems, derived for the most part from malignant cells, provide an excellent model for studying ferritin regulation in proliferating malignant cells and their differentiated (and less malignant) counterparts. In many of these model systems ferritin H transcription is selectively induced, leading to H-rich ferritin protein over the time course of differentiation. However, some of the reports infer transcriptional activity from the changes in steady-state levels of mRNA, without either assessment of transcription rates, mRNA stability, or evaluation of transcription from heterologous promoters. It should also be noted that inducers of cellular differentiation trigger a complex process often spanning many days, and the proximal regulators of ferritin alterations observed in many of the experiments described below have often not been identified.
HL60 promyelocytic cells reproducibly demonstrate a shift toward the accumulation of H-rich ferritin protein and mRNA with differentiation.120,121 In HL60 cells induced to differentiate into macrophages with the phorbol ester PMA, ferritin H mRNA levels increased up to 16-fold in 3 days; in cells induced to differentiate into neutrophils with dimethylsulfoxide (DMSO) there was a more modest 3-fold increase. In an HL60 cell line variant which produced phenotypes of intermediate differentiation, ferritin H and L were expressed at different stages of differentiation: H ferritin mRNA was expressed in the most differentiated promyelocytic cells, whereas early in the differentiation process L subunit induction was observed.122
Erythroleukemia cell lines have also been investigated extensively for their ability to express ferritin during differentiation and on exposure to hemin. Much of this work has been performed using Friend erythroleukemia cells, which are mouse erythroleukemia cells that differentiate no further along the erythropoietic lineage than proerythroblasts. In response to the differentiation inducer DMSO, a biphasic transcriptional induction of ferritin H and L mRNA was observed; however, no corresponding increase in ferritin protein synthesis was detected.123,124 Hemin increased the concentration of ferritin H mRNA 10-fold and ferritin protein 20-fold in Friend leukemia cells. Protoporphyrin IX increased ferritin H mRNA but not ferritin protein, possibly due to its iron chelation effect. Ferric ammonium citrate was a less potent inducer of both ferritin H mRNA and ferritin protein than hemin, in both Friend erythroleukemia cells125 and in fibroblasts.126 Although the differentiation inducers DMSO and hexamethylenbisacetamide (HMBA) had only a 2-fold effect on ferritin mRNA or protein synthesis when used alone as differentiation inducers, when hemin was added to these inducers, a synergistic effect on ferritin was seen. The effect was both transcriptional and translational, with ferritin H and L mRNA induction of approximately 15-fold as well as a 20-fold to 25-fold increase in ferritin protein. Desferrioxamine had no effect on ferritin mRNA accumulation. Interestingly, the induction of ferritin was not associated with a decrease in transferrin receptor expression, as might be predicted through IRP-mediated posttranscriptional mechanisms. Rather, hemin and protoporphyrin IX transcriptionally induced both ferritin H and L and transferrin receptor genes. A similar finding of induction of ferritin genes associated with an increase, not decrease, in transferrin receptor was seen in erythropoietin-induced differentiation of J2E erythroid cells.127 In murine and human erythroleukemic cells, erythropoietin treatment modulated IRP, resulting in IRP activation,128 possibly via induction of a signal transduction cascade and phosphorylation of IRP.51 Taken in aggregate, these experiments with erythroleukemia model systems show that either differentiation inducers or hemin result in a transcriptional induction of both H and L ferritin genes. It is interesting that a wide range of inducers, which can direct differentiation along different hematopoietic lineages, regulate the coordinate induction of ferritin genes.
The molecular details of transcriptional regulation of human ferritin in the mouse Friend erythroleukemia system have been investigated.129 The minimum region of the ferritin H promoter that was able to confer transcriptional regulation by heme was 77 bp upstream of the TATA box. This region binds a protein complex referred to as the heme responsive factor, which was identified as NF-Y, an ubiquitous transcription factor. The CCAAT element in this region is critical, since a point mutation abolished binding to the heme responsive factor and transcriptional activation. The pathway of hemin activation was not defined, but the induction of ferritin H and L transcription proceeded in both normal and cAMP protein kinase–deficient murine erythroleukemia (MEL) cells, suggesting the cAMP pathway is not involved in this induction.130
In addition to transcriptional regulation and posttranscriptional IRP-mediated regulation, altered ferritin mRNA stability has also been documented in hematopoetic cells. Using K562 cells in which hemin was added, the level of ferritin H and L mRNA increased 2-fold to 5-fold or 2-fold to 3-fold over 24 hours, respectively, whereas the protein increased 10-fold to 30-fold. This mRNA increase was not inhibited by desferrioxamine, suggesting that it was not mediated through chelatable iron. Further, transcription assays for ferritin H and L genes were unchanged. Although mRNA stability was not directly measured, these results led to the suggestion that changes in mRNA stability explained the ferritin mRNA rise with hemin treatment.131,132Similar conclusions were reached in the human monocytic cell line THP-1, which can be induced to differentiate into macrophages by treatment with the phorbol ester PMA; PMA treatment elicited an induction of ferritin H.133 Subsequent studies that directly investigated the stability of ferritin H mRNA found evidence that PMA stabilized ferritin H mRNA. This effect was mediated by pyrimidine-rich sequences within the 3′ UTR of the ferritin gene.134
New roles for ferritin in heme synthesis?
