A drug that specifically inhibits erythropoiesis would be clinically useful. The erythropoietin (Epo) mutant Epo (R103A) could potentially be used for this purpose. Epo (R103A) has a single amino acid substitution of alanine for arginine at position 103. Because of this mutation, Epo (R103A) is only able to bind to one of the 2 subunits of the erythropoietin receptor (EpoR) homodimer and is thus a competitive inhibitor of Epo activity. To produce large quantities of Epo (R103A) to test in animal models of thalassemia and sickle cell disease, we expressed and purified recombinant Epo (R103A) from the yeast Pichia pastoris. Using this method milligram quantities of highly purified Epo (R103A) are obtained. The yeast-expressed Epo (R103A) is properly processed and glycosylated and specifically inhibits Epo-dependent cell growth and125I-Epo binding. Epo (R103A) does not, however, directly induce apoptosis in 32D cells expressing EpoR. Epo (R103A) inhibits erythropoiesis of human CD34+ hematopoietic cells and completely blocks erythroid burst-forming unit formation in normal human bone marrow colony assays. Yeast-expressed Epo (R103A) is a specific inhibitor of primary erythropoiesis suitable for testing in animal models.
Introduction
Blood transfusion is commonly used in the treatment hemoglobinopathies. One of the goals of blood transfusion in these diseases is the suppression of abnormal endogenous erythropoiesis. For instance, thalassemia major is associated with ineffective erythropoiesis and massive bone marrow expansion. This bone marrow expansion has numerous adverse effects, including hypersplenism, bone abnormalities, growth retardation, increased susceptibility to infection, and endocrine abnormalities.1 Transfusion therapy regimens can dramatically ameliorate these problems.
Patients with sickle cell disease may have a relative erythrocytosis such that oxygen delivery begins to decrease at hematocrits as low as 30% to 35%.2 This decreased oxygen delivery is a consequence of the abnormal rheology of sickle red blood cells.3 The resulting hyperviscosity is associated with serious clinical conditions such as acute chest syndrome and acute multiorgan damage syndrome.4 5 These complications require treatment with exchange blood transfusions.
In both thalassemia and sickle cell disease chronic transfusion therapy can have important complications including iron overload, viral hepatitis, and alloimmunization. Drug therapy that specifically and safely inhibits erythropoiesis would be desirable for these patients.
A potential drug for specific inhibition of erythropoiesis is the erythropoietin (Epo) mutant Epo (R103A). Epo (R103A) has a mutation of alanine for arginine at amino acid position 103. Epo (R103A) was discovered during mutational studies to define important amino acid residues required for Epo activity.6 Epo (R103A) completely lacks the ability to support mitogenesis in cells expressing the erythropoietin receptor (EpoR).6 7
Biochemical and crystallographic studies have shown that Epo has nonidentical binding sites for each of the 2 identical EpoR subunits in the preformed EpoR homodimer.8,9 Epo binding site 1 interacts with the first EpoR subunit with nanomolar affinity.10 Epo binding site 2 interacts with the second subunit of the EpoR homodimer with micromolar affinity.10The resulting subnanomolar affinity of Epo for the EpoR dimer is the consequence of the 2 binding interactions. The interaction of Epo with the receptor homodimer results in a conformational shift of the subunits such that cytoplasmic signal transduction is activated. Arginine 103 of Epo is the primary residue involved in the second EpoR binding surface.11 Mutation of arginine 103 to alanine disrupts this binding interaction. Epo (R103A) is thus unable to induce activation of the EpoR. Because it retains the ability to bind the first EpoR subunit, Epo (R103A) is a competitive inhibitor of Epo activity.12
To study the in vivo activity of Epo (R103A) in animal models of thalassemia and sickle cell disease, we have expressed Epo (R103A) in the yeast Pichia pastoris. This methanotropic yeast allows high-level expression of properly glycosylated and folded recombinant proteins.13 We describe here the purification and biochemical characterization of yeast-expressed Epo (R103A). In addition, we demonstrate the ability of Epo (R103A) to inhibit erythropoiesis of primary human hematopoietic cells.
Materials and methods
Expression and isolation of yeast Epo antagonist
Epo (R103A) complementary DNA (cDNA; a kind gift of Dr Frank Bunn, Harvard Medical School, Boston, MA) was subcloned into the yeast expression vector pPICZα (Invitrogen, Carlsbad, CA). This vector allows insertion of the Epo (R103A) sequence into the alcohol oxygenase locus of the methanotropic yeast P pastoris. Expression of Epo (R103A) can thus be induced by growth in methanol. This vector includes a C-terminal polyhistidine tag (for purification of recombinant fusion protein on Ni2+ agarose beads) and a myc tag. Addition of a polyhistidine tag to the C-terminus does not effect the biologic activity of Epo.14 Secretion of the protein into the yeast-conditioned media is facilitated by the yeast α-mating factor signal peptide.
