In the search for genes expressed in hematopoietic stem cells, we identified that the expression of Gfi-1B (growth factor independence-1B) is highly restricted to hematopoietic stem cells, erythroblasts, and megakaryocytes. Gfi-1 and Gfi-1B are zinc finger proteins that share highly conserved SNAG and 6 zinc finger domains.Gfi-1 has been characterized as an oncogene involved in lymphoid malignancies in mice. In contrast, role of Gfi-1B in hematopoiesis has not been well characterized. In this study, we analyzed its function in human hematopoiesis. Enforced expression ofGfi-1B in human CD34+ hematopoietic progenitors induced a drastic expansion of erythroblasts in an erythropoietin-independent manner. Expression ofGfi-1B did not promote erythroid commitment, but enhanced proliferation of immature erythroblasts. Erythroblasts expanded by exogenous Gfi-1B, however, failed to differentiate beyond proerythroblast stage and showed massive apoptosis. These biologic effects of Gfi-1B were mediated through its zinc finger domain, but not by the SNAG or non–zinc finger domain. Proliferation of erythroblasts was associated with sustained expression of GATA-2 but not of GATA-1, indicating a potential link between Gfi-1B and GATA family regulators. Importantly, the function of Gfi-1B to modulate transcription was dependent on promoter context. In addition, activation of transcription of an artificial promoter was mediated through its zinc finger domain. These findings establish Gfi-1B as a novel erythroid regulator and reveal its specific involvement in the regulation of erythroid cell growth through modulating erythroid-specific gene expression.
Introduction
Gfi-1 and Gfi-1B are highly related zinc finger proteins whose expression is exclusive to hematopoietic cells.1,2 Both genes share highly conserved SNAG (Snail/Gfi-1) and 6 C2-H2 zinc finger domains in the N and C terminus, respectively. The SNAG domain is also found in Snail/Slug zinc finger proteins and has been characterized as a transcriptional repression domain.3,4 Random oligonucleotide selection studies demonstrated that Gfi-1 and Gfi-1B bind the same DNA sequence, TAAATCAC(A/T)GC(A/T).2,5 So far only a limited number of direct target genes have been identified, Bax for Gfi-1 andp21Cip1/Waf,1 Socs1, andSocs3 for Gfi-1B. In these cases, Gfi-1 and Gfi-1B act as transcriptional repressors.2,6 7
The Gfi-1 gene was originally identified as a common integration site of Moloney murine leukemia virus (MoMuLV) in rat T-cell–lymphoma cell lines.1 Gfi-1 allows interleukin 2 (IL-2)–dependent T cells to escape G1 arrest induced by IL-2 withdrawal in culture.1 Gfi-1 has also been reported to be involved in oncogenesis. Lymphomas arising in MoMuLV-inoculated Eμ/myc and Eμ/pim-1 transgenic mice harbor a provirus insertion in the Gfi-1 locus, indicating thatGfi-1 acts as a dominant oncogene and collaborates withc-myc, an HLH-LZ transcription factor, andpim-1, a cytoplasmic serine/threonine kinase for the induction of retrovirus-induced lymphomas in mice.8Recently, Gfi-1 was found to directly interact with PIAS3, an inhibitory regulator of signal transducers and activators of transcription 3 (STAT3), and affects its function, leading to an enhancement of STAT3-mediated transcriptional activation.9This finding may explain in part the positive effect of Gfi-1 on cell proliferation and its oncogenic properties.
The Gfi-1B gene was cloned from its high degree of nucleotide sequence similarity to the Gfi-1gene.2 Gfi-1B shares the SNAG and 6 zinc finger domains with Gfi-1 (86% and 88% identity at the amino acid level, respectively). However, the interviewing region between the 2 conserved domains shows a very low similarity to Gfi-1 (21% identity at the amino acid level). Gfi-1B is expressed in the bone marrow and spleen in mice whereas the chicken Gfi-1B homologue is reportedly restricted to erythroid cells.10 Sustained expression of Gfi-1B inhibits IL-6–induced G1 arrest and myeloid differentiation of the myelomonocytic cell line M1 through direct transcriptional repression of the cyclin-dependent kinase inhibitor p21Cip1/Waf1.2 In contrast with Gfi-1, however, information on the function of Gfi-1B is limited and has not yet been characterized in primary hematopoietic cells.
In the search for genes expressed in hematopoietic stem cells, we identified that Gfi-1B is preferentially expressed in hematopoietic stem cells, erythroblasts, and megakaryocytes. To further understand the role of Gfi-1B in hematopoiesis, we used pseudo-type retroviruses to introduce Gfi-1B into human primary hematopoietic progenitors. Gfi-1B expression induced a drastic expansion of erythroblasts in an erythropoietin (EPO)–independent manner as well as inhibition of myeloid cell differentiation. These effects were mediated through the highly conserved zinc finger domain. In addition, proliferation of erythroblasts was associated with a strong transactivating capacity of Gfi-1B mediated through its zinc finger domain. These observations imply novel roles of Gfi-1B in erythroid expansion in the early stage of erythropoiesis.
Materials and methods
Plasmids
Mouse Gfi-1, Gfi-1B, and Bcl-xL cDNAs were obtained by polymerase chain reaction (PCR) amplification using mouse bone marrow cDNA as a template. Gfi-1 and Gfi-1B cDNAs were FLAG-tagged at their N terminus. A series of Gfi-1B mutant cDNAs with a FLAG tag at the N terminus were also generated by PCR amplification. They were designated as follows: the SNAG domain deletion (ΔSNAG; Δ1-20), the zinc finger domain deletion fused to nuclear localization signals of simian virus 40 large T antigen (ΔZn-NLS; Δ161-343), the non–zinc finger domain deletion (Δnon-Zn; Δ21-160), and the SNAG and non–zinc finger domain deletion (Zn; Δ1-160). The Gfi-1 mutants, ΔZn-NLS (Δ253-435) and Zn (Δ1-252) were similarly generated. All cDNAs were subcloned into the pcDNA3 mammalian expression vector (Invitrogen, Groningen, Netherlands).
Production of retrovirus
The retroviral vector GCsam (pGCsam), with a long terminal repeat (LTR) derived from murine stem cell virus (MSCV), has intact splice donor and splice acceptor sequences for generation of subgenomic mRNA.11 The murine Gfi-1, Gfi-1B, and a series of Gfi-1B mutant cDNAs were subcloned into a site upstream of an internal ribosome entry site (IRES)–enhanced green fluorescent protein (EGFP) construct in pGCsam. The murine Bcl-xL cDNA followed by IRES-nerve growth factor receptor truncated in the cytoplasmic domain (tNGFR) was subcloned into pGCsam. To produce the recombinant retrovirus, plasmid DNA was transfected into 293gp cells (293 cells containing the gag and pol genes but lacking an envelope gene) along with the 10A1 env expression plasmid (pCL-10A1)12 by CaPO4 coprecipitation, and supernatant from the transfected cells was collected to infect cells.
