Abstract
The zinc-finger protein Ikaros plays an important role in lymphoid homeostasis, and loss of Ikaros expression through germline disruption impairs lymphoid development. However, the role played by Ikaros after commitment to the T-cell lineage is unclear. To address this question, this study used the lck proximal promoter to drive the expression in T-cell progenitors of a naturally occurring short Ikaros isoform (IK5), which lacks the DNA-binding domain, reasoning that IK5 will form heterodimers with long isoforms and perturb their function. The IK5 transgene led to a selective and dramatic decrease in extrathymic intestinal intraepithelial lymphocytes (IELs) and natural killer 1.1+ T (NK T) cells with little effect on conventional αβ T cells, which resembles the T-cell phenotype of interleukin-15 receptor α chain (IL-15Rα) and IL-2/IL-15 receptor β chain (IL-2Rβ) knockout mice. The expression of IL-2Rβ on double-negative T-cell progenitors of bi-5 was reduced, but enforced expression of IL-2Rβ did not rescue IELs or NK T cells in bi-5 transgenic mice, suggesting that Ikaros or Ikaros family members regulate the expression of additional genes that are essential for the development of IELs and NK T cells. The study concludes that modest changes in the ratio of short to long Ikaros isoforms can substantially perturb T-cell development, and the development of IELs and NK T cells is particularly sensitive to such changes.
Introduction
Ikaros and its related family members, Aiolos and Helios, are DNA-binding proteins that appear to regulate the development and function of lymphoid cells at multiple stages (reviewed in Cortes et al1). Ikaros is expressed throughout lymphoid development, in some myeloid lineages, and even in the earliest hematopoietic stem cells (HSCs).2-4 Alternative splicing of Ikaros messenger RNA (mRNA) leads to formation of multiple different isoforms, which differ in biologic activity. Long Ikaros isoforms bind to a core DNA-binding motif, whereas short isoforms do not.5 However, short Ikaros isoforms form heterodimers with long isoforms and may thereby inhibit the ability of long isoforms to engage DNA.5 The predominant isoforms expressed throughout T-cell development are the long isoforms (IK1 and IK2),4 whereas short isoforms appear to be relatively more abundant in some myeloid lineages and short-term engrafting HSCs.3
Ikaros is critically important for the normal development of all lymphoid cells. Ikaros null mice (IK−/−) lack B, natural killer (NK), and lymphoid dendritic cells and are markedly deficient in γδ T cells. Conventional αβ T-cell development is almost completely absent in the youngest IK−/− mice, with about 100-fold fewer thymocytes at birth.1,4 Thymocyte number increases in older mice such that the total can approach wild-type numbers after 6 weeks of age. The αβ thymocytes that develop in these mice are aberrant, as characterized by extreme expansion of CD4+ cells and a hyperproliferative response to T-cell receptor (TCR) stimulation.4 Mice in which only the DNA-binding domain of Ikaros is disrupted (IK-DNA−/−mice) have an even more severe phenotype, in that all lymphoid cells are missing.3 The more severe phenotype in IK-DNA−/− mice is thought to reflect dominant-negative inhibition by short Ikaros isoforms of long Ikaros isoforms and other members of the Ikaros family of proteins.
Germline disruption of Ikaros may preclude a clear assessment of its role at later stages by altering the development potential of HSCs for both myeloid and lymphoid development.6 7 Thus, to explore the role of Ikaros in committed lymphoid progenitors independent of effects in HSCs and to determine the importance of the predominance of long isoforms in developing T cells, we generated mice in which the short Ikaros isoform IK5 was expressed exclusively in T-cell progenitors. IK5 lacks the ability to bind the Ikaros core DNA-binding motif but still contains the C-terminal zinc fingers required to form dimers. Although IK5 transgene expression was modest and less than the aggregate expression of the endogenous Ikaros isoforms, the development of NK T cells and extrathymically derived intraepithelial lymphocytes (IELs) was markedly and selectively impaired. These results indicate that even after commitment to the T-lymphocyte lineage, the predominance of long relative to short Ikaros isoforms is essential for normal T-cell development.