The resulting construct, Epo (R103A)/pPICZα, was verified for accuracy by DNA sequencing. Epo (R103A)/pPICZα was electroporated into the yeast P pastoris, strain GS115. Positive clones were selected in zeocin as described by the manufacturer. True positives were further selected by Northern blot for Epo (R103A) messenger RNA expression. Several clones were screened for level of expression of Epo (R103A) protein by Western blot using polyclonal anti-Epo antibodies (prepared from rabbits immunized with human Epo). The highest expressing clone was chosen for subsequent protein purification.
This clone, Epo (R103A)/GS115 clone no. 4, was inoculated into 2 × 500 mL 1% yeast extract, 2% peptone, 0.1 M potassium phosphate, pH 6.0, 1.34% yeast nitrogen base, 4 × 10−5% biotin, and 1% glycerol in 2-L baffled flasks. After 2 days of growth at 30°C, the yeast was collected by centrifugation and resuspended in 2 × 100 mL 1% yeast extract, 2% peptone, 0.1 M potassium phosphate, pH 6.0, 1.34% yeast nitrogen base, 4 × 10−5% biotin, and 0.5% methanol in 500-mL baffled flasks. The cultures were grown for 5 days at 30°C. Methanol (0.5 mL) was added to the flasks every day to replace methanol consumed by the yeast.
After 5 days of culture in methanol, the yeast-conditioned media were collected by centrifugation. The 200 mL yeast-conditioned media were concentrated to 20 mL with Centriprep 10 YM concentrators (Millipore, Bedford, MA) and dialyzed overnight into 5 mM imidazole, 0.5 M NaCl, 20 mM Tris, pH 8.0 (binding buffer). A 1-mL solution (50% slurry) of Ni-NTA resin (Qiagen, Valencia, CA) was washed twice with water and then twice with binding buffer in a 50-mL conical centrifuge tube. The dialyzed yeast-conditioned media was added to the washed Ni-NTA resin and incubated with rocking for 4.5 hours at 4°C. The beads were collected by centrifugation and washed twice in 10 mL 0.5 M NaCl, 20 mM Tris, pH 8.0 (wash buffer). Enough wash buffer was then added to produce a 50% slurry. The slurry was packed into a 4-mL plastic chromatography column. The column was washed with wash buffer until the absorbance at 280 nm was less than 0.01. The bound protein was eluted with 0.25 M imidazole, 0.5 M NaCl, 20 mM Tris, pH 8.0. Fractions of 1 mL were collected. Fractions with an absorbance at 280 nm more than 0.1 were collected and pooled. The pooled fractions were dialyzed overnight into 0.15 M NaCl, 20 mM Tris, pH 7.4. The dialyzed Epo (R103A) was sterilized using a syringe filter and the final protein concentration determined using the DC Protein Assay (Bio-Rad, Hercules, CA). Wild-type human Epo was expressed and purified from P pastoris exactly as described above for Epo (R103A).
Purified Epo (R103A) was subjected to N-glycanase digestion as suggested by the manufacturer (New England Biolabs, Beverly, MA). Carbohydrate stains on nitrocellulose membranes were performed with the Glycan Detection Kit (Roche, Indianapolis, IN) as described by the manufacturer.
Cell culture
The 32D cells were maintained in Iscoves modified Dulbecco medium, 10% fetal calf serum, containing 10% WEHI media for parental 32D cells, or 2 U/mL Epo (equivalent to 15.5 ng/mL) for 32D/EpoR cells, in 5% CO2 and 95% humidified air at 37°C. 32D parental cells were stably transfected with the full-length EpoR (32D/EpoR WT) or the cytoplasmically truncated EpoR15 found in familial erythrocytosis (32D/EpoR FE). We have previously characterized these cell lines with respect to growth characteristics in Epo and interleukin 3 (IL-3), as well as EpoR number by Scatchard analysis.15 These cells express physiologically relevant numbers of functional cell surface EpoR (200-500 sites/cell).
125I-Epo binding assays were performed with125I-Epo (Amersham, Piscataway, NJ) as previously described.16 Cell growth and viability were measured using methyl-thiazol-diphenyl-tetrazolium (MTT) reagent (Sigma, St Louis, MO) or 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2- (4-sulfophenyl)-2H-tetrazolium (MTS) reagent (Promega, Madison, WI) as described by the manufacturers. Conversion of commercial recombinant Epo from U/mL to μg/mL was performed using the published value of 129 000 U/mg.17Apoptosis was assessed by flow cytometry using Alexa 488–labeled annexin V (Molecular Probes, Eugene, OR) as described by the manufacturer.