Purification of human primary cells
Human umbilical cord blood samples were obtained, with informed consent, from placentas of full-term healthy newborn infants. Peripheral blood (PB) samples were obtained from healthy volunteers who had given informed consent. After isolation of mononuclear cells by density gradient centrifugation, CD34+hematopoietic progenitors were obtained using magnetic bead separation (Miltenyi Biotech, Bergisch Gladbach, Germany). In all experiments, 95% of purified cells were positive for CD34 as assessed by specific antibodies.11 Informed consent was provided according to the Declaration of Helsinki. This study was approved by the institutional review committees of the University of Tsukuba.
Transduction of CD34+ cells
CD34+ cells were prestimulated in Iscove modified Dulbecco medium (IMDM; Sigma, St Louis, MO) supplemented with 10% fetal bovine serum (FBS), 50 ng/mL stem cell factor (SCF), 50 ng/mL thrombopoietin (TPO; kindly provided by KIRIN, Tokyo, Japan), 50 ng/mL interleukin 6 (IL-6; Peprotech, Rocky Hill, NJ), and 50 ng/mL Flt-3 ligand (Peprotech) for 20 hours. After replating onto recombinant fibronectin fragment-coated culture dishes (Takara Shuzo, Otsu, Japan) containing virus supernatant and 5 μg/mL protamine sulfate (Sigma), cells were centrifuged at 1000g for 30 minutes. Transduction was repeated with fresh virus supernatant every 12 hours, 3 times. At 60 hours after the first transduction, cells were stained with phycoerythrin (PE)-conjugated anti-CD34 (PharMingen, San Diego, CA). Cells positive for both CD34 and EGFP, or for CD34, EGFP, and NGFR, were selected by cell sorting on a FACS Vantage (Becton Dickinson, San Jose, CA) and subjected to subsequent analyses. At this time point, more than or equal to 85% of the cells were still positive for CD34.11 To detect the expression of tNGFR on the cell surface, cells were stained with mouse anti–human NGFR (Chemicon, Temecula, CA) followed by PE-conjugated rabbit anti–mouse immunoglobulin (DAKO, Glostrup, Denmark).
Colony assay and in vitro liquid culture
CD34+ cells transduced with indicated retroviruses were plated in methylcellulose medium (Stem Cell Technologies, Vancouver, BC, Canada) or cultured in IMDM with 10% FBS supplemented with 50 ng/mL SCF, IL-3, granulocyte macrophage–colony-stimulating factor (GM-CSF; Peprotech), and 5 U/mL erythropoietin (EPO; kindly provided by Chugai Pharmaceutical, Tokyo, Japan). The culture dishes were incubated at 37°C in a 5% CO2 atmosphere. Colony numbers were counted on day 14. Erythroid cell maturation was evaluated by benzidine staining.
Flow cytometric analysis
Expression of cell-surface antigens and cytoplasmic proteins was analyzed on a FACS Vantage. To detect cell-surface antigens, cells were stained with allophycocyanin (APC)–conjugated anti–human CD11b (PharMingen), PE-conjugated anti–human CD15 (Immunotech, Marseille, France), CD14, and Glycophorin A (PharMingen). Cells that stained with propidium iodide (PI) were gated out as dead cells.
Cells and ex vivo generation of erythroid progenitor cells
After isolation of mononuclear cells from bone marrow (BM) and PB, human hematopoietic cells were fractionated by cell sorting on a FACS Vantage. Murine hematopoietic cells fractionation and ex vivo differentiation of human erythroid progenitors were performed as previously described.13 14 In brief, human granulocyte colony-stimulating factor (G-CSF; Chugai Pharmaceutical and Kyowa Hakko Pharmaceutical, Tokyo, Japan) was administered to healthy Japanese volunteers who had previously signed consent forms approved by the Hokkaido University School of Medicine and the Hokkaido Red Cross Blood Center Committee for the Protection of Human Subjects. Mobilized PB CD34+ cells were isolated using immunomagnetic beads. Cells were then cryopreserved and stored until use in liquid nitrogen. Frozen PB CD34+ cells were thawed and cultured in serum-containing medium with IL-3 at 100 U/mL, SCF at 100 ng/mL, and EPO at 4 U/mL at 37°C in a 5% CO2.
Reverse transcription–polymerase chain reaction (RT-PCR)
Total RNA was isolated using ISOGEN-LS solution (Nippon Gene, Tokyo, Japan) and reverse-transcribed using the ThermoScript RT-PCR system (Gibco BRL, Gaithersburg, MD) and oligo-dT primer. The amount of cDNA was normalized by the quantitative PCR using TaqMan rodent glyceraldehyde phosphate dehydrogenase (GAPDH) control reagent (Perkin-Elmer Applied Biosystem, Foster City, CA). Semiquantitative RT-PCR reactions were then carried out for 36 cycles using normalized cDNAs and recombinant Taq DNA polymerase. Cycling parameters were denaturation at 94°C for 20 seconds, annealing at 60°C for 20 seconds, and extension at 72°C for 30 seconds. PCR products were separated on agarose gels and visualized by ethidium bromide staining. The primer sequences were: human Gfi-1B sense primer 5′-AAC TTC GAG TGC CGC ATG TGC-3′, antisense primer 5′-TGA TGA GGT TGG AGC TCT GGC-3′; human Gfi-1 sense primer 5′-TAC AAG TGC ATC AAG TGC AGC-3′, antisense primer 5′-TTG TGA AGC TTC TCA CCA GTG-3′; humanSCL sense primer 5′-ATG ACC TCC TGC AAG ACG TGC-3′, antisense primer 5′-CTG ATC TCC TAC TGG TAC AGG-3′; humanGATA-1 sense primer 5′-TCA ATT CAG CAG CCT ATT CC-3′, antisense primer 5′-TTC GAG TCT GAA TAC CAT CC-3′; humanGATA-2 sense primer 5′-TGT TGT GCA AAT TGT CAG ACG-3′, antisense primer 5′-CAT AGG TGC CAT GTG TCC AGC-3′; humanBcl-xL sense primer 5′-TTC AGT GAC CTG ACA TCC CAG-3′, antisense primer 5′-TGC ATT GTT CCC ATA GAG TTC C-3′: human hypoxanthine phosphoribosyltransferase (Hprt) sense primer 5′-AAG GAC CCC ACG AAG TGT TGG-3′, antisense primer 5′-AGG GAA CTG ATA GTC TAT AG-3′; mouse Gfi-1B sense primer 5′-TTG GAG CAG CAT ACT CAC GTC-3′, antisense primer 5′-AGA TTG TGT TGA CTC TCA CG-3′; mouse Gfi-1 sense primer 5′-AGA GCT TCA AGA GGT CAT CC-3′, antisense primer 5′-TTG AGT CCA TGC TGA GTC TC-3′; mouseHprt sense primer 5′-GCT GGT GAA AAG GAC CTC T-3′, antisense primer 5′-CAC AGG ACT AGA ACA CCT GC-3′.
