The glycoprotein Ib (GPIb) complex is composed of GPIbα covalently attached to GPIbβ and noncovalently complexed with GPIX and GPV. Patients with Bernard-Soulier syndrome demonstrate that mutations in either GPIbβ or GPIX result in an absence of platelet GPIbα. This occurs through the interaction of GPIX with GPIbβ. The precise sites of interaction of GPIbβ with GPIX are not known. To characterize the interaction of GPIbβ and GPIX, we developed an anti-GPIbβ monoclonal antibody MBC 257.4, whose epitope was in the N-terminal region of GPIbβ. N-terminal truncations of GPIbβ were expressed in mammalian cells. N-terminal truncations of GPIbβ, missing the first 14, 26, or 31 amino acids, were surface-expressed but did not enable coexpressed GPIX to be surface expressed, suggesting that the site of interaction with GPIX was modified by these deletions. GPIbβ and GPIX chimeras corresponding to predicted boundaries were used to define the sites of interaction of GPIbβ with GPIX. Replacing the N-terminal disulfide loops of GPIbβ (amino acids 1-14) with the corresponding disulfide loops of GPIX (amino acids 1-22) resulted in surface expression of coexpressed wildtype GPIX. However, when the N terminus of GPIbβ was replaced to residue 32 with the N terminus of GPIX (amino acids 1-36), GPIX did not surface express with this chimera. These results suggest that the cysteine knot region of GPIbβ in the N terminus is critical for the conformation of GPIbβ that interacts with GPIX and further suggests that a critical interaction of GPIbβ with GPIX involve residues 15 through 32 of GPIbβ.
Introduction
The platelet membrane glycoprotein Ib-IX-V (GPIb-IX-V) complex has a critical role in platelet adhesion mediating hemostasis. The complex is composed of 4 type 1 membrane-spanning proteins: GPIbα, GPIbβ, GPIX, and GPV. Each of these glycoproteins is encoded by a single copy gene. The genes for GPIbα and GPIbβ are located on chromosomes 17 and 22, respectively,1,2while the genes for GPIX and GPV are located on chromosome 3.3,4 GPIbα is disulfide linked to GPIbβ5,6; GPIX and GPV are noncovalently associated with the complex.7
It is known that GPIbβ and GPIX are essential for the surface expression of the complex from experiments in cell lines and naturally occurring mutations8,9 and that the interaction of GPIbβ with GPIX is essential for the surface expression of GPIbα. There are no data on how GPIbβ interacts with GPIX for the ordered surface expression of the complex. The von Willebrand factor (VWF) binds to the extracellular N-terminal domain of GPIbα (His1–Glu282),10,11 which contains 7 tandem leucine-rich repeats (LRRs) and has flanking disulfide looped sequences. GPIbβ and GPIX are related to GPIbα and are very similar to GPIbα in their extracellular domains. Both GPIbβ and GPIX have a single LRR. The amino- and carboxyl-terminal sequences flanking the LRR of GPIbβ and GPIX are very similar to each other and to GPIbα. In both of these regions, cysteines are conserved, suggesting conserved disulfide-loop structures. However, GPIbβ and GPIX have 4 cysteines N-terminal to the LRR, as compared with 2 in GPIbα. On the basis of homology with other LRR proteins, it has been suggested that these 4 cysteines form 2 disulfide loops with a loop-within-a-loop structure.5 To explore the site(s) of interaction between GPIX and GPIbβ, we developed a novel series of antibodies to GPIbβ. We used site-directed mutagenesis to produce N-terminal truncations of GPIbβ as well as GPIβ-IX chimeras to study the surface expression of the complex in mammalian cell lines.
Materials and methods
Monoclonal antibodies and reagents
Antibodies were produced as previously described.12 Briefly, BALB/c mice were immunized with acrylamide slices excised from sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gels containing reduced denatured platelet GPIbβ immunopurified as a disulfide-linked complex with GPIbα by means of the monoclonal antibody (MoAb) AP-1. The anti-GPIbα antibody AP-1 blocks VWF binding to GPIbα. From one fusion, a panel of 6 clones was obtained from which 4 were chosen since they reacted strongly with reduced platelet GPIbβ. The MoAb MBC 257.4 immunoblots both reduced and nonreduced platelet GPIbβ and was used in the studies described below.