Recently, a novel mitochondrial ferritin gene has been reported.14 This intronless gene contains a mitochondrial localization signal and is expressed in the mitochondrial matrix. It exhibits more than 75% sequence identity to the ferritin H gene, and appears to sequester iron more avidly than cytosolic H-rich ferritins. Northern blot analysis revealed that expression of mitochondrial ferritin is normally restricted to testicular tissue. However, use of a specific antibody demonstrated that mitochondrial ferritin can also be expressed in erythroblasts from patients with X-linked sideroblastic anemia. Although work on this new form of ferritin is still in its early stages, these results suggest that mitochondrial ferritin may be induced under conditions of pathologic iron accumulation in heme-synthesizing cells.
In another area of intense recent investigation, a novel role for ferritin H in hemoglobin switching has also been proposed. Using gel mobility shift assays in K562 erythroleukemic cells, Broyles and coworkers demonstrated specific binding of a protein with properties of ferritin H to a conserved CAGTGC motif in the beta globin promoter.135 Transient transfection assays revealed that ferritin H repressed synthesis of beta globin, suggesting that ferritin may play a role in hemoglobin switching. Although a nuclear distribution and function for ferritin has not been unequivocally documented, the accumulation of reports of nuclear ferritin localization needs careful attention,136,137 particularly given the report of an E coli protein structurally related to ferritin that binds to and protects DNA from oxidative damage.138
Ferritin regulation by oxidants
One of the major functions of ferritin is to limit Fe(II) available to participate in the generation of oxygen free radicals (ROS). Oxidant stress is an ever-present threat to organismal survival, both from exogenous and endogenous cellular sources; it is therefore not surprising that oxidant stress activates multiple pathways of ferritin regulation. How these pathways interact is just beginning to be understood, as is the role of ferritin in the substantial cassette of gene and protein alterations that coordinately limit oxidant toxicity.
There is strong experimental support for ferritin as a protectant against oxidant stress. Early studies demonstrated that exposure to heme induced ferritin synthesis in endothelial cells and concordantly reduced their cytotoxic response to hydrogen peroxide.139In tumor cell lines, sensitivity to oxidants was inversely correlated with ferritin protein levels; modulation of ferritin levels with hemin could alter oxidant sensitivity.140 These results are consistent with more recent observations that increased ferritin levels reduce the low molecular weight (“labile” or “regulatory”) iron pool.30 They are also consistent with observations that a reduction in ferritin sensitizes cells to pro-oxidant cytotoxicity,141 that overexpression of ferritin reduces oxidant species in cells challenged with oxidants142,143 and reduces oxidant toxicity,144 as well as the importance of ferritin H ferroxidase activity142 in limiting oxidant toxicity.
Both transcriptional and posttranscriptional mechanisms have been implicated in ferritin induction by oxidants. Oxidants induce ferritin transcription by directly targeting conserved regions of ferritin genes.145 Transcriptional induction of ferritin H and L genes was also observed in rat livers after injection with phorone, which reduces glutathione concentration and therefore limits free radical defense mechanisms.146 Oxidative stress can also contribute to ferritin induction by inactivating IRP1 through reversible oxidation of critical cysteine residues.147 However, oxidant-mediated inactivation of IRP1 is not always seen. In fact, in other experimental systems, oxidants had the opposite effect: hydrogen peroxide activated the iron responsive protein (IRP1), possibly through induction of a signaling pathway that mobilizes iron from the 4Fe-4S cubane cluster.148 This results in reduced ferritin synthesis posttranscriptionally, potentially leaving the cell more susceptible to oxidative injury.149 These observations are difficult to reconcile with a postulated role for ferritin in the protection against oxidative stress.