Primary cell cultures and proliferation and viability assays
Human umbilical cord blood (UCB) was obtained from the Department of Obstetrics and Gynecology of Duke University Medical Center through an Institutional Review Board–approved protocol after normal full-term pregnancy. Peripheral blood mononuclear cells were collected from healthy volunteers. Cord blood units and cells were diluted with 50 mL phosphate-buffered saline containing 0.8% bovine serum albumin and then submitted to Ficoll density gradient separation. The mononuclear cells were harvested and enriched for CD34+ cells by positive selection using anti-CD34 antibody and magnetic cell sorting on Mini-MACS columns (CD34 isolation kit, Miltenyi Biotec, Auburn, CA). CD34+ enriched cells were cultured in growth medium containing 100 ng/mL human stem cell factor (SCF; Sigma, St Louis, MO), 7.5 ng/mL recombinant human Epo (Ortho Biotech, Raritan, NJ), 40 ng/mL insulinlike growth factor (Sigma), 10−6 M dexamethasone (Sigma), and 10−6 M β-estradiol (Sigma) as described previously.18 On day 9 of culture, cells were analyzed for surface expression of CD71 by flow cytometry to confirm the purity of the erythroid cell population. Staining of the cells using monoclonal, fluorescein isothiocyanate (FITC)–conjugated antihuman CD71 (Sigma) and FITC-mouse isotype control followed by flow cytometry analysis confirmed more than 99% CD71 positivity of the erythroid cell population. Morphologic examination of the Wright stains of cells cytocentrifuged onto glass slides confirmed the presence of a homogeneous population of erythroid cells with proerythroblast morphology. Cells were washed with serum-free culture medium and then placed in differentiation medium containing 7.5 ng/mL Epo and 1 μg/mL recombinant human insulin (Humulin-R, Lilly, Indianapolis, IN) as described to induce further expansion during terminal differentiation.18 For dose-response experiments in the presence of Epo (R103A), 106 cells were placed in 3 mL culture medium with various concentrations of Epo (R103A) and cells were counted daily using a hemocytometer. Day 9 erythroblasts were plated in 96-well plates (5000-40 000 cells/well) and maintained in differentiation medium with various concentrations of added Epo (R103A). At 5 days, medium was aspirated from the wells and MTT reagent (1 mg/mL) was added to each well. The cells were then incubated for 1 hour in 37°C, lysed with isopropanol and read in an enzyme-linked immunosorbent assay (ELISA) plate reader at 570 to 650 nm. Assays were performed in triplicate. Results were analyzed for statistical significance using the Student t test.
Bone marrow colony assays were performed using bone marrow aspirates from healthy volunteers via a protocol approved by the University of Alabama at Birmingham Institutional Review Board. Bone marrow mononuclear cells (1 × 105/mL) were plated in Methocult complete methylcellulose media (Stem Cell Technologies, Vancouver, BC) containing SCF (50 ng/mL), granulocyte-macrophage colony-stimulating factor (GM-CSF; 10 ng/mL), and IL-3 (10 ng/mL). Some cultures also contained Epo (23 ng/mL) or Epo (R103A) or both as indicated. Cultures were maintained for 14 days in 5% CO2at 37°C and then all colonies on the plate were counted.
Results
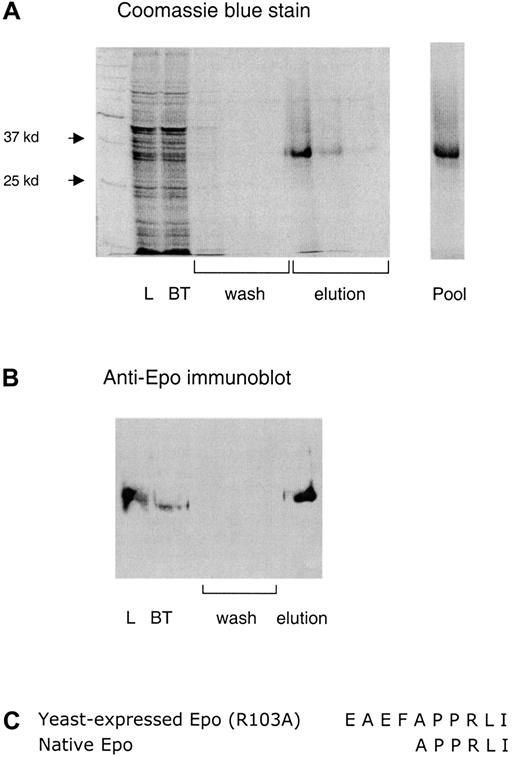
Recombinant Epo (R103A) protein was expressed and purified from the yeast P pastoris. The human Epo (R103A) cDNA sequence (including C-terminal polyhistidine tag) was subcloned into the yeast expression vector pPICZα. The subsequent construct was used to transform P pastoris strain GS115. Following induction of protein expression driven by the alcohol oxidase promoter during 6 days' growth in methanol, Epo (R103A) is secreted into the growth medium using the yeast α-mating factor signal sequence. The mature Epo (R103A) protein was purified from conditioned media by Ni2+ column chromatography (Figure1A,B). Using this method we obtain 1 to 2 mg/L yeast culture of purified Epo (R103A).
Expression and purification of Epo (R103A).