Anti-GPIX MoAbs FMC 25 and GRP were purchased from Harlan Bioproducts (Indianapolis, IN). AK1, a MoAb that recognizes an epitope on GPIX that requires the intact GPIb-IX complex,13 was a generous gift of Dr Michael C. Berndt (Baker Medical Research Institute, Central Melbourne, Victoria, Australia). We have previously shown that this antibody requires the presence of GPIbβ in cells stably transfected with GPIbα and GPIX to recognize the complex.14
Construction of expression vectors
We have previously described a Bernard-Soulier syndrome (BSS) patient who had a deletion of nucleotide 336 in GPIbβ causing a frameshift and premature termination (GPIbβ-trunc).9This patient has no detectable GPIbα on the surface of his platelets and has the classical BSS phenotype. We used recombinantly expressed protein containing this mutation to identify the epitope for MBC 257.4. Full-length expression vectors containing GPIbβ and full-length GPIX have previously been described.9 N-terminal truncations of GPIbβ were created by means of a strategy based on the type IIS restriction enzyme BsmBI.15 16 Since BsmBI creates a staggered cut on opposite strands of double-stranded DNA at 1 and 5 bases, respectively, 3′ of its nonpalindromic recognition sequence (CGTCTC), any desired 4-base overhang can be created by placing sites at the 5′-end of polymerase chain reaction (PCR) primers. The nonhomologous recognition sequence is removed when digested. Thus left- and right-half pairs of PCR products designed with uniquely compatible cohesive ends can be merged seamlessly.
Full-length GPIbβ was used as the amplification template in creating N-terminal truncations. The left-half PCR extended 3′ from the flanking pCIneo sequence (pCIneo961 5′CTTGCGTTTCTGATAGGCACCT) through the signal peptide of GPIbβ. The right-half PCR extended from the point of the desired truncation through the remainder of GPIbβ and into the other side of pCIneo (pCI1158 5′-ATTAACCCTCACTAAAGGGAAG (Table1). The numbering system used in the tables and “Results” refers to the first amino acids following the signal peptide.
PCR products were ligated into PCRII and sequenced to confirm fidelity. Then right- and left-half constructs were digested with XhoI and BsmBI and subcloned into the pCIneo expression vector at the XhoI/XbaI site, in a 3-fragment ligation.
GPIX/GPIbβ chimeras were created with the use of wildtype plasmids as templates in a strategy in which 2 overlapping first-round PCRs are combined as templates for second-round reamplification to allow extension of a full-length complementary DNA. The first-round PCR products were amplified as follows: the N-terminal portion of the chimera was amplified from GPIX by means of a plasmid primer paired with a chimeric antisense primer containing approximately 20 bases of GPIX sequence followed by approximately 20 bases of GPIbβ sequence, to produce a first-round product consisting of GPIX sequence up to the desired splice point, followed by a 20-base GPIbβ overhang. The 3′ (C-terminal) portion was amplified from a GPIbβ plasmid in a similar manner with the use of a chimeric sense primer containing approximately 20 bases of GPIX overhang followed by GPIbβ sequence complementary to the 20–base pair overhang produced in the previous PCR. The 2 first-round products anneal at the overlapping chimeric GPIX/β overhangs to serve as templates for second-round amplification. The final construct is amplified by means of flanking plasmid primers to produce a chimera consisting of GPIX and GPIbβ (Table 2).
Transient expression
For transient expression studies, HEK293T cells were used. The parent HEK293T cell line is a human renal epithelial cell, transformed with simian virus–40 large T antigen.17 HEK293T cells were maintained at 37°C in a 5% CO2 humidified chamber in modified Eagle media (Sigma, St Louis, MO) supplemented with 10% fetal calf serum. Expression plasmids were transfected into HEK293T cells in the presence of Lipofectamine and Lipfectamine Plus (GIBCO BRL, Rockville, MD) as described.9
Flow cytometry studies
Transfected cells were detached from tissue-culture plates with 3 mM EDTA, centrifuged at 250g, and resuspended in Hanks Balanced Salt Solution with 2% bovine serum albumin. Then, 3 × 105 cells were transferred to each well of a 96-well V-bottom plate (Dynatech, Chantilly, VA) and incubated with either anti-GPIbβ MoAb 257.4 (5 μg/mL); the anti-GPIX MoAbs FMC-25 or GRP (5 μg/mL); or the complex-specific MoAb AK1 (ascites 1:1200). The cells were then centrifuged, resuspended, and incubated for an additional 30 minutes in a darkened room with a 1:100 dilution of phycoerythrin-conjugated affinity-purified F(ab′)2 donkey anti–mouse immunoglobulin-G (IgG) (Jackson Immunoresearch Laboratories, West Grove, PA). The cells were then centrifuged and resuspended in 2% paraformaldehyde, allowed to incubate at least 1 hour at 4°C, and analyzed in a Becton Dickinson (San Jose, CA) FACScan flow cytometer.