Materials and methods
Mice strains and transgenic mouse construction
A full-length Ikaros complementary DNA (cDNA) was provided by K. Georgopoulos (Massachusetts General Hospital, Charlestown, MA). The IK5 isoform was made from full-length cDNA by polymerase chain reaction (PCR) with splice overlap extension to remove exons 4 to 6 and was sequenced to verify that there were no mutations. The IK5 cDNA was inserted into the BamHI site of the p1026 promoter (lck proximal promoter plus Eμ enhancer) obtained from R. Perlmutter.8 Transgenic mice were generated as described previously.9 Mice were screened by PCR of tail DNA by using primers that amplify across the junction between the lck proximal promoter and the 5′ coding region of Ikaros. Mice expressing the murine interleukin-2 receptor β (IL-2Rβ) chain under the control of the CD2 promoter/enhancer have been described previously.10Mice expressing a bcl-xL transgene under the lck proximal promoter11 were a gift from S. Korsmeyer (Dana-Farber Cancer Center, Boston, MA). C57BL/6 and β2M−/− mice were obtained from Taconic. CD1−/− mice, which had been backcrossed 4 times onto C57BL/6 before being used, were obtained from M. Grusby (Harvard School of Public Health, Boston, MA).12
Northern blot analysis
Cells were isolated from thymus, spleen, and bone marrow. After ammonium chloride lysis, total RNA was extracted from the cells using Trizol (Life Technologies). A probe containing exon 7 of Ikaros was used to identify transgene expression. The Northern blot was stripped and reprobed using an elongation factor-1α cDNA probe, as previously described.13
Western blot analysis
Cells were collected and lysed in TNT buffer (50 mM Tris-HCL, 150 mM NaCl,1% Triton X-100).14 Total protein was quantitated using a Coomassie protein assay (Pierce). Each lane received either 40 μg protein or the lysates from 100 000 cells for the cell equivalent Western blots. The samples were separated by a 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel. After transfer to nitrocellulose by using a semidry transfer apparatus (Ellard Instrumentation), Ikaros was detected by using 1:1000 anti–C-terminal Ikaros antisera (provided by S. Smale, University of California, Los Angeles) and goat antirabbit–horseradish peroxidase at 1:3000 (Zymed). The enhanced chemiluminence system (New England Nuclear) was used to visualize bands.
IEL preparations
The small intestine was removed and Peyer patches were excised. After removing fecal matter, the intestines were first cut longitudinally and then cut into 1-cm pieces. The pieces were digested by using 1 mM EDTA in Hanks balanced salt solution (HBSS) at 37°C with repeated vortexing to separate lymphocytes from the epithelial sheathes. Aliquots were removed over time as the epithelial sheathes settled to the bottom. The first 2 aliquots were replaced with EDTA/HBSS, and subsequent aliquots were replaced with 5% fetal bovine serum in HBSS. Removed aliquots were pooled and concentrated before running over a nylon wool (Sigma) column. A 40%/75% Percoll (Sigma) gradient was used to further enrich for IELs before analysis.
Cell staining and processing
Cells were stained using anti-CD3ε, CD4, CD5, CD8α, CD24, CD43, IL-2Rγ, IL-2Rβ, Vβ8-TCR, TCRβ, TCRγδ (Pharmingen), B220, NK1.1, CD8β, CD25, and CD44 (CalTag, South San Francisco, CA). Antibodies to IL-7Rα were obtained from Andy Farr (University of Washington, Seattle). The antibodies were directly conjugated to phycoerythrin (PE), fluorescein isothiocyanate (FITC), Tricolor (TC), CyChromeC (CyC), PharRed, Allophycocyanin (APC), or biotin. Those antibodies labeled with biotin were secondarily stained with streptavidin (SA)–TC. Bead depletions were performed following the manufacturer's instructions by using Dynal SA-magnetic beads and biotinylated antibodies.
Results
Generation of transgenic mice that express the Ikaros isoform IK5
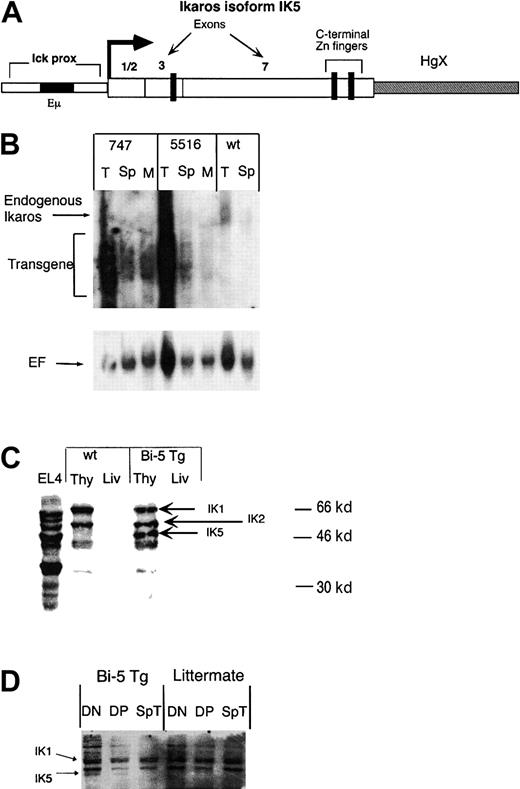
Transgenic mice were generated in which IK5 was expressed under the control of the immunoglobulin heavy chain enhancer (Eμ and the lck proximal promoter (Figure1A),8 which hereafter are referred to as bi-5 mice. The transgene mRNA was expressed in thymocytes, with reduced expression in splenocytes and total bone marrow (Figure 1B). After normalizing transgene expression to an elongation factor control, the 2 lines of bi-5 mice (lines 747 and 5516) appeared to express transgene mRNA in thymocytes in approximately similar amounts. By reverse transcriptase (RT)–PCR, expression of the transgene mRNA in NK cells and in mature T cells was approximately 5- to 10-fold less than in CD4+CD8+double-positive (DP) thymocytes (data not shown). This is consistent with previous data, indicating that expression directed by this promoter/enhancer is highest in T-cell progenitors.8 15Western blot analysis (Figure 1C) indicated that the abundance of IK5 protein in the thymus of bi-5 mice was similar to or somewhat less than the abundance of the full-length isoform IK1 and the slightly shorter isoform IK2. IK5 protein was expressed in CD4−CD8− (double- negative [DN]) and DP thymocytes of the bi-5 mice but was not detected in splenic T cells of bi-5 mice or in thymocytes or splenic T cells of controls (Figure 1D). Ikaros protein was not detected in the livers of bi-5 or littermate control mice (Figure 1C). IK5 mRNA was detected by sensitive isoform-specific RT-PCR in thymocytes from control mice, indicating that this isoform is expressed normally but in low abundance relative to the long isoforms (data not shown).