Apoptosis assay
Cells were stained with PI and biotinylated anti–Annexin V (PharMingen), followed by APC-conjugated streptavidin (PharMingen). Apoptosis was measured on a FACS Vantage as PI-negative and Annexin V–positive cells.
Reporter construct and luciferase assay
A tetramer oligonucleotide containing a consensus Gfi-1 binding site (CACACCAAATCACTGC)2 was subcloned into the enhancerless pT81 luciferase vector in front of a minimal herpes simplex thymidine kinase promoter.15 The SOCS-1 promoter (bp −2438 to +639) was kindly provided by Dr H. Wu.7 A cytomegalovirus (CMV)–driven Renilla luciferase control reporter plasmid (pRL-CMV; Promega, Madison, WI) was used as an internal control for transfection efficiency in all transfection experiments. Suspension cells (1 × 107cells/transfection) were transiently transfected by electroporation as previously described.16 Briefly, transfection was carried out by electroporation using 5 μg reporter construct, 5 μg each expression construct or empty vector, and 10 ng of an internal control plasmid (pRL-CMV), with the total amount of DNA brought up to 20 μg with carrier plasmid. K562 and U937 cells were electroporated at 250 volts, 960 μF, and 300 volts, 960 μF, respectively. NIH3T3 cells were transiently transfected using Lipofectamine PLUS reagent (Gibco BRL). Cells (3 × 104) were plated in 24-well tissue culture plates 16 hours before transfection. The cells were then transfected with 200 ng reporter construct, plus or minus 40 ng of each expression construct or empty vector, 200 pg internal control plasmid (pRL-CMV), 1 μL Lipofectamine, and 4 μL PLUS reagent for 3 hours in serum-free medium. After 3 hours, serum was added to a final concentration of 10%. The cells were harvested 24 hours after the beginning of transfection. Firefly luciferase activities from the reporter plasmids and Renilla luciferase activities from the internal control plasmid pRL-CMV were determined using the Dual-Luciferase Reporter Assay System (Toyo Inki, Tokyo, Japan). Firefly luciferase values were normalized to the Renillaluciferase values of pRL-CMV.
Results
Restricted expression of Gfi-1B in erythroid and megakaryocytic lineages
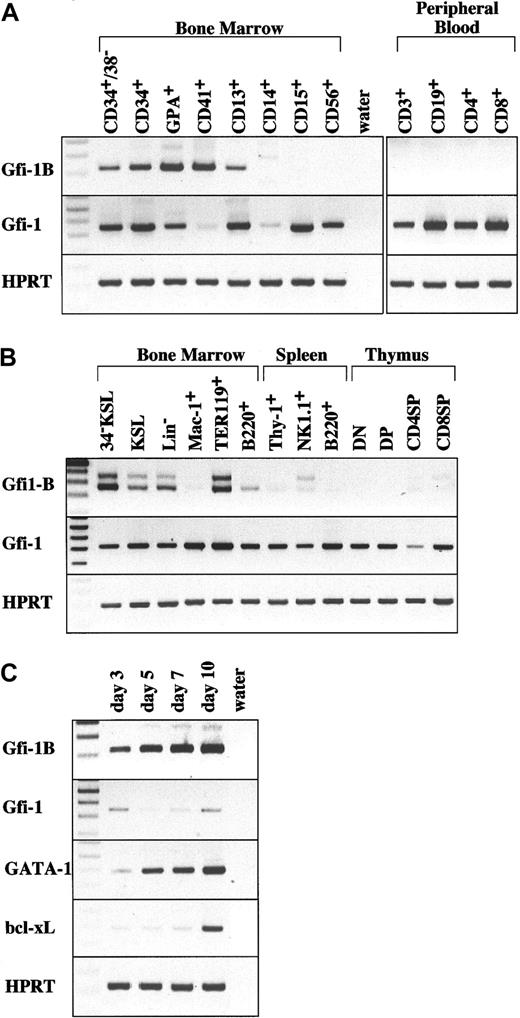
While searching for transcriptional regulators expressed in mouse hematopoietic stem cells, we identified Gfi-1B as a candidate gene. RT-PCR analysis revealed that a significant amount ofGfi-1B mRNA is expressed in mouse hematopoietic stem cells and erythroid cells. This expression profile of Gfi-1B was quite similar to those of SCL and GATA-2, important regulators for hematopoiesis (Figure 1B). In humans,Gfi-1B was up-regulated in both erythroid and megakaryocytic lineages (Figure 1A). In an ex vivo model of expansion of human erythroblasts (see “Materials and methods”), Gfi-1Bexpression was up-regulated as the population of erythroid cells increased during culture, and was maintained throughout successive differentiation stages from erythroid–blast-forming units (BFU-Es) to mature erythroblasts (Figure 1C). Notably, the expression profile ofGfi-1B was well correlated with that of GATA-1 during erythroid differentiation. On the other hand, expression of the related family member Gfi-1 was relatively ubiquitous in hematopoietic cells (Figure 1A-B). However, Gfi-1 was not expressed in human megakaryocytes, and its expression was much weaker than that of Gfi-1B in human erythroblasts (Figure1A,C).
Restricted expression of Gfi-1B in hematopoietic cells detected by RT-PCR.