Immunoblotting
Cell lysates were separated by SDS-PAGE on a 4% to 20% gradient gel according to Laemmli.18 The separated proteins were electroblotted onto a polyvinylidine difluoride membrane (Novex, San Diego, CA) as described by Towbin et al,19blocked in phosphate-buffered saline (PBS) containing 5% powdered milk, and then incubated overnight with the anti-GPIbα MoAb MBC 257.4. The membrane was washed 3 times with PBS containing 5% powdered milk, incubated with a goat anti–mouse IgG conjugated with horseradish peroxidase, washed again 3 times, developed with SuperSignal Chemiluminescent Substrate (Pierce, Rockford, IL), and visualized by luminescent radiography.
Results
Characterization of GPIbβ MoAb MBC 254.7
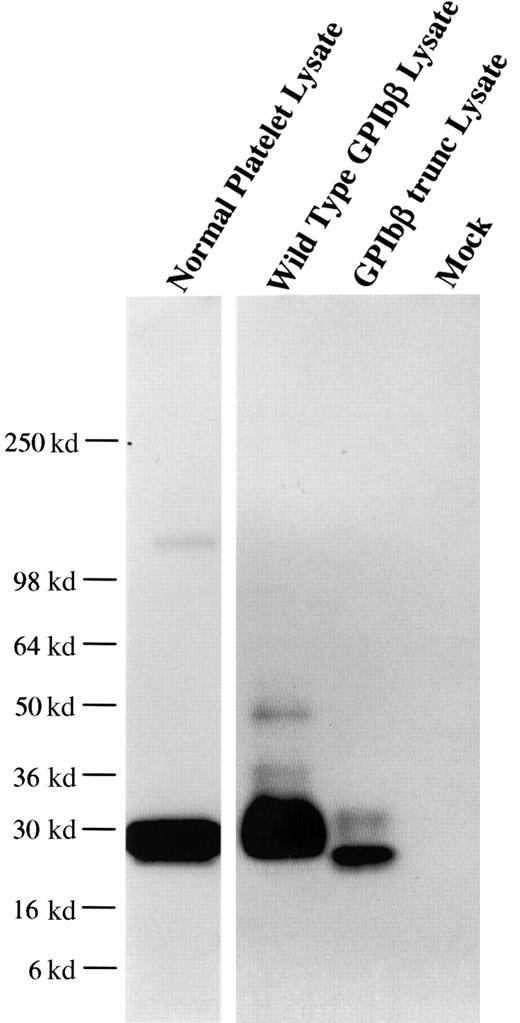
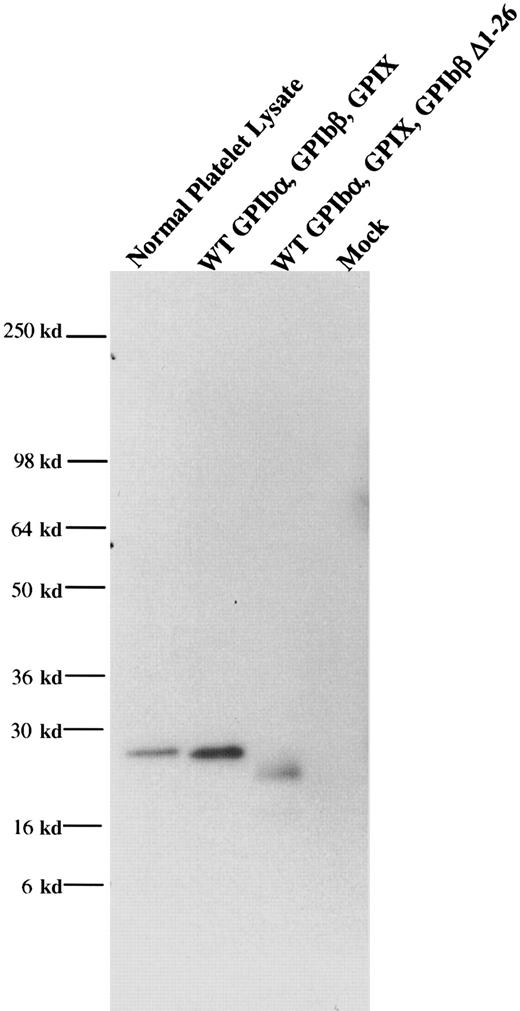
From one anti-GPIbβ fusion, a panel of 6 anti-GPIbβ clones were obtained, of which 4 reacted strongly with reduced platelet GPIbβ by Western blot. The MoAb MBC 257.4 was selected on the basis of its ability to immunoblot both reduced and nonreduced GPIbβ. The epitope of MBC 257.4 was evaluated in a series of experiments expressing known recombinant proteins. In a BSS patient we have previously characterized, we identified a single nucleotide deletion within the codon for Ala80 in GPIbβ. This mutation causes a translational frameshift, which encodes for 86 altered amino acids and predicts stop codon 15 amino acids short of the length of the wildtype protein. HEK293T cells were transfected with this construct, lysed, run on SDS-PAGE, and reacted with MBC 257.4. MBC 257.4 recognized this expressed protein (Figure 1), suggesting that the epitope for MBC 257.4 is within the N-terminal 80 amino acids of GPIbβ.