(A) A 10% sodium dodecyl sulfate gel of a typical purification is shown. Load indicates the yeast-conditioned media loaded on the Ni2+ column. BT indicates the breakthrough fraction, wash indicates the 3 wash fractions, elution indicates the three 1-mL fractions eluted with 0.25 M imidazole, and pool indicates the final pooled product. (B) An immunoblot of the Epo (R103A) preparation using polyclonal rabbit anti-Epo antibody. (C) N-terminal protein sequence of yeast-expressed Epo (R103A).
Expression and purification of Epo (R103A).
(A) A 10% sodium dodecyl sulfate gel of a typical purification is shown. Load indicates the yeast-conditioned media loaded on the Ni2+ column. BT indicates the breakthrough fraction, wash indicates the 3 wash fractions, elution indicates the three 1-mL fractions eluted with 0.25 M imidazole, and pool indicates the final pooled product. (B) An immunoblot of the Epo (R103A) preparation using polyclonal rabbit anti-Epo antibody. (C) N-terminal protein sequence of yeast-expressed Epo (R103A).
The purified yeast-expressed Epo (R103A) was biochemically characterized. To demonstrate proper processing by the yeast secretory pathway, the N-terminal sequence of the purified protein was determined (Figure 1C). The sequence demonstrates proper cleavage of the α-mating factor signal peptide. As predicted for the previously reported processing of this signal peptide in yeast,19there are 4 residual amino acid residues from the signal peptide (Glu-Ala-Glu-Phe) followed by the mature human Epo sequence. Neither these additional 4 N-terminal amino acids, nor the C-terminal myc tag, alter the activity of wild-type Epo (data not shown).
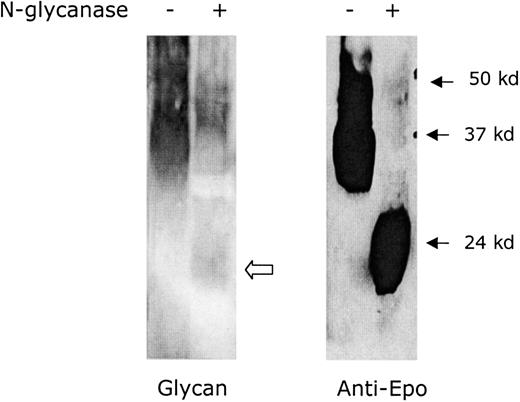
The degree of glycosylation of Epo (R103A) was determined using N-glycanase digestion (Figure 2). The Epo (R103A) peptide has a predicted molecular weight of 21 000. The 32 000 molecular weight purified Epo (R103A) is reduced to 22 000 molecular weight following digestion with N-glycanase. This is consistent with 10 000 molecular weight of N-linked glycosylation. A small amount of carbohydrate staining remains following N-glycanase digestion, consistent with 1000 molecular weight of residual O-linked sugars (Figure 2, open arrow). The yeast-expressed Epo (R103A) thus has amounts of N- and O-linked glycosylation similar to mammalian-expressed Epo.20
Glycosylation of yeast-expressed Epo (R103A).
Epo (R103A) was digested with N-glycanase to determine the extent of N-linked glycosylation. N-glycanase treatment is indicated with + and no treatment is indicated with −. The left gel shows glycosylation detected using glycan stain. The open arrow indicates residual glycan staining due to O-linked glycosylation. The right gel shows a Western blot using anti-Epo antibody. The blot was overloaded to demonstrate complete digestion.
Glycosylation of yeast-expressed Epo (R103A).
Epo (R103A) was digested with N-glycanase to determine the extent of N-linked glycosylation. N-glycanase treatment is indicated with + and no treatment is indicated with −. The left gel shows glycosylation detected using glycan stain. The open arrow indicates residual glycan staining due to O-linked glycosylation. The right gel shows a Western blot using anti-Epo antibody. The blot was overloaded to demonstrate complete digestion.
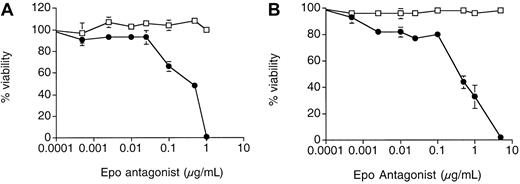
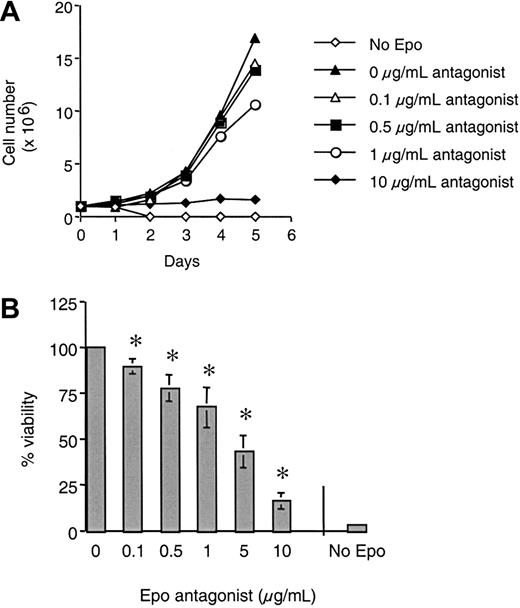
As previously reported for mammalian-expressed Epo (R103A),12 yeast-expressed Epo (R103A) is a potent antagonist of Epo growth (Figure 3A). Addition of Epo (R103A) caused a dose-dependent inhibition of growth of 32D cells expressing the full- length human EpoR (32D/EpoR WT cells) with half-maximal inhibition at approximately 0.5 μg/mL (equivalent to 15.6 nM). At 1 μg/mL Epo (R103A) added (approximately 80-fold excess with regard to the Epo concentration used) there was a complete absence of cell growth. Trypan blue staining revealed that all cells incubated for 3 days in 1 μg/mL Epo (R103A) were nonviable (data not shown). Epo (R103A) had no effect on IL-3–dependent growth of these cells (open squares). Similar results were seen with 32D cells expressing the murine EpoR and with BaF3 cells expressing the human EpoR (data not shown).