(A) Expression of human Gfi-1B. Cells analyzed are bone marrow CD34+CD38− stem cells, CD34+ progenitor cells, glycophorin A (GPA)+erythroblasts, CD41+ megakaryocytes, CD13+myeloid progenitors, CD14+ macrophage, CD15+granulocytes, CD56+ NK cells, and peripheral blood CD3 T+ cells, CD19+ B cells, CD4+ T cells, and CD8+ T cells. (B) Expression of murineGfi-1B. Cells analyzed are bone marrow CD34−c-Kit+Sca-1+ lineage marker− stem cells (34–KSL), KSL progenitors, lineage marker− cells (Lin−), Mac-1+ myeloid cells, TER119+ erythroblasts, B220+ B cells, spleen Thy-1+ T cells, NK1.1+ natural killer (NK) cells, B220+ B cells, and thymic CD4−CD8− T cells (DN), CD4+CD8+ T cells (DP), CD4+CD8− T cells (CD4SP), and CD4−CD8+ (CD8SP). (C) Expression of human Gfi-1B during erythroid differentiation. Human peripheral blood CD34+ cells mobilized by G-CSF were collected and cultured under conditions that preferentially drive erythroid differentiation (details in “Materials and methods”). After incubation for the number of indicated days, cells were collected and subjected to RT-PCR analysis. The percentages of GPA+cells, BFU-Es, and CFU-Es in cells on the indicated days are as follows: day 3 (18.1%, 17.2%, 10.8%), day 5 (51.4%, 5.5%, 37.6%), day 7 (80.4%, 0%, 24.8%), and day 10 (91.6%, 0%, 2.1%). “Water” represents the negative control without template. RT-PCR was performed on normalized cDNA templates. PCR products were electrophoresed on agarose gels and visualized by ethidium bromide staining.
Restricted expression of Gfi-1B in hematopoietic cells detected by RT-PCR.
(A) Expression of human Gfi-1B. Cells analyzed are bone marrow CD34+CD38− stem cells, CD34+ progenitor cells, glycophorin A (GPA)+erythroblasts, CD41+ megakaryocytes, CD13+myeloid progenitors, CD14+ macrophage, CD15+granulocytes, CD56+ NK cells, and peripheral blood CD3 T+ cells, CD19+ B cells, CD4+ T cells, and CD8+ T cells. (B) Expression of murineGfi-1B. Cells analyzed are bone marrow CD34−c-Kit+Sca-1+ lineage marker− stem cells (34–KSL), KSL progenitors, lineage marker− cells (Lin−), Mac-1+ myeloid cells, TER119+ erythroblasts, B220+ B cells, spleen Thy-1+ T cells, NK1.1+ natural killer (NK) cells, B220+ B cells, and thymic CD4−CD8− T cells (DN), CD4+CD8+ T cells (DP), CD4+CD8− T cells (CD4SP), and CD4−CD8+ (CD8SP). (C) Expression of human Gfi-1B during erythroid differentiation. Human peripheral blood CD34+ cells mobilized by G-CSF were collected and cultured under conditions that preferentially drive erythroid differentiation (details in “Materials and methods”). After incubation for the number of indicated days, cells were collected and subjected to RT-PCR analysis. The percentages of GPA+cells, BFU-Es, and CFU-Es in cells on the indicated days are as follows: day 3 (18.1%, 17.2%, 10.8%), day 5 (51.4%, 5.5%, 37.6%), day 7 (80.4%, 0%, 24.8%), and day 10 (91.6%, 0%, 2.1%). “Water” represents the negative control without template. RT-PCR was performed on normalized cDNA templates. PCR products were electrophoresed on agarose gels and visualized by ethidium bromide staining.
Enforced expression of Gfi-1Bsupports EPO-independent erythropoiesis
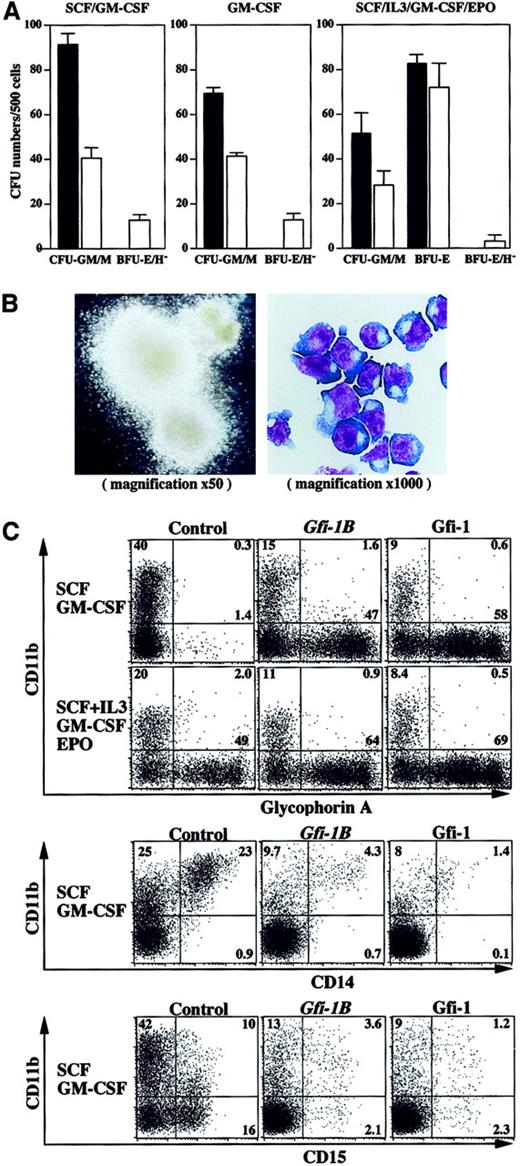
Preferential expression of Gfi-1B in the erythroid lineage is consistent with the finding that the chickenGfi-1B homologue is restricted to erythroid cells.10 However, the function of Gfi-1B in hematopoiesis has not yet been characterized. To understand the roles of Gfi-1B in hematopoiesis, we used retroviruses to introduce Gfi-1B into human primary CD34+ hematopoietic progenitors. We transduced cells with the retroviral vector GCsam-Gfi-1B-IRES-EGFP, which drives expression of both Gfi-1B and EGFP from a single bicistronic message. In our earlier studies using the same retroviral transduction system, we demonstrated that both erythroid and myeloid progenitor cells are transduced with similar efficiencies, and that the expression of transduced genes are high in both lineages as judged by EGFP expression.17 After transduction, cells positive for both CD34 and EGFP were selected by cell sorting and subjected to in vitro assays. Unexpectedly, CD34+ cells transduced with Gfi-1B gave rise to erythroblast colonies in the absence of EPO (Figure 2A). Although the frequency of these erythroblast colonies was 5-fold lower than that of control erythroid bursts generated in the presence of EPO (Figure2A, left and middle panels vs right panel), they were much larger than the control erythroid bursts and were barely hemoglobinized (Figure 2B, left panel). Cytologic analysis revealed that they comprised immature erythroblasts arrested at the proerythroblast stage (Figure 2B, right panel). In the presence of EPO, Gfi-1B–transduced cells gave rise to a comparable number of erythroid bursts to the control (Figure 2A, right panel), indicating that Gfi-1B does not affect erythroid commitment. However, they still formed some large erythroblast colonies that were poorly hemoglobinized even in the presence of EPO (data not shown). Generation of erythroblast colonies from Gfi-1B–transduced cells was completely dependent on GM-CSF (Figure 2A) and to a lesser extent on IL-3 (data not shown). In contract, Gfi-1B–transduced cells gave rise to fewer myeloid colonies than the control (Figure 2A). In addition, the myeloid colonies from Gfi-1B–transduced cells were much smaller than control (data not shown).