MoAb 257.4 recognition of an epitope in the N terminus 80 residues of GPIbβ.
The recombinant proteins GPIbβ and GPIbβ-trunc9 were expressed in HEK293T cells. The cells were lysed, analyzed by SDS-PAGE, and immunobloted with MBC 257.4. Lane 1 shows platelet lysate; lane 2, recombinant wildtype GPIbβ; lane 3, recombinant GPIbβ-trunc.9 The epitope for MBC 257.4 is within the N terminus 80 residues of GPIbβ.
MoAb 257.4 recognition of an epitope in the N terminus 80 residues of GPIbβ.
The recombinant proteins GPIbβ and GPIbβ-trunc9 were expressed in HEK293T cells. The cells were lysed, analyzed by SDS-PAGE, and immunobloted with MBC 257.4. Lane 1 shows platelet lysate; lane 2, recombinant wildtype GPIbβ; lane 3, recombinant GPIbβ-trunc.9 The epitope for MBC 257.4 is within the N terminus 80 residues of GPIbβ.
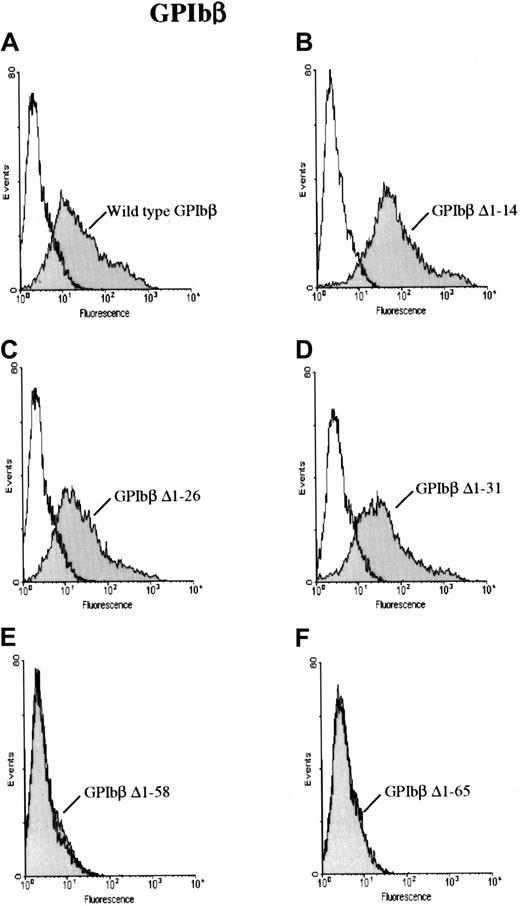
To further characterize the antibody, HEK 293T cells were transfected with a construct for GPIbβ with sequential N-terminal deletions. The cells were then reacted with MoAb 257.4 and analyzed by flow cytometry. GPIbβ constructs truncated at residues 14, 26, or 31 were readily detectable on the surface of cells transfected with the respective constructs. However GPIbβ truncated at residues 58 and 65 was not detectable with MoAb 257.4 (Figure2). When the cells transfected with the N-terminal deletions 58 and 65 were reacted with an anti-GPIbβ polyclonal antibody, there was no increase in surface fluorescence, further suggesting that these constructs were not surface expressed.
GPIbβ with N-terminal deletions to residue 31 and N-terminal deletions of GPIbβ distal to residue 58.
GPIbβ with N-terminal deletions to residue 31 is expressed on the cell surface and recognized by MoAb 257.4, whereas N-terminal deletions of GPIbβ distal to residue 58 are not detected on the cell surface. HEK293T cells were transfected with wildtype GPIbβ alone (panel A) and with GPIbβ truncated at residue 14 (panel B), 26 (panel C), 31 (panel D), 58 (panel E), and 65 (panel F). The cells were then reacted with MoAb 257.4. There is a significant increase in fluorescence on the cells expressing wildtype GPIbβ and GPIbβ truncated at residues 14, 26, and 31. There is no increase in surface fluorescence in cells transfected with GPIbβ truncated at residue 58 or 65 compared with mock-transfected cells. Thus, N-terminal truncations to residue 31 appear to be expressed while truncations to 58 do not.