Generation of transgenic mice expressing the Ikaros isoform IK5.
(A) Representation of the construct used to generate IK5 (bi-5) transgenic mice. The construct contains the lck proximal promoter and the Eμ immunoglobulin heavy-chain enhancer to direct expression of IK5. The IK5 cDNA lacks exons 4 to 6, which eliminates 3 of 4 possible DNA-binding zinc fingers, yet retains all of the C-terminal Zn++ fingers. The HgX minigene has been shown to enhance the expression of transgenes and was part of the original p1026 promoter construct.9 (B) Northern blot of the 2 different bi-5 lines, 747 and 5516, using a probe containing exon 7 of Ikaros. Both endogenous Ikaros and transgene mRNA are marked in the figure. Cells used include thymocytes (T), splenocytes (Sp), and total bone marrow (M). The blot is slightly overexposed to better visualize marrow and spleen expression of the transgene. The HgX minigene, although not capable of being translated, contains both introns and exons; therefore, the transgene mRNA is expressed as multiple, overlapping bands. The lower panel shows the blot after stripping and reprobing for elongation factor-1α (EF) to account for loading differences. (C) Western blot using antisera against the C-terminal portion of Ikaros, which recognizes all Ikaros isoforms. Samples include protein isolated from the thymus (Thy) and liver (Liv) of bi-5 transgenic line 747 and a littermate control (wt). (D) Cell equivalent Western blot using 100 000 sorted cells per lane. Samples include DN and DP thymocytes and splenic T cells (Sp T) from a representative bi-5 mouse (Bi-5 Tg) and littermate control (littermate).
Generation of transgenic mice expressing the Ikaros isoform IK5.
(A) Representation of the construct used to generate IK5 (bi-5) transgenic mice. The construct contains the lck proximal promoter and the Eμ immunoglobulin heavy-chain enhancer to direct expression of IK5. The IK5 cDNA lacks exons 4 to 6, which eliminates 3 of 4 possible DNA-binding zinc fingers, yet retains all of the C-terminal Zn++ fingers. The HgX minigene has been shown to enhance the expression of transgenes and was part of the original p1026 promoter construct.9 (B) Northern blot of the 2 different bi-5 lines, 747 and 5516, using a probe containing exon 7 of Ikaros. Both endogenous Ikaros and transgene mRNA are marked in the figure. Cells used include thymocytes (T), splenocytes (Sp), and total bone marrow (M). The blot is slightly overexposed to better visualize marrow and spleen expression of the transgene. The HgX minigene, although not capable of being translated, contains both introns and exons; therefore, the transgene mRNA is expressed as multiple, overlapping bands. The lower panel shows the blot after stripping and reprobing for elongation factor-1α (EF) to account for loading differences. (C) Western blot using antisera against the C-terminal portion of Ikaros, which recognizes all Ikaros isoforms. Samples include protein isolated from the thymus (Thy) and liver (Liv) of bi-5 transgenic line 747 and a littermate control (wt). (D) Cell equivalent Western blot using 100 000 sorted cells per lane. Samples include DN and DP thymocytes and splenic T cells (Sp T) from a representative bi-5 mouse (Bi-5 Tg) and littermate control (littermate).
NK T cells are reduced in the bi-5 mice
On the basis of the pattern of expression, the IK5 transgene was predicted to target developing T cells. Yet, contrary to the observations in Ikaros−/− and IK-DNA−/−mice, the bi-5 transgene had little effect on the development of conventional CD4+ and CD8+ αβ T cells, thymic γδ T cells, NK cells, or B cells. The proportions of these cell populations in the spleen, thymus, and bone marrow were similar in bi-5 transgenic mice and littermate controls (Figure2). There was a slight reduction in thymocyte numbers in bi-5 mice, which averaged 75% of normal (Figure 2D).
Representative flow cytometry dot plots of cells from bi-5 mice and littermate controls.
(A) Thymus, (B) spleen, and (C) bone marrow. Scattergram plot (D) of the percentage of normal cell number for spleen (Spl) and thymus (Thy). Each data point on the plot represents the percentage of total thymocyte number that a bi-5 mouse had to a littermate control harvested on the same day. (E) Splenic NK cell number versus mouse age. NK cells were scored by NK1.1+/CD3−staining.
Representative flow cytometry dot plots of cells from bi-5 mice and littermate controls.
(A) Thymus, (B) spleen, and (C) bone marrow. Scattergram plot (D) of the percentage of normal cell number for spleen (Spl) and thymus (Thy). Each data point on the plot represents the percentage of total thymocyte number that a bi-5 mouse had to a littermate control harvested on the same day. (E) Splenic NK cell number versus mouse age. NK cells were scored by NK1.1+/CD3−staining.