Enforced expression ofGfi-1B supports EPO-independent erythropoiesis.
(A) Growth and differentiation of Gfi-1B–transduced CD34+ cells in vitro. CD34+ cells were transduced with either empty vector (black bars) or Gfi-1B(white bars). After transduction, cells positive for both EGFP and CD34 were selected and cultured in methylcellulose medium supplemented with indicated cytokines. Cell growth and differentiation was evaluated by counting colony-forming units (CFUs) on day 14. BFU-E/H– indicates poorly hemoglobinized erythroid burst-forming unit. Results are shown as mean ± SD of 3 representative experiments. (B) Morphology of erythroblast colonies formed from Gfi-1B–transduced CD34+ cells in the absence of EPO. A typical erythroblast colony observed under a phase-contrast microscope (left panel). The colony was recovered, cytospun onto a slide glass, then subjected to May-Grüenwald-Giemsa staining (right panel). (C) Flow cytometric profiles of transduced cells cultured in liquid culture in the presence of indicated cytokines for 10 days. Control represents cells transduced with empty vector. Results are representative of 5 experiments.
Enforced expression ofGfi-1B supports EPO-independent erythropoiesis.
(A) Growth and differentiation of Gfi-1B–transduced CD34+ cells in vitro. CD34+ cells were transduced with either empty vector (black bars) or Gfi-1B(white bars). After transduction, cells positive for both EGFP and CD34 were selected and cultured in methylcellulose medium supplemented with indicated cytokines. Cell growth and differentiation was evaluated by counting colony-forming units (CFUs) on day 14. BFU-E/H– indicates poorly hemoglobinized erythroid burst-forming unit. Results are shown as mean ± SD of 3 representative experiments. (B) Morphology of erythroblast colonies formed from Gfi-1B–transduced CD34+ cells in the absence of EPO. A typical erythroblast colony observed under a phase-contrast microscope (left panel). The colony was recovered, cytospun onto a slide glass, then subjected to May-Grüenwald-Giemsa staining (right panel). (C) Flow cytometric profiles of transduced cells cultured in liquid culture in the presence of indicated cytokines for 10 days. Control represents cells transduced with empty vector. Results are representative of 5 experiments.
We next analyzed the differentiation of transduced cells in liquid culture (Figure 2C). As expected from the colony assay data,Gfi-1B–transduced cells showed an expansion of glycophorin A+ erythroblasts in an EPO-independent manner. Even in the presence of EPO, they generated more erythroblasts than the control. On the contrary, development of CD15+ granulocytes and CD14+ monocytes/macrophages were repressed by enforcedGfi-1B expression. Colony assays of cells cultured in EPO-free media at days 10 and 14 revealed thatGfi-1B–transduced cells still contained a significant number of BFU-Es and erythroid–colony-forming units (CFU-Es) compared with the control (data not shown). Enforced expression ofGfi-1 showed biologic effects similar to those ofGfi-1B, indicating a redundancy in function of the 2 molecules in vitro (Figure 2C).
Gfi-1B promotes erythroblast proliferation but not differentiation
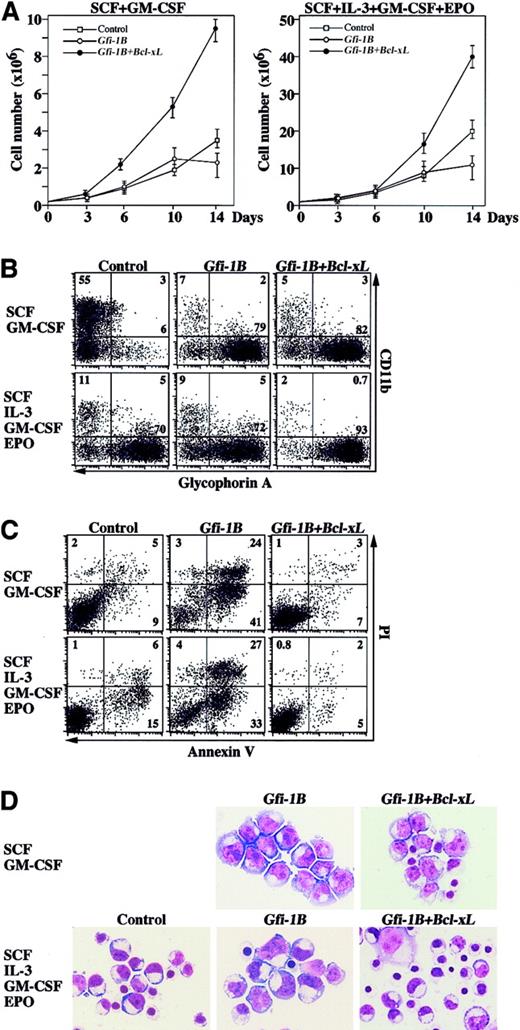
The observation that Gfi-1B did not affect erythroid commitment prompted us to check the effect of Gfi-1B on cell growth. In the presence of SCF and GM-CSF, Gfi-1B–transduced cells showed slightly elevated cell growth compared with the control, but the cell number reached its plateau around day 10 of culture (Figure3A). As shown in Figure 2,Gfi-1B–transduced cells preferentially generated erythroblasts at the proerythroblast stage, whereas the control cells differentiated into myeloid cells (Figure 3B). Unexpectedly,Gfi-1B–expressing cells demonstrated more apoptosis than the control, and apoptotic cell death became prominent after day 10 of culture (Figure 3C). During ex vivo erythroid differentiation from CD34+ progenitors, Bcl-xL expression was up-regulated on day 10 of culture (Figure 1C). However, erythroblasts derived from Gfi-1B–transduced CD34+ cells failed to express a significant amount of Bcl-xL mRNA. Therefore, we cotransduced Bcl-xL with Gfi-1Binto CD34+ cells. Exogenous Bcl-xL efficiently rescued cells from apoptosis and facilitated increased proliferation ofGfi-1B–expressing erythroblasts (Figure 3A-C). In addition, a significant number of rescued erythroblasts differentiated beyond the proerythroblast stage (Figure 3D). On the other hand, addition of EPO was not effective in preventing apoptosis but promoted differentiation (Figure 3C-D). In contrast, cotransduction of Bcl-xL in combination with EPO supplementation efficiently rescued apoptosis and further promoted differentiation (Figure 3C-D).
Gfi-1B promotes proliferation of erythroblasts.