Epo (R103A) inhibits Epo-dependent growth of 32D/EpoR cells.
32D/EpoR WT cells (A) or 32D/EpoR FE cells (B) were grown were grown for 4 days in Epo (15.5 ng/mL, solid circle) or IL-3 (2 ng/mL, open square) in the presence of the indicated concentration of Epo (R103A). Initial cell number was 5 × 103 cells/well in 150 μL culture media. Cell viability after 4 days of growth was determined using MTS reagent. Each point was done in quadruplicate with SE bars shown.
Epo (R103A) inhibits Epo-dependent growth of 32D/EpoR cells.
32D/EpoR WT cells (A) or 32D/EpoR FE cells (B) were grown were grown for 4 days in Epo (15.5 ng/mL, solid circle) or IL-3 (2 ng/mL, open square) in the presence of the indicated concentration of Epo (R103A). Initial cell number was 5 × 103 cells/well in 150 μL culture media. Cell viability after 4 days of growth was determined using MTS reagent. Each point was done in quadruplicate with SE bars shown.
We have previously shown that 32D cells expressing a cytoplasmically truncated EpoR associated with the disease familial erythrocytosis (FE) are hypersensitive to Epo-induced growth.15 21 As shown in Figure 3B, Epo (R103A) also effectively blocked Epo-induced growth of 32D expressing the EpoR FE (32D/EpoR FE cells). In our hands, the inhibition of EpoR FE routinely requires about 5-fold more Epo (R103A) compared to inhibition of wild-type EpoR (compare inhibition of viability at 1 μg/mL in Figure 3). This right-shift of the inhibition curve is likely a reflection of the hypersensitivity of the truncated EpoR for Epo (eg, it takes more complete blockade of the EpoR FE to obtain equivalent biologic effect).
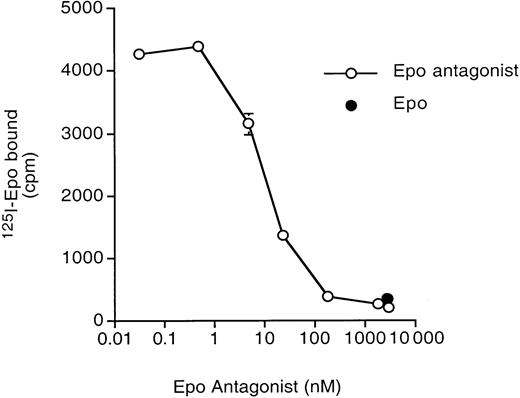
Epo (R103A) is a competitive inhibitor of 125I-Epo binding (Figure 4). The apparent Kiwas 10 nM (approximately 0.32 μg/mL) under the conditions of this experiment. This is similar to the half-maximal inhibition by Epo (R103A) of Epo-induced growth (0.5 μg/mL) described above. At concentrations above 1000 nM, only nonspecific binding of125I-Epo occurred (compare binding in the presence of excess unlabeled Epo, closed circle).
Epo (R103A) inhibits 125I-Epo binding.
125I-Epo (at a final concentration of 0.132 nM) was incubated with 32D/EpoR WT cells in the presence of increasing amounts of unlabeled Epo (R103A). Total cell-associated 125I-Epo was measured. Binding in the presence of 3000-fold excess unlabeled Epo (to determine nonspecific binding) is shown in the solid circle. Determinations were done in quadruplicate with SD shown.
Epo (R103A) inhibits 125I-Epo binding.
125I-Epo (at a final concentration of 0.132 nM) was incubated with 32D/EpoR WT cells in the presence of increasing amounts of unlabeled Epo (R103A). Total cell-associated 125I-Epo was measured. Binding in the presence of 3000-fold excess unlabeled Epo (to determine nonspecific binding) is shown in the solid circle. Determinations were done in quadruplicate with SD shown.