A more striking difference was found in the percentage and numbers of NK T cells. These cells have both NK and T-cell markers and are capable of developing in thymectomized mice.16 Thymic NK T cells are defined as HSAlo/−, NK1.1+, TCRint, CD8−, and CD44hi,12,17,18 with 60% CD4+ and the remainder coreceptor negative. The TCR repertoire of NK T cells is highly limited, comprising the invariant α chain Vα14Jα281 paired either with Vβ8, which is greatly favored,19 or with Vβ7 or Vβ2. Thymic NK T cells were markedly reduced in the bi-5 mice, as indicated by an approximately 5-fold decrease in the percentage of Vβ8intCD44hi and NK1.1+CD44hi cells after depletion of CD8+ and HSA+ cells (Figure3A). To estimate the severity of the phenotype, bi-5 mice were compared with mouse strains known to lack NK T cells. Development of NK T cells requires positive selection on the nonclassical major histocompatibility complex (MHC) class I–like molecule CD1.12,20 After depletion of HSA+cells, CD1−/− and bi-5 mice had similar percentages of the Vβ8intCD44hi cells (Figure 3B), whereas wild-type mice had a much greater amount. The β2M−/− mice, which lack MHC class I expression and CD1 expression, had a greater reduction in this population than CD1−/− and bi-5 mice, most likely because of the lack of a TCRintNK1.1− population that does not require CD1 for positive selection.18
NK T cells are reduced in bi-5 mice.
(A) Flow cytometric analysis of thymocytes from a representative bi-5 mouse and littermate control that were depleted of HSA+ and CD8+ cells and analyzed in parallel. Unless otherwise stated, all data are from line 747. (B) Flow cytometric analysis of thymocytes depleted of HSA+ cells. This figure compares the bi-5 mice to littermate controls, CD1−/−, and β2M−/− mice. The greater percentage of NK T cells in mice shown in (B) than in (A) is due in part to age-dependent differences (NK T-cell numbers increase with age) and to experimental variability. The mean percentage of HSA−/CD8−cells before bead depletion was 1.7% ± 0.8% (mean ± SD) for bi-5 and 2.8% ± 1.6% for littermate controls. Results are representative of those obtained in 4 or more independent experiments.
NK T cells are reduced in bi-5 mice.
(A) Flow cytometric analysis of thymocytes from a representative bi-5 mouse and littermate control that were depleted of HSA+ and CD8+ cells and analyzed in parallel. Unless otherwise stated, all data are from line 747. (B) Flow cytometric analysis of thymocytes depleted of HSA+ cells. This figure compares the bi-5 mice to littermate controls, CD1−/−, and β2M−/− mice. The greater percentage of NK T cells in mice shown in (B) than in (A) is due in part to age-dependent differences (NK T-cell numbers increase with age) and to experimental variability. The mean percentage of HSA−/CD8−cells before bead depletion was 1.7% ± 0.8% (mean ± SD) for bi-5 and 2.8% ± 1.6% for littermate controls. Results are representative of those obtained in 4 or more independent experiments.
Extrathymically derived IELs are severely reduced
Like NK T cells, IELs are derived extrathymically, at least in part.16,21 Normal mice have relatively similar numbers of γδ TCR+ IELs (γδ-IEL) and αβ TCR+IEL (αβ-IEL). The overwhelming majority of γδ-IELs express the CD8αα coreceptor as a homodimer, whereas αβ-IELs may express CD8αα, CD8αβ, CD4, or no coreceptor.22 There is considerable evidence to suggest that CD8αα expression only occurs on extrathymically derived IELs.23 24 The bi-5 mice had a drastic reduction in the percentage and number of γδ-IELs and a corresponding increase in the percentage of αβ-IELs (Figure4A). Among the αβ-IELs, there was a decrease in the percentage of cells using CD8αα and an increase in the percentage of cells using CD8αβ cells in bi-5 mice compared with littermate controls (Figure 4B). Thus, bi-5 mice had reductions both in NK T cells and in extrathymically derived IELs.
IEL populations in the bi-5 mice.
Flow cytometric analysis of the IELs isolated from bi-5+and littermate control mice for (A) γδ TCR versus αβ TCR and (B) CD8α versus CD8β gated on αβ−TCR+ cells. Bi-5 γδ-IELs averaged 8.3% ± 3.3% (mean ± SD), whereas littermate γδ-IELs averaged 23.5% ± 8.6% (P = .001 by unpaired Studentt test). IELs recovered from bi-5 mice were approximately 68% (range, 20%-100%) of the numbers recovered in parallel from littermate controls. The average number of IELs recovered from bi-5 mice was 4.2 × 105, which is 68% (range, 20%-100%) of the numbers recovered in parallel from littermate controls with considerable variation between experiments in cell recovery because of differences in age and the multiple processing steps. Results are representative of a minimum of 6 or more independent experiments with mice between 3 and 15 weeks of age.
IEL populations in the bi-5 mice.