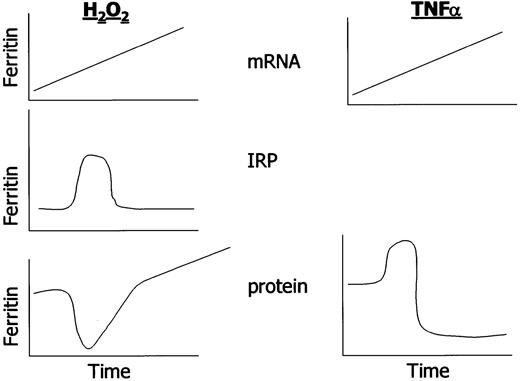
Recent work has offered a model that permits observations regarding IRP activation, ferritin induction, and the protection from oxidative stress to be resolved. In cultured BNLCL2 mouse liver cells following acute oxidant challenge, IRP was activated only transiently, and thus ferritin translation was only transiently inhibited. At the same time, a sustained increase in ferritin transcription was induced. The ultimate result was an increase in ferritin protein in oxidant-treated cells, a condition that permits reduction in oxidant-induced cell injury.143 145 These results emphasize that transcriptional and translational controls collaborate in the determination of ultimate cellular ferritin content (see Figure3).
Complexities in ferritin regulation.
The schematic highlights temporal changes in ferritin transcription and translation in response to oxidants and cytokines in cultured cells. Note that hydrogen peroxide induces both H and L ferritin; TNF induces H ferritin but not L ferritin in cells of mesenchymal lineage. IRPs have not been investigated temporally after TNFα treatment.
Complexities in ferritin regulation.
The schematic highlights temporal changes in ferritin transcription and translation in response to oxidants and cytokines in cultured cells. Note that hydrogen peroxide induces both H and L ferritin; TNF induces H ferritin but not L ferritin in cells of mesenchymal lineage. IRPs have not been investigated temporally after TNFα treatment.
Oxidants may also alter ferritin transcription and translation through release of iron from cellular proteins. Oxidants, including ROS86,146,150 and nitric oxide,151 may release iron from ferritin, IRP1, or hemoglobin,152 either directly or through heme oxygenase (in the case of heme-containing proteins).39 This can lead to ferritin induction through IRP inhibition (above), and perhaps through direct iron-mediated transcriptional regulation of ferritin.153,154 Assessment of alterations in cellular iron due to breakdown of ferritin, aconitase iron clusters, heme-containing proteins, and other proteins will be immeasurably aided by recently described methods to directly measure a “labile” iron pool.31
Although much experimental work has centered on hydrogen peroxide–mediated generation of ROS, other forms of oxidant stress also alter ferritin. For example, UV irradiation, which produces oxygen-free radicals and damages DNA, induced ferritin H mRNA, and protein.155,156 Menadione, a synthetic vitamin K derivative, and its water soluble form, menadione sodium bisulfite, were shown to induce ferritin in the rat liver, an effect which was preceded by heme oxygenase induction, as well as hydrogen peroxide generation and changes in the intrahepatic GSH pool. Menadione sodium bisulfite was also shown to decrease both IRP1 and aconitase activity in B6 fibroblasts.157 Another oxidant, oxidized lipoproteins, dramatically stimulated ferritin L in the THP-1 macrophage line.158 Various components of oxidized LDL were able to recapitulate this response, which may be mediated by peroxisome proliferator–activated receptor γ (PPARγ).159 However, other studies using oxidized LDL in THP-1 cells failed to show either ferritin H or ferritin L induction.5
The role of ferritin in xenobiotic stress, the antioxidant response, and chemoprevention
A major stress faced by hematopoietic and other cells and organisms is exposure to oxidant radicals induced by xenobiotic pro-oxidants. These include a broad range of agents, including environmental toxicants such as cigarette smoke, herbicides, pesticides, ozone, and many others. Exposure to these agents leads to the induction of a group of antioxidant protective enzymes (“phase II” enzymes), including glutathione S transferase, MnSOD, heme oxygenase, NAD(P)H quinone oxidoreductase (NQO1), gamma glutamyl cysteine synthetase, and others, that collectively act to mitigate cell damage mediated by these toxicants. Chemopreventives are synthetic or naturally occurring compounds that are intended to deliberately elicit this protective response without exerting toxic effects of their own (reviewed in Kensler et al160). Intriguingly, they are frequently mild oxidants, and as such, might be expected to induce ferritin. Indeed, Primiano et al161 showed that rats treated with the chemopreventive dithiolethione increased levels of ferritin in the liver. Further, the induction of phase II enzymes by chemopreventives is transcriptionally mediated via an “EpRE” (electrophilic response element, also termed antioxidant response element, [ARE] or oxidative stress response element [OSRE]). A sequence search revealed the presence of a putative EpRE in the murine ferritin L gene; when ligated to a reporter construct, the sequence was shown to be inducible by tert butyl hydroquinone (tBHQ), a model phase II enzyme inducer, in HEPG2 cells.162 Recent evidence indicates that ferritin H and L are induced by oltipraz, a chemopreventive agent currently in clinical trials, in murine and human liver cells.163 An induction of ferritin has also been observed in animals treated with oltipraz by gavage.164