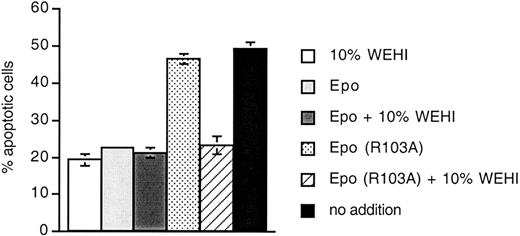
A GM-CSF mutant with structural and antagonistic properties similar to Epo (R103A) has been reported to directly activate apoptosis in myeloid precursor cells.22 We tested if Epo (R103A) is able to directly activate apoptosis. 32D/EpoR cells were treated with 10% WEHI (as a source of IL-3), Epo, or Epo (R103A) for 24 hours. The cells were harvested and apoptosis was determined by measuring annexin V binding (Figure 5). In this rapidly growing cell line, about 20% of cells are undergoing apoptosis at steady state under normal growth conditions (10% WEHI, Epo, or Epo plus 10% WEHI). In contrast, about 50% of cells cultured in Epo (R103A) are undergoing apoptosis (dotted bar). This is similar to the degree of apoptosis in the absence of any cytokine (black bar). Thus, Epo (R103A) is unable to support antiapoptotic pathways. However, addition of 10% WEHI to cells cultured in Epo (R103A) (hatched bar) returns the degree of apoptosis to the background level. Thus, Epo (R103A) does not directly induce apoptosis in this system.
Epo (R103A) does not induce apoptosis in 32D/EpoR cells.
32D/EpoR cells (300 000 cells/well) were cultured in triplicate in 24-well plates. Cultures contained 10% WEHI-conditioned media (as a source of IL-3), Epo (15.5 ng/mL), or Epo (R103A) (1 μg/mL) as indicated. Cells were harvested 24 hours later and apoptosis was measured using flow cytometry to detect exposure of annexin V binding sites.
Epo (R103A) does not induce apoptosis in 32D/EpoR cells.
32D/EpoR cells (300 000 cells/well) were cultured in triplicate in 24-well plates. Cultures contained 10% WEHI-conditioned media (as a source of IL-3), Epo (15.5 ng/mL), or Epo (R103A) (1 μg/mL) as indicated. Cells were harvested 24 hours later and apoptosis was measured using flow cytometry to detect exposure of annexin V binding sites.
Epo (R103A) effectively inhibits erythropoiesis of primary human hematopoietic cells (Figure 6). UCB CD34+ cells were used in an in vitro erythropoiesis culture system to obtain normal proerythroblasts.18Proerythroblasts were incubated for 5 days in the presence of increasing concentrations of Epo (R103A). There was a dose-dependent inhibition of the ability of Epo to support expansion of these proerythroblasts (Figure 6A). This inhibition of proerythroblast expansion was associated with a dose-dependent inhibition of cellular viability by Epo (R103A) (Figure 6B).
Epo (R103A) inhibits primary human erythroblasts.
(A) Inhibition of primary erythroblast proliferation by Epo (R103A). UCB CD34+ cells were expanded as described in “Materials and methods.” Day 9 proerythroblasts were cultured for 5 days in differentiation medium containing recombinant Epo (7.5 ng/mL) in the absence or presence of increasing concentrations of Epo (R103A) as indicated (range, 0-10 μg/mL). As a negative control, cells were cultured in the absence of any added recombinant Epo or Epo (R103A) (no Epo). Comparable results were obtained in 3 independent experiments. (B) Epo (R103A) results in decreased viability of primary erythroblasts. Day 9 proerythroblasts were cultured in triplicate in differentiation medium in 96-well plates (10 000 cells/well) in the absence or presence of increasing concentrations or Epo (R103A) (range, 0-10 μg/mL). MTT colorimetric assay was performed after 5 days as described in “Materials and methods.” Viability of cells was expressed as a percentage of maximum viability in the absence of any Epo antagonist. As a negative control, cells were cultured in differentiation medium without added recombinant Epo and without any Epo antagonist (no Epo). Asterisks indicate inhibition of viability that is significantly different from viability in Epo alone (P < .05). Comparable results were obtained in several experiments.
Epo (R103A) inhibits primary human erythroblasts.
(A) Inhibition of primary erythroblast proliferation by Epo (R103A). UCB CD34+ cells were expanded as described in “Materials and methods.” Day 9 proerythroblasts were cultured for 5 days in differentiation medium containing recombinant Epo (7.5 ng/mL) in the absence or presence of increasing concentrations of Epo (R103A) as indicated (range, 0-10 μg/mL). As a negative control, cells were cultured in the absence of any added recombinant Epo or Epo (R103A) (no Epo). Comparable results were obtained in 3 independent experiments. (B) Epo (R103A) results in decreased viability of primary erythroblasts. Day 9 proerythroblasts were cultured in triplicate in differentiation medium in 96-well plates (10 000 cells/well) in the absence or presence of increasing concentrations or Epo (R103A) (range, 0-10 μg/mL). MTT colorimetric assay was performed after 5 days as described in “Materials and methods.” Viability of cells was expressed as a percentage of maximum viability in the absence of any Epo antagonist. As a negative control, cells were cultured in differentiation medium without added recombinant Epo and without any Epo antagonist (no Epo). Asterisks indicate inhibition of viability that is significantly different from viability in Epo alone (P < .05). Comparable results were obtained in several experiments.