Flow cytometric analysis of the IELs isolated from bi-5+and littermate control mice for (A) γδ TCR versus αβ TCR and (B) CD8α versus CD8β gated on αβ−TCR+ cells. Bi-5 γδ-IELs averaged 8.3% ± 3.3% (mean ± SD), whereas littermate γδ-IELs averaged 23.5% ± 8.6% (P = .001 by unpaired Studentt test). IELs recovered from bi-5 mice were approximately 68% (range, 20%-100%) of the numbers recovered in parallel from littermate controls. The average number of IELs recovered from bi-5 mice was 4.2 × 105, which is 68% (range, 20%-100%) of the numbers recovered in parallel from littermate controls with considerable variation between experiments in cell recovery because of differences in age and the multiple processing steps. Results are representative of a minimum of 6 or more independent experiments with mice between 3 and 15 weeks of age.
Reduced expression of IL-2Rβ on thymocytes of bi-5 mice
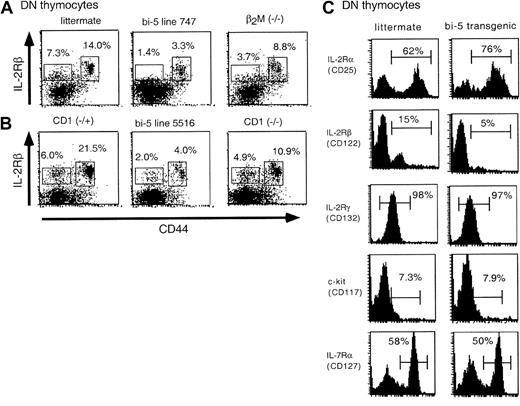
IL-15 signals through IL-2Rβ and the common cytokine receptor γ chain.25,26 For IL-15–specific binding, IL-15Rα is also required.26 Like bi-5 mice, knockouts of IL-2Rβ and IL-15Rα or IRF-1 (a molecule required for IL-15 to be produced) have severely reduced extrathymically derived IELs and NK T cells but very mild defects in conventional αβ T-cell development.27,28 Because IL-2Rβ expression is restricted to hematopoietic cells, whereas IL-15Rα and IL-15 have more ubiquitous expression patterns,25,29 we first evaluated IL-2Rβ expression on progenitors common to both αβ and NK T cells.17 IL-2Rβ is expressed on DN thymocytes and is down-regulated as cells mature to the DP stage.9 For this reason, thymocytes were gated on the DN population and analyzed for the expression of IL-2Rβ on CD44+ (pro T1/pro T2) and CD44− (pro T3/pro T4) cells.30 The expression of IL-2Rβ on CD44+ and CD44− DN thymocytes from both lines of bi-5 mice was substantially reduced compared with control mice (Figure 5A). Among CD44− DN thymocytes, IL-2Rβ expression was restricted to the proT4 (CD44−CD25−) subset and was substantially reduced on cells from bi-5 mice compared with controls (data not shown).
Expression of IL-2Rβ is decreased on CD4−CD8− (DN) thymocytes of bi-5 mice.
(A) Expression of IL-2Rβ on thymocytes from bi-5 (lines 747 and 5516), β2M−/−, wild-type (littermate for 747 line), CD1−/−, and CD1−/+ mice. Thymocytes were stained with CD8-biotin, CD4-biotin, IL-2Rβ-PE, and CD44-FITC and SA-APC and were gated on the CD4−CD8− population. Results are representative of the decrease in IL-2Rβ observed in the DN population of bi-5 thymocytes in 6 or more separate experiments. (B) Expression of IL-2Rβ on CD3−/CD4−/CD8−/NK1.1−/B220−thymocytes. Thymocytes were stained with CD3-APC, CD4-CyC, CD8-FITC, NK1.1-biotin, B220-biotin, SA-PharRed, and IL-2Rβ-PE. The results are representative of 2 or more separate experiments. (C) DN thymocytes from bi-5 mice and littermate controls were evaluated for the expression of a variety of cytokine receptors.
Expression of IL-2Rβ is decreased on CD4−CD8− (DN) thymocytes of bi-5 mice.
(A) Expression of IL-2Rβ on thymocytes from bi-5 (lines 747 and 5516), β2M−/−, wild-type (littermate for 747 line), CD1−/−, and CD1−/+ mice. Thymocytes were stained with CD8-biotin, CD4-biotin, IL-2Rβ-PE, and CD44-FITC and SA-APC and were gated on the CD4−CD8− population. Results are representative of the decrease in IL-2Rβ observed in the DN population of bi-5 thymocytes in 6 or more separate experiments. (B) Expression of IL-2Rβ on CD3−/CD4−/CD8−/NK1.1−/B220−thymocytes. Thymocytes were stained with CD3-APC, CD4-CyC, CD8-FITC, NK1.1-biotin, B220-biotin, SA-PharRed, and IL-2Rβ-PE. The results are representative of 2 or more separate experiments. (C) DN thymocytes from bi-5 mice and littermate controls were evaluated for the expression of a variety of cytokine receptors.