(A) Cell growth of transduced cells in liquid culture supplemented with cytokines as indicated. CD34+ cells were transduced with either empty vector (control) or Gfi-1B, or cotransduced with Gfi-1B and Bcl-xL. Results are shown as mean ± SD of triplicate cultures. (B) Flow cytometric profiles of transduced cells cultured for 10 days in the presence of indicated cytokines. Results are representative of 3 experiments. (C) Massive apoptosis of Gfi-1B–expressing erythroblasts. Apoptosis was detected by staining cells with PI and anti–Annexin V on day 10 of culture. Results are representative of 3 experiments. (D) Morphology of Gfi-1B–expressing erythroblasts. CD34+ cells transduced with Gfi-1B or cotransduced with Gfi-1B and Bcl-xL were cultured in the presence of the indicated cytokines. On day 10 of culture, glycophorin A+ erythroblasts were purified by cell sorting and subjected to morphologic analysis by May-Grüenwald-Giemsa staining. Original magnification × 1000.
Gfi-1B promotes proliferation of erythroblasts.
(A) Cell growth of transduced cells in liquid culture supplemented with cytokines as indicated. CD34+ cells were transduced with either empty vector (control) or Gfi-1B, or cotransduced with Gfi-1B and Bcl-xL. Results are shown as mean ± SD of triplicate cultures. (B) Flow cytometric profiles of transduced cells cultured for 10 days in the presence of indicated cytokines. Results are representative of 3 experiments. (C) Massive apoptosis of Gfi-1B–expressing erythroblasts. Apoptosis was detected by staining cells with PI and anti–Annexin V on day 10 of culture. Results are representative of 3 experiments. (D) Morphology of Gfi-1B–expressing erythroblasts. CD34+ cells transduced with Gfi-1B or cotransduced with Gfi-1B and Bcl-xL were cultured in the presence of the indicated cytokines. On day 10 of culture, glycophorin A+ erythroblasts were purified by cell sorting and subjected to morphologic analysis by May-Grüenwald-Giemsa staining. Original magnification × 1000.
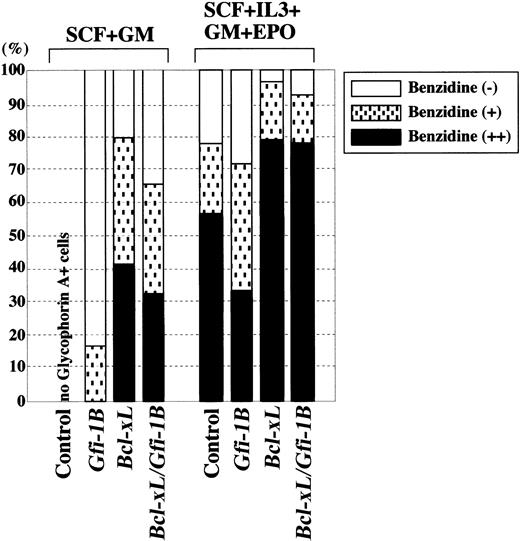
Next, we further analyzed the effect of Gfi-1B on erythroid differentiation. Glycophorin A+ erythroblasts were collected by cell sorting, and then hemoglobin production was evaluated by benzidine staining (Figure 4). Erythroblasts differentiated from Gfi-1B–transduced CD34+ cells barely produced hemoglobin in the absence of EPO, whereas prevention of apoptosis by Bcl-xL allowed differentiation and hemoglobin production. Addition of EPO also facilitated differentiation of erythroblasts from both control andGfi-1B–expressing cells. These findings indicate that Gfi-1B does not have an apparent effect on erythroid differentiation.
Gfi-1B does not affect erythroid differentiation.
CD34+ cells were transduced with empty vector (control),Gfi-1B or Bcl-xL, or cotransduced withGfi-1B and Bcl-xL. Cells were cultured in the presence of indicated cytokines. With exogenous Bcl-xL, a small but significant number of erythroblasts developed even in the absence of EPO (data not shown). On day 10 of culture, glycophorin A+ erythroblasts were purified by cell sorting, and differentiation was evaluated by benzidine staining.
Gfi-1B does not affect erythroid differentiation.
CD34+ cells were transduced with empty vector (control),Gfi-1B or Bcl-xL, or cotransduced withGfi-1B and Bcl-xL. Cells were cultured in the presence of indicated cytokines. With exogenous Bcl-xL, a small but significant number of erythroblasts developed even in the absence of EPO (data not shown). On day 10 of culture, glycophorin A+ erythroblasts were purified by cell sorting, and differentiation was evaluated by benzidine staining.
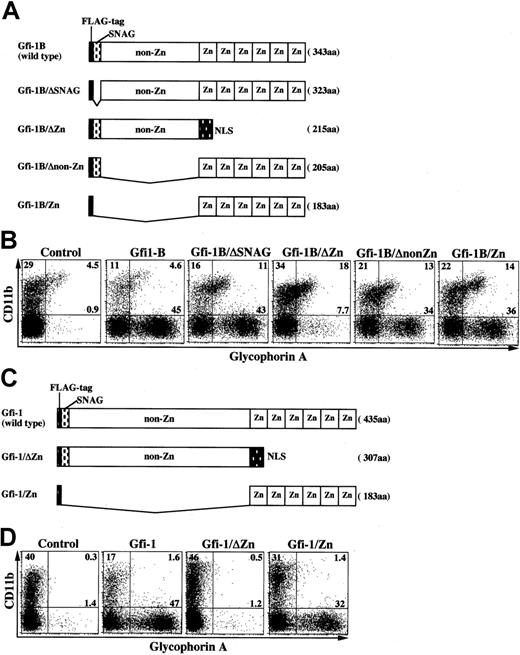
The Gfi-1B zinc finger domain is essential for erythroid expansion
Among the 3 Gfi-1B domains, the SNAG domain has been implicated in the transcriptional repression. To determine the domain responsible for erythroid expansion, we generated a series of Gfi-1B mutants (Figure5A) and transduced them into CD34+ progenitor cells. As shown in Figure 5B, the zinc finger domain was essential for erythroid expansion. On the other hand, the SNAG and non–zinc finger domains did not have any significant effects on this process. The zinc finger domain is highly conserved between Gfi-1B and Gfi-1. As expected, the Gfi-1 domain responsible for erythroid expansion was also localized to its zinc finger domain (Figure 5C-D).
The Gfi-1B zinc finger domain is essential for erythroid expansion.