GPIbβ with N-terminal deletions to residue 31 and N-terminal deletions of GPIbβ distal to residue 58.
GPIbβ with N-terminal deletions to residue 31 is expressed on the cell surface and recognized by MoAb 257.4, whereas N-terminal deletions of GPIbβ distal to residue 58 are not detected on the cell surface. HEK293T cells were transfected with wildtype GPIbβ alone (panel A) and with GPIbβ truncated at residue 14 (panel B), 26 (panel C), 31 (panel D), 58 (panel E), and 65 (panel F). The cells were then reacted with MoAb 257.4. There is a significant increase in fluorescence on the cells expressing wildtype GPIbβ and GPIbβ truncated at residues 14, 26, and 31. There is no increase in surface fluorescence in cells transfected with GPIbβ truncated at residue 58 or 65 compared with mock-transfected cells. Thus, N-terminal truncations to residue 31 appear to be expressed while truncations to 58 do not.
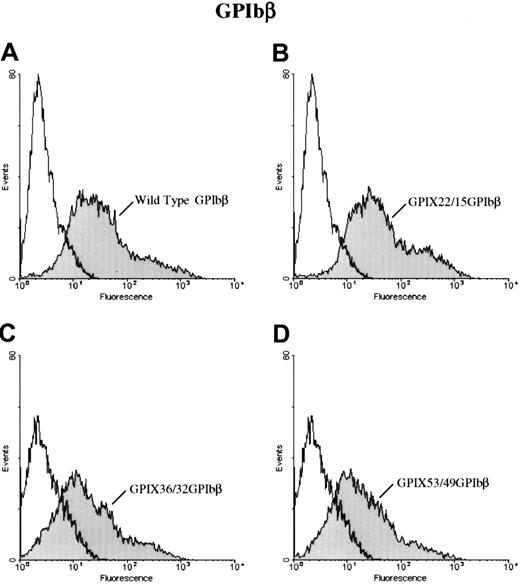
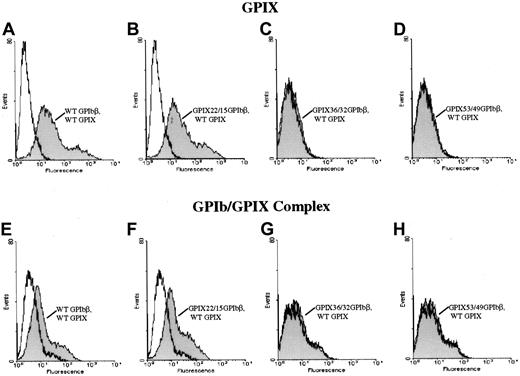
To further define the epitope for 257.4, we analyzed the binding of 257.4 to a series of GPIbβ/GPIX chimeras by flow cytometry. Chimeras were created where sequential N-terminal deletions of GPIbβ were replaced with the corresponding structural domain of GPIX. This approach allowed us to more precisely define the binding region for MoAb 257.4. The cysteine knot at the N terminus of GPIbβ (residues 1-14) was spliced and replaced with the cysteine knot region of GPIX, residues 1 through 22, to create chimera GPIX22/β15. MoAb 257.4 bound to this chimera (Figure 3). The N terminus of GPIbβ was truncated more distally at residue 31 and replaced with the homologous region from GPIX, chimera GPIX36/β32. This chimera was detected on the surface of transfected cells with MoAb 257.4 (Figure 3). The N terminus of GPIbβ was truncated at residue 49 and replaced with the homologous region of GPIX, chimera GPIX53/β49. This chimera was recognized by MoAb 257.4 (Figure 3). Since GPIbβ was replaced to residue 49 in these chimeras and the MoAb 257.4 reacted strongly with this chimera, this suggests that the epitope for the antibody lies distal to residue 49. This antibody also recognizes a recombinant mutant GPIbβ with a translational frameshift at residue 80. Together, these results suggest that the epitope for MBC 257.4 is between residues 50 and 80 of GPIbβ. As discussed above, these truncations defined the epitope for MBC 257.4 and allowed us to explore more precisely the interactions of GPIbβ with GPIX.
Surface expression of chimeras of GPIX and GPIbβ.
HEK 293T cells transfected with wildtype GPIbβ (panel A) or the chimeras GPIX22ñ β15, (panel B) GPIX36/β32 (panel C), and GPIX53/49 (panel D) were reacted with MoAb 257.4. There is a significant increase in fluorescence in cells transfected with chimeras GPIX22ñ β15 and GPIX36/β32 and GPIX53/49. Thus, chimeras of GPIX and GPIbβ are surface expressed.