Because NK T cells are known to express IL-2Rβ, and some are coreceptor negative, reduced expression of IL-2Rβ on DN thymocytes might reflect a reduction in NK T cells that express this protein rather than a reduction in expression of this protein on T-cell progenitors.17 To address this possibility, 2 different approaches were taken. First, CD1−/− and β2M−/− mice were studied, because these mice have substantially reduced NK T cell numbers (Figure 3B) but no known defect in IL-2Rβ expression. The expression of IL-2Rβ on CD44+ and CD44− DN thymocytes from both lines of bi-5 mice was substantially reduced compared with wild type, CD1−/−, or β2M−/− mice (Figure 5A). As a second approach, we excluded NK T cells, γδ T cells, B cells, and NK cells by gating on CD3−/CD4−/CD8−/B220−/NK1.1−thymocytes. Again, the expression of IL-2Rβ was reduced on this population of T-cell progenitors in bi-5 transgenic mice compared with littermate controls (Figure 5B). Together, these findings suggest that the reduction in IL-2Rβ on DN thymocytes does not result solely from the loss of NK T cells or a population of mature hematopoietic cells for which this receptor is a marker. The decrease in IL-2Rβ expression for bi-5 transgenic mice appeared to be selective, because the expression of IL-2Rα, γc, c-kit, or IL-7Rα by DN thymocytes of bi-5 mice and littermate controls was similar (Figure5C), as was the abundance of IL-15Rα mRNA (data not shown).
Partial restoration of T-cell development in bi-5 mice by a bcl-xL transgene but not by an IL-2Rβ transgene
These findings suggested that the bi-5 phenotype might result, at least in part, from reduced expression of IL-2Rβ expression on T-cell progenitors. Two approaches were used to address this possibility.
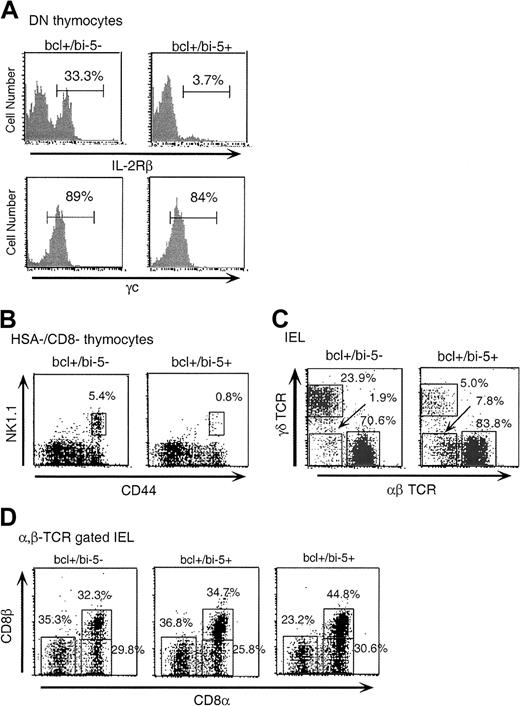
IL-2Rβ and γc transduce multiple signals in response to IL-2 or IL-15, which lead to T-cell proliferation and differentiation and are mediated in part through STAT3, STAT5, JAK1, and JAK3,10,31 and which counter apoptosis by inducing bcl-xL or bcl-2.32-35 The enforced expression of bcl-2 or bcl-xL in lymphoid progenitors substantially restores development of conventional αβ T cells in mice lacking IL-7Rα (or γc) but does not restore development of γδ T cells, B cells, or NK cells 11,36-38; NK T cells were not addressed in these studies. Thus, if IL-2Rβ acts in an analogous manner for extrathymic and NK T-cell development as does IL-7Rα for intrathymic T-cell development, enforced expression of bcl-xL in the appropriate T-cell progenitors should restore NK T cells and CD8αα+ αβ-IELs but not γδ-IELs in bi-5 mice. To explore these possibilities, bi-5 mice were crossed to mice that expressed bcl-xL under the control of the lck proximal promoter (bcl+ mice).11 As predicted, because bcl-xL is downstream of IL-2Rβ, the bi-5+/bcl+ mice still had reduced IL-2Rβ expression and normal γc expression on DN thymocytes (Figure 6A). Enforced expression of bcl-xL did not restore NK T cells (Figure 6B) or γδ-IELs (Figure 6C) in bi-5 mice, but CD8αα+αβ-IELs were rescued, because the ratio of CD8αα- to CD8αβ-expressing cells within the αβ-IELs of bcl+/bi-5+ mice and bcl+/bi-5 littermates was similar (Figure 6D). These findings are similar to those obtained in mice that are IL-15 deficient because of disruption of the IRF-1 gene, in which enforced expression of bcl-2 restored CD8+ T-cell numbers but did not restore NK cells, NK T cells, or γδ-IELs.39
A bcl-xL (bcl+) transgene rescued CD8αα, αβ-TCR IELs in bi-5 mice, but not γδ-IELs or NK T cells.
(A) IL-2Rβ and γc expression in bcl+/bi5+ mice. IL-2Rβ was reduced in bi-5+/bcl+ pro-T thymocytes, but γc was expressed at normal levels. (B) NK T cells and (C) γδ-IELs were not rescued by bcl-xl in bi-5+ mice. (D) CD8α versus CD8β on αβ−TCR+IELs. Bcl-xL appeared to restore the number of CD8αα αβTCR+ IELs in the bi-5 mice. The ratio of CD8αα to CD8αβ IELs was similar for both bcl+/bi-5+and bcl+/bi-5− mice. Two plots of bcl+/bi-5+ are shown to demonstrate the degree of variability. There was also a substantial increase in the numbers of α,β TCR+CD8− T cells in bcl-xLtransgenic mice. These could be either CD4+ T cells or coreceptor–negative T cells.