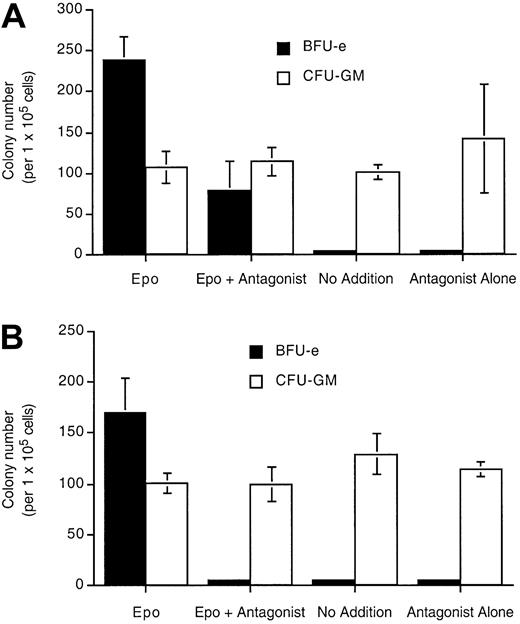
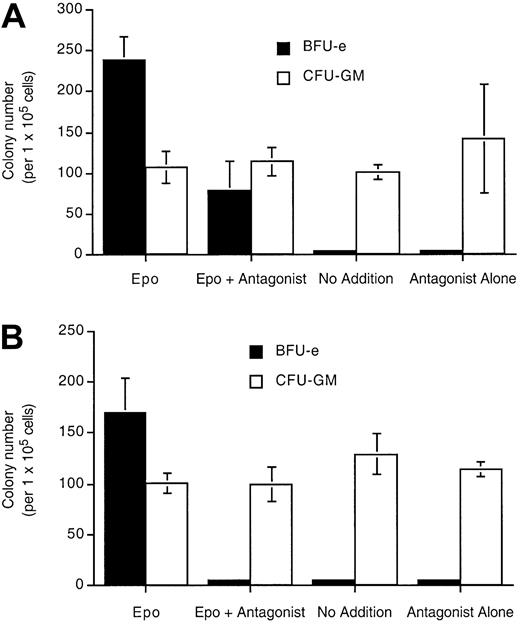
Epo (R103A) is also a specific inhibitor of day 14 erythroid burst-forming units (BFU-Es) in human bone marrow cultures (Figure7). In the presence of Epo and 1 μg/mL Epo (R103A), the number of BFU-Es was decreased to about 33% of the number in Epo alone (Figure 7A). In the presence of Epo and 10 μg/mL Epo (R103A) (a concentration sufficient to completely saturate the EpoR, Figure 4), BFU-E formation was completely blocked (Figure 7B). There was no effect on nonerythroid colony number (GM-CFU) with Epo (R103A) treatment. Epo (R103A) alone was unable to support BFU-E viability and differentiation.
Epo (R103A) specifically inhibits erythroid precursors in human bone marrow culture.
Bone marrow mononuclear cells (1.2 × 106) were cultured in 1 mL methylcellulose culture medium containing Epo (23 ng/mL) or Epo (R103A) or both at 1 μg/mL (A) or 10 μg/mL (B). Cultures also contain IL-3, GM-CSF, and SCF. BFU-E and CFU-GM colonies were counted at day 14. “No addition” indicates that neither Epo nor Epo antagonist was added. Cultures were done in triplicate with SD indicated with the bars.
Epo (R103A) specifically inhibits erythroid precursors in human bone marrow culture.
Bone marrow mononuclear cells (1.2 × 106) were cultured in 1 mL methylcellulose culture medium containing Epo (23 ng/mL) or Epo (R103A) or both at 1 μg/mL (A) or 10 μg/mL (B). Cultures also contain IL-3, GM-CSF, and SCF. BFU-E and CFU-GM colonies were counted at day 14. “No addition” indicates that neither Epo nor Epo antagonist was added. Cultures were done in triplicate with SD indicated with the bars.
Discussion
The present work was undertaken with the long-range goal of determining if Epo (R103A) may have useful activity as a drug for the suppression of erythropoiesis. Previous studies of Epo (R103A) have used unpurified conditioned media from transfected mammalian cells6,7,12 or polyhistidine tagged Epo (R103A) purified from mammalian cell conditioned media.23 Levels of secreted Epo (R103A) obtained in these studies ranged from 0.01 to 1 μg/mL. In the only study in which Epo (R103A) was purified, the final yield was not described and the purity of the isolated protein was not demonstrated.23 These previously reported preparations of Epo (R103A) are thus not sufficiently characterized for animal studies.