Surface expression of chimeras of GPIX and GPIbβ.
HEK 293T cells transfected with wildtype GPIbβ (panel A) or the chimeras GPIX22ñ β15, (panel B) GPIX36/β32 (panel C), and GPIX53/49 (panel D) were reacted with MoAb 257.4. There is a significant increase in fluorescence in cells transfected with chimeras GPIX22ñ β15 and GPIX36/β32 and GPIX53/49. Thus, chimeras of GPIX and GPIbβ are surface expressed.
Complex formation of GPIbβ with GPIX
Expression of GPIX with N-terminal truncations of GPIbβ.
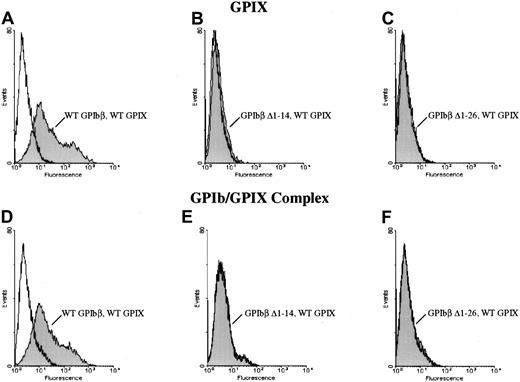
To investigate the interaction of GPIbβ with GPIX, plasmids encoding GPIbβ and GPIX or wildtype GPIX with serial N-terminal truncations of GPIbβ were transiently transfected into HEK293T cells. These cells were then incubated with an anti-GPIX MoAb, FMC25, followed by a phycoerythrin-conjugated donkey antimouse antibody, and then analyzed by flow cytometry.
When wildtype GPIbβ and wildtype GPIX were simultaneously cotransfected into HEK 293T cells, GPIX were readily detectable on the cell surface (Figure 4). However, when GPIbβ truncated at residues 14 or 26 was cotransfected into HEK293T cells, GPIX was not detectable on the cell surface (Figure 4B-C), nor was there any increase in fluorescence when the cells were reacted with the complex-specific antibody AK1 (Figure 4E-F). To determine if these truncations of GPIbβ interacted with GPIbα, GPIbβ truncated at residue 26 and wildtype GPIbα were simultaneously transfected into HEK293T cells. The cells were immunoprecipitated with the anti-GPIbα antibody and immunoblotted with MBC 257.4. Since GPIβΔ1-26 is present, this demonstrates that it is complexed with GPIbα (Figure5).
The role of the N terminus of GPIbβ in the coexpression of GPIX.
The N terminus of GPIbβ is essential for the coexpression of GPIX. Wildtype GPIbβ and wildtype GPIX were simultaneously cotransfected into HEK 293T cells (panels A, D). GPIbβ truncated at residues 14 (B, E) or 26 (C, F) were cotransfected into HEK293T cells with GPIX. Cells were then reacted with the anti-GPIX antibody FMC 25 (panels A-C) or the complex-specific antibody AK1 (panels D-F). N-terminal truncations of GPIbβ do not bring GPIX to the surface.
The role of the N terminus of GPIbβ in the coexpression of GPIX.
The N terminus of GPIbβ is essential for the coexpression of GPIX. Wildtype GPIbβ and wildtype GPIX were simultaneously cotransfected into HEK 293T cells (panels A, D). GPIbβ truncated at residues 14 (B, E) or 26 (C, F) were cotransfected into HEK293T cells with GPIX. Cells were then reacted with the anti-GPIX antibody FMC 25 (panels A-C) or the complex-specific antibody AK1 (panels D-F). N-terminal truncations of GPIbβ do not bring GPIX to the surface.
GPIbβΔ1-26 complexed with GPIbα.
GPIbβ truncated at residue 26 and wildtype GPIbα were simultaneously transfected into HEK293T cells. The cells were immunopreciptated with an anti-GPIbα antibody and immunoblotted with an anti-GPIbβ polyclonal antibody. This demonstrated that GPIβΔ1-26 is complexed with GPIbα.
GPIbβΔ1-26 complexed with GPIbα.
GPIbβ truncated at residue 26 and wildtype GPIbα were simultaneously transfected into HEK293T cells. The cells were immunopreciptated with an anti-GPIbα antibody and immunoblotted with an anti-GPIbβ polyclonal antibody. This demonstrated that GPIβΔ1-26 is complexed with GPIbα.
Expression of GPIX with GPIbβ/GPIX chimeras.