A bcl-xL (bcl+) transgene rescued CD8αα, αβ-TCR IELs in bi-5 mice, but not γδ-IELs or NK T cells.
(A) IL-2Rβ and γc expression in bcl+/bi5+ mice. IL-2Rβ was reduced in bi-5+/bcl+ pro-T thymocytes, but γc was expressed at normal levels. (B) NK T cells and (C) γδ-IELs were not rescued by bcl-xl in bi-5+ mice. (D) CD8α versus CD8β on αβ−TCR+IELs. Bcl-xL appeared to restore the number of CD8αα αβTCR+ IELs in the bi-5 mice. The ratio of CD8αα to CD8αβ IELs was similar for both bcl+/bi-5+and bcl+/bi-5− mice. Two plots of bcl+/bi-5+ are shown to demonstrate the degree of variability. There was also a substantial increase in the numbers of α,β TCR+CD8− T cells in bcl-xLtransgenic mice. These could be either CD4+ T cells or coreceptor–negative T cells.
The restoration of CD8αα+ αβ-IELs in b-5 mice by the bcl-xL transgene suggested that loss of a survival signal provided through IL-2Rβ might contribute to the bi-5 phenotype. However, enforced expression of IL-2Rβ10failed to rescue CD8αα+ αβ-IELs, γδ-IELs, or NK T cells in bi-5 mice (data not shown). There was a substantial reduction in the number of NK T cells in the IL-2Rβ single transgenic mice and a substantial reduction in total thymic cellularity and the percentage of DP thymocytes in bi-5/IL-2Rβ double transgenic mice, which may have masked our ability to detect an effect of the IL-2Rβ transgene on T-cell development in bi-5 mice. Nonetheless, these observations suggest that more than the loss of IL-2Rβ during T-cell development led to the phenotype observed in the bi-5 mice.
Discussion
Comparison between bi-5 mice and mice with Ikaros gene disruptions
Our studies demonstrate that enforced expression of IK5 in committed T-cell progenitors of bi-5 mice alters cell fate by selectively impairing the development or survival of NK T cells and extrathymically derived IELs. This phenotype was observed in 2 independent lines of bi-5 mice and differs greatly from those of IK−/− and the IK-DNA−/− mice. T-cell number is markedly reduced and CD4+ T cells are enriched in adult IK−/− mice,4 and T cells are absent in IK-DNA−/− mice.3 The much more profound defects in IK−/− and IK-DNA−/− mice compared with bi-5 and IK-DNA+/− mice likely reflect differences both in the magnitude and in the timing of the defect in Ikaros expression. In IK-DNA−/− mice, long Ikaros isoform expression is abolished and only short isoforms are expressed beginning in HSCs and continuing thereafter in their cellular progeny, whereas in bi-5 mice IK5 transgene expression commences in committed T-cell progenitors and declines in mature T cells (Figure 1B,D), paralleling the normal pattern of activity of the lck proximal promoter.8
Mice that are heterozygous for the Ikaros DNA-binding domain disruption (IK-DNA+/− mice) have extremely hyperproliferative T cells and eventually develop clonal T-cell lymphomas and leukemias.40 Like bi-5 mice, IK-DNA+/− mice have increased expression of short relative to long Ikaros isoforms, so the differences in phenotype between these mice likely result primarily from differences in the timing of altered Ikaros expression. It is also possible that differences in the nature of the short isoform contribute, because the predominant short isoform expressed in IK-DNA+/− mice is IK7, whereas bi-5 mice express the IK5 isoform. However, there is at present no evidence for functional differences between these short isoforms. Together the phenotypes of the bi-5 mice and the IK-DNA+/− mice support the notion that modest changes in the relative abundance of Ikaros isoforms can differentially affect cell fate and that the effects observed reflect processes most sensitive to modest changes in Ikaros biologic activity at that stage of development. Similarly, when the ratio between short and long Ikaros isoforms was altered in human CD34+ HSCs by retrovirally mediated overexpression of IK7, the development of lymphoid dendritic cells but not myeloid dendritic cells was selectively impaired.41
Reduced IL-2Rβ expression on T-cell progenitors of bi-5 mice
The impaired development of IELs and NK T cells in bi-5 mice was associated with impaired expression of IL-2Rβ on DN T-cell progenitors. Like bi-5 mice, the numbers of NK T cells and γδ-IELs are dramatically reduced in IL-2Rβ and IL-15Rα knockout mice,27,42 suggesting that reduced expression of IL-2Rβ might account for these effects of the bi-5 transgene. Similarly, although the genes downstream of Ikaros that contribute to the striking phenotypes observed in Ikaros knockout mice have not been clearly defined, recent studies suggest that impaired expression of the flt-3 receptor and of c-kit ligand on HSCs and early T-cell progenitors may contribute.1,7,41 Consistent with this notion, the impaired development of lymphoid dendritic cells from human CD34+ progenitors that overexpress IK7 was associated with and may have resulted in part from reduced flt-3 receptor expression.7,41 It is possible that the reduced expression of IL-2Rβ on T-cell progenitors in bi-5 mice, and of flt-3 receptor and c-kit ligand on hematopoietic progenitors in the studies with Ikaros knockout mice,1,7,41 may be due in part to reductions in progenitor populations that express these receptors and cytokines. However, our studies on IL-2Rβ (Figure 5) and the effects of retrovirally transduced IK7 on flt-3 receptor in vitro41suggest that this is unlikely to be the sole explanation. Rather, these findings collectively suggest that Ikaros may regulate the expression of cytokine and cytokine receptor genes that play sequential and necessary roles in the proliferation, survival, and differentiation of lymphoid progenitors.43 Consistent with this model, there are 2 potential Ikaros binding motifs (GGGAA) in close proximity within the promoter region of human IL-2Rβ, and deletions that eliminate these sites greatly reduced reporter expression in CAT transcription assays.44 45 There is also at least one GGGAA motif within the promoter of murine IL-2Rβ (S.N.T., unpublished observations, May 2000).