(A) Schematic representation of Gfi-1B mutants. Zn indicates zinc finger domain; NLS, nuclear localization signal. (B) Mapping of the Gfi-1B domain responsible for erythroid expansion. CD34+ progenitors transduced with each Gfi-1B mutant were cultured in the presence of SCF and GM-CSF for 10 days, then analyzed by flow cytometry. (C) Schematic representation of Gfi-1 mutants. (D) Mapping of the Gfi-1 domain responsible for erythroid expansion. CD34+ progenitors transduced with each Gfi-1 mutant were cultured in the presence of SCF and GM-CSF for 10 days, then analyzed by flow cytometry.
The Gfi-1B zinc finger domain is essential for erythroid expansion.
(A) Schematic representation of Gfi-1B mutants. Zn indicates zinc finger domain; NLS, nuclear localization signal. (B) Mapping of the Gfi-1B domain responsible for erythroid expansion. CD34+ progenitors transduced with each Gfi-1B mutant were cultured in the presence of SCF and GM-CSF for 10 days, then analyzed by flow cytometry. (C) Schematic representation of Gfi-1 mutants. (D) Mapping of the Gfi-1 domain responsible for erythroid expansion. CD34+ progenitors transduced with each Gfi-1 mutant were cultured in the presence of SCF and GM-CSF for 10 days, then analyzed by flow cytometry.
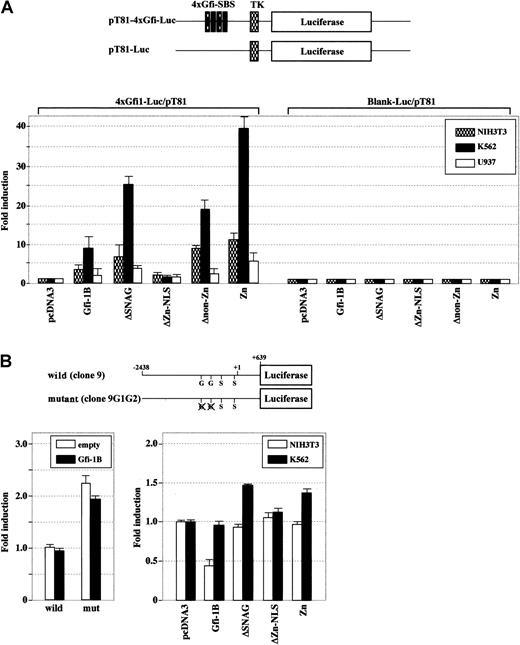
The zinc finger domain confers transactivating activity in erythroid cells
Although Gfi-1B has been characterized as a transcriptional repressor,2 functions of transcriptional regulators in general largely rely on the context, including cell type and sequences flanking the promoter. To analyze its ability to modulate transcription, we used a reporter plasmid with 4 Gfi-1 binding sites in front of a basal thymidine kinase promoter. Gfi-1B transactivated the promoter 7.8-fold above the basal level in an erythroid cell line, K562, 2.5-fold in NIH3T3 fibroblasts, and 1.9-fold in a myeloid cell line, U937 (Figure 6A). Transfection efficiencies were approximately 9% for both K562 and U937 cells and more than 80% for NIH3T3 cells (data not shown). Analysis of Gfi-1B mutants demonstrated that the zinc finger domain acts as an activation domain whereas the SNAG and non–zinc finger domains serve as repression domains. Notably, Gfi-1B activated transcription preferentially in erythroid cells. As expected, Gfi-1 exhibited similar profiles of transactivation of this artificial promoter (data not shown).
The zinc finger domain confers transactivating activity in erythroid cells.
(A) NIH3T3, K562, and U937 cells were transiently transfected with a reporter plasmid containing 4 Gfi-1–specific binding sequences (4xGfi-1-SBS) in front of a basal thymidine kinase promoter (TK) along with either wild-type Gfi-1B or a series ofGfi-1B mutants. (B) K562 cells were transiently transfected with either wild-type or mutant SOCS-1 promoter along with wild-type Gfi-1B (left panel). NIH3T3 and K562 cells were transiently transfected with the wild-type SOCS-1 promoter along with either wild-typeGfi-1B or a series of Gfi-1B mutants (right panel). Luciferase activities were determined 24 hours after transfection and normalized to the Renilla luciferase activities of the internal control plasmid pRL-CMV. The fold increase in promoter activity was measured relative to that obtained with empty vector. The results are shown as the mean ± SD of triplicate transfections. G indicates Gfi-1B binding site; S, STAT binding site.
The zinc finger domain confers transactivating activity in erythroid cells.
(A) NIH3T3, K562, and U937 cells were transiently transfected with a reporter plasmid containing 4 Gfi-1–specific binding sequences (4xGfi-1-SBS) in front of a basal thymidine kinase promoter (TK) along with either wild-type Gfi-1B or a series ofGfi-1B mutants. (B) K562 cells were transiently transfected with either wild-type or mutant SOCS-1 promoter along with wild-type Gfi-1B (left panel). NIH3T3 and K562 cells were transiently transfected with the wild-type SOCS-1 promoter along with either wild-typeGfi-1B or a series of Gfi-1B mutants (right panel). Luciferase activities were determined 24 hours after transfection and normalized to the Renilla luciferase activities of the internal control plasmid pRL-CMV. The fold increase in promoter activity was measured relative to that obtained with empty vector. The results are shown as the mean ± SD of triplicate transfections. G indicates Gfi-1B binding site; S, STAT binding site.
As reported, however, Gfi-1B behaved as a repressor of the SOCS-1 promoter that contains 2 Gfi-1B binding sites (Figure6B).7 In K562 cells that express endogenous Gfi-1B, exogenous Gfi-1B did not affect the SOCS-1 promoter activity, but the introduction of mutations in both Gfi-1B binding sites led to higher SOCS-1 promoter activity (more than 2-fold). In contrast, exogenous Gfi-1B clearly repressed the SOCS-1 promoter in NIH3T3 cells, which do not express endogenous Gfi-1B. Analysis of Gfi-1B mutants demonstrated that the SNAG domain serves as a transcriptional repression domain. Deletion of the SNAG domain resulted in loss of repression in NIH3T3 cells and in partial derepression in K562 cells. The zinc finger domain did not activate transcription of the SOCS-1 promoter. These data establish that the function of Gfi-1B to modulate transcription is dependent on both the promoter and cell type context.