Since N-terminal truncations of GPIbβ allowed the surface expression of GPIbβ alone but did not bring GPIX to the surface, we explored the interaction of GPIbβ with GPIX with a series of GPIbβ/GPIX chimeras. To control for potential conformational changes induced by deleting the N terminus of GPIbβ, we generated a series of chimeras between GPIbβ and GPIX at defined boundaries. The cysteine knot at the N terminus of GPIbβ (residues 1-14) was spliced and replaced with the cysteine knot region of GPIX (residues 1-22) to create chimera GPIX22/β15. This chimeric construct was cotransfected simultaneously with wildtype GPIX. The cells were then reacted with the complex-specific antibody AK1. Chimera GPIX22/β15 expressed on the cell surface (Figure 3). When chimera GPIX22/β15 was cotransfected into HEK293T cells simultaneously with GPIX, there was a significant increase in fluorescence when the cells were reacted with the complex-specific antibody AK1, suggesting that chimera GPIX22/β15 interacts with GPIX to bring GPIX to the surface (Figure6).
N-terminal 14 amino acids of GPIbβ and surface expression of GPIX.
The N-terminal 14 amino acids of GPIbβ are essential for the surface expression of GPIX. The chimeras GPIX22/β15, GPIX36/β32, and GPIX53/49 were cotransfected with GPIX into HEK293T cells and reacted with the anti-GPIX antibody FMC 25 (panels A-D) or the complex-specific antibody AK1 (panels E-H). Panels A and E are wildtype beta and wildtype IX. Panels B and F are the chimera GPIX22/β15, which brings GPIX to the surface and is complexed to GPIX22/β15 since there is a significant increase in surface fluorescence in these cells when reacted with the antibody AK1 as compared with mock-transfected cells. In contrast, when cells simultaneously transfected with chimeras GPIX36/β32 (panels C, G) and GPIX53/49 (panels D, H) and GPIX are reacted with either the anti-GPIX (panels C, D) antibody or AK1 (panels G, H), there is no increase in fluorescence compared with mock-transfected cells. Thus, the N-terminal 14 residues of GPIbβ are essential for the surface expression of GPIX.
N-terminal 14 amino acids of GPIbβ and surface expression of GPIX.
The N-terminal 14 amino acids of GPIbβ are essential for the surface expression of GPIX. The chimeras GPIX22/β15, GPIX36/β32, and GPIX53/49 were cotransfected with GPIX into HEK293T cells and reacted with the anti-GPIX antibody FMC 25 (panels A-D) or the complex-specific antibody AK1 (panels E-H). Panels A and E are wildtype beta and wildtype IX. Panels B and F are the chimera GPIX22/β15, which brings GPIX to the surface and is complexed to GPIX22/β15 since there is a significant increase in surface fluorescence in these cells when reacted with the antibody AK1 as compared with mock-transfected cells. In contrast, when cells simultaneously transfected with chimeras GPIX36/β32 (panels C, G) and GPIX53/49 (panels D, H) and GPIX are reacted with either the anti-GPIX (panels C, D) antibody or AK1 (panels G, H), there is no increase in fluorescence compared with mock-transfected cells. Thus, the N-terminal 14 residues of GPIbβ are essential for the surface expression of GPIX.
In another chimeric construct, the N terminus of GPIbβ was truncated more distally at residue 31 (N terminus to the LRR region) and replaced with the homologous region from GPIX, chimera GPIX36/β32. This chimera surface expresses (Figure 3), but does not interact with, GPIX (Figure 6) since it is not detected with AK1.
The N terminus of GPIbβ was truncated at residue 49 (middle of the leucine-rich motif) and replaced with the homologous region of GPIX, chimera GPIX53/β49; this chimera did surface express (Figure 3), but did not complex with, GPIX (Figure 6), as shown by the negative response when the cells are reacted with AK1. Thus, the residues 15 through 32 of GPIbβ are critical for the interaction with GPIX.
Discussion
It has previously been shown, in experiments using recombinant expression in mammalian cell lines and in naturally occurring mutations, that the interaction of GPIbβ with GPIX is essential for the normal surface expression of GPIbα.8 9The precise sites of interaction of GPIbβ and GPIX are not known. To define the sites of interaction of GPIbβ with GPIX, we made a new series of antibodies, which recognize GPIbβ. We demonstrated that one of these antibodies recognizes an epitope between residues 50 and 80 of GPIbβ. We then constructed a series of truncations of GPIbβ at the amino terminus. These experiments suggested that the amino terminus was important for the interaction between GPIbα and GPIbβ. We then generated a series of chimeras, replacing the N terminus of GPIbβ with the corresponding domain of GPIX, and demonstrate that the cysteine knot region of GPIbβ is critical for the conformation of GPIbβ that interacts with GPIX. Furthermore, we have shown that residues 15 through 32 of GPIbβ are critical for the interaction of GPIbβ with GPIX.