Although enforced expression of IL-2Rβ failed to rescue IELs or NK T cells in bi-5 transgenic mice, this finding does not exclude a role for reduced IL-2Rβ expression in the phenotype of bi-5 mice. It does suggest that Ikaros or Ikaros family members regulate the expression of additional genes that are essential for the development of IELs and NK T cells. This is consistent with current models, which suggest that Ikaros plays a broad role in the regulation of gene expression in lymphoid cells through multiple mechanisms.
How might the IK5 transgene perturb expression of IL-2Rβ and other genes that affect T-cell fate?
Ikaros has been proposed to play both positive and negative roles in the regulation of gene expression in lymphoid cells. The dichotomous effects of Ikaros may result in part from the complexity of Ikaros and Ikaros family isoforms and their molecular interactions with each other and with other regulatory proteins. The Ikaros family of proteins interact with each other through C-terminal zinc fingers, whereas the N-terminal zinc fingers are required for direct DNA binding.46,47 The IK5 isoform expressed in bi-5 mice lacks 3 of 4 N-terminal Zn++ fingers required to bind the Ikaros core binding motif but still contains the C-terminal Zn++fingers.5 Similarly, the truncated Ikaros isoforms expressed in IK-DNA−/− and IK-DNA+/− mice lack N-terminal Zn++ fingers. On the basis of studies performed in vitro,46 these short isoforms can form dimers with and function as dominant-negative inhibitors of long isoforms that contain all 4 N-terminal Zn++ fingers.
Although originally proposed to regulate transcription directly, most recent studies suggest that Ikaros regulates transcription primarily by recruitment to target genes of macromolecular complexes, including the Mi-2/NURD (nucleosome remodeling and histone deacetylation), mSIN3A (histone deacetylase), and SWI/SNF chromatin remodeling complexes.2,48,49 SWI/SNF complexes may help to open chromatin and facilitate transcription, whereas the Mi-2/NURD and mSIN3A compact chromatin and impede transcription.50 Thus, in bi-5 mice, IK5 could function as a dominant-negative inhibitor by partially sequestering the DNA-binding isoforms away from the promoters of genes required for proper T-cell development, thereby impeding the ability of endogenous longer Ikaros isoforms to facilitate transcription through the recruitment of SWI/SNF chromatin-remodeling complexes to these loci. An alternative explanation is that Ikaros represses the transcription of genes in developing T cells, either by recruitment of the Mi-2/NURD and mSIN3A complexes1,50 or by direct competition with transcription factors needed for gene expression, as it does at the λ5 locus in developing B lymphocytes51 and at the TdT locus in DP thymocytes in vitro.52 In this model, the short Ikaros isoforms in bi-5, IK-DNA−/−, and IK-DNA+/− mice would facilitate repression mediated by long isoforms, perhaps through the assembly into multimeric complexes with long isoforms bound to pericentromeric foci.52 It is also possible that the actions of short Ikaros isoforms are context dependent, acting as dominant-negative inhibitors when long Ikaros isoforms are limiting, as in the IK-DNA−/− mice, but forming multimers and acting in concert with long isoforms when long isoform abundance is not compromised.52 The complexity of the Ikaros system may account in part for the difficulty in defining specific target genes that account for the phenotypes observed in Ikaros mutant mice in this and other studies.
We thank Stephen Smale for discussions and Ikaros antisera; Michael Grusby for the CD1−/− mice; Katia Georgopoulos for the Ikaros cDNA; R. Perlmutter for the p1026 promoter; Tadatsugu Taniguchi, Michael Bevan, Brad Nelson, Mark Groudine, and Michael Farrar for review and discussions; Zandrea Ambrose for the IEL preparation protocol; Ben Jacobson for the transgenic injections; and Kathryn Allen for flow cytometry support.
Supported in part by grants HD18184 and AI37107 and by grant T32CA09537 (S.N.T.) from the National Institutes of Health.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Christopher B. Wilson, Dept of Immunology, University of Washington, Campus Box 357650, 1959 NE Pacific St, Seattle, WA 98195; e-mail: cbwilson@u.washington.edu.