Ferritin regulation during hypoxia-ischemia and hyperoxia
Tissue ischemia and cellular hypoxia have been modeled in a number of conditions and changes in ferritin documented. Hypoxia in neonatal rat oligodendrocytes and human oligodendrogliomas induced the synthesis of ferritin. This effect was not inhibited by actinomycin D, nor did mRNA levels of ferritin H mRNA change. The effect in oligodendrocytes was recapitulated by exogenous iron and blocked by desferrioxamine.165-167 Similar effects on ferritin induction were observed in a rat model of acute hypoxic/ischemic insult. Shortly after the hypoxic-ischemic insult, ferritin-positive microglia accumulate in subcortical white matter. In addition, the ratio of H to L ferritins shifts toward H-rich ferritins, especially in the hemisphere in which both hypoxic and ischemic insult was applied.3 In another ischemic-reperfusion model of the rat brain, induction of both ferritin H and L mRNAs occurred in the ischemic hemisphere, beginning 12 hours after 60 minutes of ischemia and lasting 14 days. Protein levels, as determined by immunohistochemistry, paralleled rises in mRNA; surprisingly, although overall induction of H and L mRNA was equal, the distribution of H and L ferritin mRNA as determined by in situ hybridization of rat brains was completely different.4
Ferritin changes in hypoxia are at least in part mediated by altered regulation of the IRP proteins. IRP1 binding activity decreased under hypoxic conditions in rat hepatoma cells168; IRP1 decreased and ferritin levels increased in hypoxic mouse macrophages.169 In contrast, IRP2 activity was found to increase under similar conditions.170However, in 2 human cell lines, one group found an induction of IRP activity and a decrease in ferritin protein.171
The period of reperfusion after ischemia is thought to be a critical period during which oxidant damage is maximal in many tissues, including heart, brain, and other organs. During postischemic reoxygenation of rat liver, early ferritin degradation was counteracted by enhanced ferritin transcription and concomitant IRP down-regulation. It was suggested that this might act to re-establish ferritin levels and limit reperfusion damage.153 Similarly, in a model of transient surgically-induced segmental intestinal ischemia reperfusion in rats, cytosolic ferritin mRNA and protein (in addition to brush border enzymes) decreased after 3 hours and 6 hours of reperfusion. By 12 hours, ferritin mRNA but not protein had increased to higher than normal levels. Ferritin appeared to be regulated both pretranslationally and translationally in response to ischemia reperfusion.172 Consistent with these findings, reoxygenation was found to induce IRP1 in a hepatoma cell line.168
Raising levels of inspired oxygen (hyperoxia) is an effective treatment of lung diseases such as the adult respiratory distress syndrome (ARDS) as well as treatment of preterm infants with inadequate pulmonary maturation. Unfortunately, lung damage on exposure to high concentrations of oxygen is a major side effect of such treatment. Exposure of the lung to high oxygen concentration can cause the production of partially reduced oxygen species such as O2− and H2O2. When rats were exposed to hyperoxic conditions (95% O2), a selective increase in mRNA for ferritin L was seen at 24 hours and 48 hours, but not earlier. This result is reminiscent of acute iron overload in the rat.154 Indeed, hypotransferrinemic mice, which have high levels of ferritin and lactoferrin, are resistant to hyperoxia-induced lung injury.173 In contrast, at early time points (4 hours) in murine peritoneal macrophages treated with 80% O2, IRP activity was increased and ferritin synthesis, both H and L, was repressed.169
Oncogenes, cancer, and ferritin
Early views of the relationship between ferritin and cancer stem from work demonstrating an increase in total ferritin as well as a shift toward acidic (H-rich) ferritins in the serum of patients with various malignancies.174 However, subsequent evaluations of ferritin levels in tumor tissue itself have revealed a complex, perhaps disease-specific picture: for example, in some cases such as colon cancer,175 testicular seminoma,176 and breast cancer,177,178 increases in ferritin in tumor tissue versus comparable normal tissue have been reported; in other cases, including liver cancer, a decrease in ferritin is seen.179 New forms of ferritin may be involved in certain situations: Moroz and coworkers have described a novel isoform of ferritin that is elevated in the serum of patients with neoplastic breast disease as well as during pregnancy and HIV infection.180-182