The secondary structures of GPIX and GPIbβ have not yet been defined. It has been suggested that these regions are homologous to GPIbα.5 GPIbα has an N-terminal flank with a single cysteine loop, 7 LRRs, and a C-terminal flanking region containing 2 disulfide loops. GPIbβ and GPIX have similar structural domains, with each containing a single LRR. It is interesting that the flanking amino acids on both sides of the LRR are homologous in the proteins of this group. It has been suggested by Hickey et al20 that the flanking amino acid sequences on both sides of the region containing the LRR may represent the product of an ancestral oligonucleotide sequence in which the flanking segments remained constant and the central LRR underwent duplication. The results of the present investigation demonstrate a critical role for the N terminus–flanking region of GPIbβ with GPIX.
Molecular characterization of the defects in BSS has helped define critical regions in the GPIb-IX complex. Our previous investigations have demonstrated that GPIbβ is essential for the normal surface expression of GPIX. There are 4 published reports of mutations within GPIbβ that cause the BSS phenotype.9,14,21,22 To date, there have been no descriptions of point mutations within the N-terminal flanking region of GPIbβ that cause the syndrome. In contrast, 2 mutations that cause BSS have been described within the C-terminal flanking region of GPIX.23,24 It is unclear why point mutations in the N-terminal flanking region of GPIX give rise to the BSS phenotype. One explanation is that the mutation produces an unstable protein that fails to express. Alternatively, if this region of GPIX can no longer interact with GPIbβ, neither GPIbα nor GPIX would be detectable on the platelet surface. In the patient described by Rivera et al,23 a homozygous T→C transition changed Cys8→Arg. The results of our experiments suggest that this region of GPIX might no longer interact with GPIbβ, and hence GPIbα would not be expressed on the cell surface. Interestingly, Rivera et al noted circulating glycocalicin in their patient's plasma. This suggests that GPIbα was being made, yet not inserted in the plasma membrane. We have noted a similar finding in another BSS patient with a mutation in GPIbβ.14 Wright et al24 described 3 patients who were doubly heterozygous for mutations within GPIX. One mutation was an A→G transition in codon 21, and the other was an A→G change in codon 45. These mutations changed an Asp to Gly in the N terminus–flanking region of GPIX and an Asn to Ser within the LRR. Again, it was not clear how these mutations caused the BSS phenotype although the authors proposed that these mutations disrupted the stable assembly of the complex. Our results support their hypothesis. Mutations in GPIX that result from changes in the codons for cysteines on either flanking region of the LRR resulted in BSS.23,25 This mutation probably disrupts the disulfide bonding pattern in the amino terminus and hence altered the secondary structure of GPIX, which disrupts the proper assembly and anchoring of the complex.23 In the present investigation, we used chimeras where the cysteine knots of both GPIbβ and GPIX were preserved within each of the subunits by swapping precise structural domains. These results demonstrate the critical importance of this region for the precise insertion of the whole complex in the plasma membrane.
Dong et al26 investigated assembly and transport of the entire GPIb complex using transfected mammalian cells. They showed that the full complex was formed within minutes in the endoplasmic reticulum. Surprisingly, they noted that GPIbα, even in the complexed form, was highly susceptible to degradation. When GPIbα was expressed as a partial complex with either GPIbβ or GPIX, more than 60% of GPIbα was degraded. In contrast, the partial complex consisting of GPIbβ and GPIX was relatively stable. From our previous investigations on patients with BSS, we have shown that it is the critical interaction of GPIbβ with GPIX that brings GPIbα to the surface. The present investigation extends our previous work and the analysis of complex assembly by Dong et al.26
GPIX mutations appear to disrupt the GPIb/IX structure, as evidenced by exposure of a cryptic site on GPIbβ23 and decreased expression of GPIX thorough its failure to interact with GPIbβ.24 Previous investigations on the role of GPIbβ have been limited by a paucity of GPIbβ antibodies. In this investigation, we made and characterized a novel anti-GPIbβ antibody. While the results of this investigation further define the role of GPIbβ in assembly of the complex, additional studies are required to further define the functional role of GPIbβ.
Supported by National Institutes of Health grants HL56027, HL44612, and HL33721 and grants from the Higher Education Authority and Enterprise Ireland.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Dermot Kenny, MACC Fund Research Center—Dept of Pediatrics, Medical College of Wisconsin, 8701 Watertown Plank Rd, PO Box 26509, Milwaukee, WI 53226-0509; e-mail: dkenny@rcsi.ie.