Because Epo (R103A) binds with a 50-fold lower affinity to the EpoR compared to wild-type Epo (10 nM compared to 200 pM for Epo), large quantities of Epo (R103A) will be required for in vivo use. To obtain the large quantities of purified protein required for animal studies, we elected to express and purify Epo (R103A) from the yeast P pastoris. Using this method we can easily and inexpensively obtain up to 12 mg purified Epo (R103A) using 6 L yeast culture. Yields of 10-fold higher or more are possible with fermentation methods.
We have biochemically characterized the yeast-expressed Epo (R103A) with respect to N-terminal sequence, glycosylation, and receptor-binding activity. N-terminal sequence and glycosylation were found to be similar to wild-type, mammalian cell–expressed Epo. Appropriate glycosylation is of particular importance to ensure adequate pharmacokinetics in vivo, because unglycosylated Epo has a first-pass clearance in the liver.17 We demonstrate that Epo (R103A) will antagonize 125I-Epo binding to the EpoR. The inhibition of 125I-Epo binding by Epo (R103A) has not previously been demonstrated. The agreement between the apparent Ki for inhibition of 125I-Epo binding and the half-maximal inhibition of cell growth (Figures 3 and 4) is consistent with a mechanism of growth inhibition by Epo (R103A) based on competitive binding to the EpoR.
We have also characterized the biologic activity of the yeast-expressed Epo (R103A). We have confirmed previous work in which Epo (R103A) was shown to be an antagonist for Epo-dependent growth of cultured cell lines.12 In addition, we show that Epo (R103A) effectively and specifically antagonizes erythropoiesis of primary human erythroid cells from bone marrow and UCB (Figures 6 and7). This inhibition of erythropoiesis is due, at least in part, to decreased viability of the cells in presence of Epo (R103A). This decreased viability is likely due to the fact that erythroid precursors have an absolute requirement for Epo to avoid apoptosis24rather than by direct activation of apoptosis by Epo (R103A) (Figure5).
The dose-dependent inhibition of in vitro erythropoiesis is about 10-fold higher than that for direct competition for125I-Epo binding. As shown in Figure 4, the apparent Ki for inhibition of 125I-Epo binding is 0.32 μg/mL (equivalent to 10 nM), whereas half-maximal inhibition of in vitro erythropoiesis occurs at about 5 μg/mL (Figure 6). This difference might represent the residual biologic activity of the very few EpoR occupied by Epo in the presence of 5 μg/mL Epo (R103A).
Epo (R103A) exhibits an inhibitory effect on in vitro erythropoiesis over a wide range of concentrations from 0.1 to 10 μg/mL (Figure 6B). The dose-dependent inhibition of viability was significant over all doses of Epo (R103A) tested. This suggests that Epo (R103A) could be effectively used in vivo to titrate erythropoietic activity. Yeast-expressed Epo (R103A) thus represents a high-yield, low-cost, and well-characterized reagent that is suitable for studies in animal models.
Epo (R103A) has several potential research and clinical uses. As a research reagent, Epo (R103A) could be useful in the study of the regulation of erythropoiesis in vivo in animals, as well as in the study of nonerythropoietic functions of Epo, such as on endothelium.25 26 Epo (R103A) could be useful in suppressing endogenous abnormal erythropoiesis in diseases such as sickle cell anemia and thalassemia major. Yeast-expressed Epo (R103A) has the potential to be immunogenic, due both to the amino acid point mutation and the yeast glycosylation. This potential immunogenicity will need to be evaluated in animal models. We do not anticipate that Epo (R103A) will be an alternative to phlebotomy for most patients with erythrocytosis because phlebotomy is inexpensive, easy, and safe to perform, and is free of substantial side effects.
Several other mutant cytokines with antagonist properties have been described, including antagonists of growth hormone,27GM-CSF,22 IL-6,28 and IL-13.29The marked specificity of these agents makes them attractive potential drugs. The availability of large quantities of characterized yeast expressed Epo (R103A) will allow further studies to determine if this agent will have clinical utility as member of this new class of drugs.
We thank Dr Frank Bunn for the kind gift of Epo (R103A). Epo was a kind gift of Ortho Biotech, Raritan, NJ. Dr Linda Bonewald performed the protein sequencing at the University of Texas Health Science Center at San Antonio Protein Core Facility. Flow cytometry was performed at the University of Alabama at Birmingham Arthritis Center flow cytometry core. Vijaya Sambandam and Latania Watson provided excellent technical assistance.
Supported by a Veterans Administration Merit Review grant (K.W.H.) and National Institutes of Health grant DK-02566 (M.O.A.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Kevin W. Harris, Division of Hematology/Oncology, University of Alabama at Birmingham, Wallace Tumor Institute 520-D, 1530 3rd Ave S, Birmingham, AL 35294; e-mail:kevin.harris@ccc.uab.edu.