Molecular explanations for the perturbations of ferritin in cancer have been slow to advance, with often conflicting conclusions. For example, Modjtahedi et al183 studied subclones of an SW 613-W human colon carcinoma cell line that differ in their ability to develop tumors in nude mice. Differential screening revealed markedly higher expression of ferritin H but not ferritin L mRNA in the most tumorigenic cell lines. These highly tumorigenic clones had a high copy number of the c-myc gene and expressed high levels ofc-myc mRNA. Similarly, in immortalized (MCF-10F) breast cancer cells, ferritin H mRNA was elevated relative to the mortal HBEC line S-130.184 A study of breast cancer tissue also showed high levels of ferritin H mRNA in malignant tissues: in an examination of human breast tissue removed at mastectomy for breast cancer, cells with ductal hyperplasia, carcinoma-in-situ, and infiltrating ductal carcinoma showed ferritin H expression by in situ hybridization, whereas normal breast tissue had the lowest levels of expression measured.184 Conversely, cancers with the highest metastatic potential showed the lowest levels of ferritin expression in rat transitional cell carcinoma of the urothelium.185Unfortunately, ferritin protein levels were not reported in any of these studies.183-185
A study published in 1993 was the first to demonstrate that a defined oncogene modulated ferritin H mRNA and protein.186 This experiment demonstrated that the E1A oncogene, an immortalizing nuclear oncogene of adenovirus that falls within the same complementation group as c-myc in transformation assays,187selectively repressed ferritin H.186 Subsequent experiments demonstrated that this was the result of E1A-dependent transcriptional repression of the ferritin H gene.117
Consistent with these findings, down-regulation of ferritin H in highly tumorigenic, clonogenic c-myc–transformed, Epstein-Barr virus–immortalized B cells was seen.188 Transcriptional induction of IRP2 was also observed, leading to the suggestion that c-myctransformation maximizes iron available for proliferation through coordinate transcriptional and translational effects on proteins of iron metabolism. A key finding in this paper was that reexpression of high levels of ferritin H reverted c-myc–induced transformation, indicating that repression of ferritin H was critical to c-myc–mediated transformation.188
Immortalization and transformation are linked to changes in the control of cellular proliferation. In some cell types, ferritin has been observed to increase in growth arrest. Thus, downregulation ofc-myc and coordinate upregulation of ferritin H was seen in U937 cells induced to differentiate with TPA (12-O-tetradecanoylphorbol 13-acetate).189 Upregulation of ferritin was also associated with induction of differentiation and growth arrest in hematopoetic systems (see above), premature replicative senescence of fibroblasts,190 and differentiation of preadipocytes to adipocytes.33 Growth suppression associated with overexpression of ferritin H has also been reported.142 However, in rat-1 fibroblast cells, microarray analysis of c-myc–responsive genes did not identify ferritin H in either the upregulated or downregulated gene profile.191
In part, conflicting findings relating to the relationship between ferritin and cancer reflect the relative paucity of experiments in this area; additional experiments focused on defined oncogenes and the pathways they elicit will no doubt clarify these issues. Perhaps some contradictions are inevitable (and accurate), given the complexity and variability of pathophysiologic processes that underlie the collection of diseases grouped together as “cancer.”
Future directions
Observations over the last few years have placed the regulation of ferritin within the broad context of cell injury and stress as well as altered growth regulation. This has led to the discovery of new hormonal and growth factors that regulate ferritin, many through transcriptional targeting of ferritin genes. The pathways that link these factors to changes in ferritin gene expression and protein levels are just beginning to be understood, but clearly involve multiple pathways, some of which are cell-type– and disease-specific: they include NFκB, cellular oxidant pathways that target the electrophilic response element, cAMP signaling pathways, as well as signals important for growth and cell cycle progression, such as the proto-oncogenec-myc and the adenovirus E1A oncoprotein. These observations are inseparably linked to classical posttranscriptional ferritin regulation, since cellular mRNA translation of ferritin genes remains dependent on cellular iron status through IRE-IRP–mediated regulation.192 How these pathways are perturbed in diseases in which ferritin dysregulation has been described, and whether changes in ferritin have a role in causation or cellular protective response, are questions that can now be reasonably posed.
Supported in part by grants DK42412 and DK57781 from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Frank M. Torti, Comprehensive Cancer Center, Wake Forest University School of Medicine, Medical Center Blvd, Winston-Salem, NC 27157; e-mail: ftorti@wfubmc.edu.