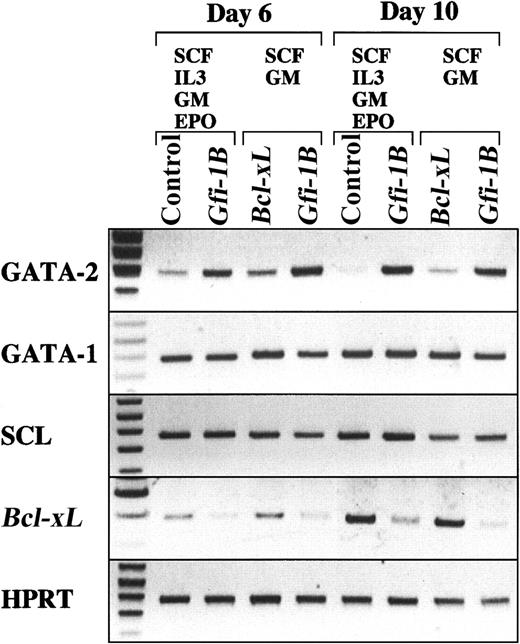
Effect of Gfi-1B on the expression of genes involved in erythropoiesis
To obtain an insight into the role of Gfi-1B in erythropoiesis, we analyzed the effect of Gfi-1B on the expression of well-known erythroid regulators. RT-PCR analysis revealed that enforced expression ofGfi-1B induces sustained expression of GATA-2 but not GATA-1 (Figure 7), indicating a potential link between Gfi-1B and GATA family regulators.
Effect of Gfi-1B on the expression of genes involved in erythropoiesis.
CD34+ cells were transduced with either Bcl-xLor Gfi-1B and were cultured in the presence of SCF and GM-CSF. In addition, CD34+ cells transduced with either empty vector (control) or Gfi-1B were cultured in the presence of SCF, IL-3, GM-CSF, and EPO. With exogenousBcl-xL, a small but significant number of erythroblasts developed even in the absence of EPO (data not shown). On day 10 of culture, glycophorin A+ erythroblasts were purified by cell sorting, then subjected to RT-PCR analysis. RT-PCR was performed on normalized cDNA templates. PCR products were electrophoresed on agarose gels and visualized by ethidium bromide staining.
Effect of Gfi-1B on the expression of genes involved in erythropoiesis.
CD34+ cells were transduced with either Bcl-xLor Gfi-1B and were cultured in the presence of SCF and GM-CSF. In addition, CD34+ cells transduced with either empty vector (control) or Gfi-1B were cultured in the presence of SCF, IL-3, GM-CSF, and EPO. With exogenousBcl-xL, a small but significant number of erythroblasts developed even in the absence of EPO (data not shown). On day 10 of culture, glycophorin A+ erythroblasts were purified by cell sorting, then subjected to RT-PCR analysis. RT-PCR was performed on normalized cDNA templates. PCR products were electrophoresed on agarose gels and visualized by ethidium bromide staining.
Discussion
In this study, we first analyzed the detailed expression ofGfi-1B and Gfi-1 in hematopoietic cells (Figure1). Both Gfi-1B and Gfi-1 are expressed in hematopoietic stem and progenitor cells. Among the various cell lineages, however, Gfi-1 is broadly expressed, whereasGfi-1B is specifically expressed in erythroid cells and megakaryocytes. Throughout human erythroid differentiation, mRNA expression of Gfi-1B is maintained at a high level. These expression profiles indicate lineage-specific involvement of Gfi-1B in hematopoiesis.
To investigate the roles of Gfi-1B in hematopoiesis, we overexpressedGfi-1B in human CD34+ hematopoietic progenitor cells. In the absence of EPO, hematopoietic progenitors did not form erythroblast colonies. In contrast, CD34+ progenitors transduced with Gfi-1B gave rise to a number of large erythroid colonies in response to GM-CSF in an EPO-independent manner (Figure 2A-B). Importantly, there was no difference in frequency of erythroid colony formation between the control andGfi-1B–transduced cells in the presence of EPO (Figure 2A, right panel), indicating that Gfi-1B does not affect erythroid commitment but rather promotes erythroblast proliferation. At a stage prior to hemoglobin synthesis, erythroid cells can be categorized as either BFU-Es or CFU-Es. BFU-Es can be further classified as immature or mature BFU-Es according to their proliferative capacity and responsiveness to EPO.18-20 Human immature BFU-Es have few EPO receptors and are not responsive to EPO, whereas mature BFU-Es express low levels of EPO receptors and are weakly responsive to EPO. These cells give rise to CFU-Es that are highly responsive to EPO.18 BFU-Es require the growth factors such as IL-3, GM-CSF, and SCF to develop into CFU-Es.19 20 Given the GM-CSF–dependent but EPO-independent proliferation of erythroblasts in response to expression of exogenous Gfi-1B, it is conceivable that Gfi-1B is involved in the mitogenic signaling pathway activated by EPO and is responsible for cell growth of early erythroblasts at the transition from mature BFU-Es to CFU-Es (Figure8). In contrast to EPO, however, the function of Gfi-1B seems to be specific to cell proliferation. We could not detect any positive effect of Gfi-1B on erythroid differentiation or survival (Figures 3 and 4). The specific function of Gfi-1B on cell proliferation evokes the possibility that Gfi-1B is involved in the JAK2/STAT signaling pathway, which is the major mitogenic signaling pathway activated by both EPO and GM-CSF.
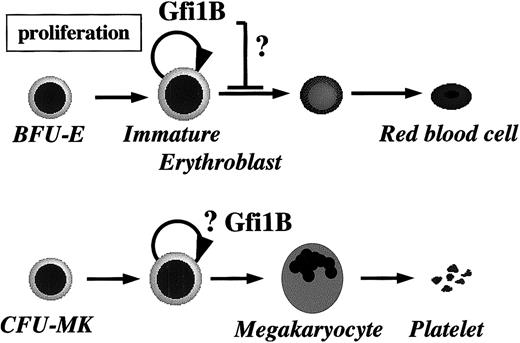
A model for the role of Gfi-1B in erythropoiesis.
As explained in “Discussion,” our findings suggest that Gfi-1B plays a role in the expansion of immature erythroblasts. However, its effect on terminal maturation remains to be determined. Specific expression of Gfi-1B also indicates its function in hematopoietic stem cells and megakaryopoiesis.
A model for the role of Gfi-1B in erythropoiesis.
As explained in “Discussion,” our findings suggest that Gfi-1B plays a role in the expansion of immature erythroblasts. However, its effect on terminal maturation remains to be determined. Specific expression of Gfi-1B also indicates its function in hematopoietic stem cells and megakaryopoiesis.
Importantly, enforced expression of Gfi-1B induced sustained mRNA expression of GATA-2, but not of GATA-1 in erythroblasts. This might be in part due to the accumulation of erythroid progenitor cells that are GATA-2–positive. However, the possibility that Gfi-1B positively regulates GATA-2 expression is intriguing to pursue. GATA-2 is preferentially expressed in hematopoietic stem and progenitor cells and is essential for their expansion.21 It promotes proliferation of erythroid progenitors and blocks their differentiation.22 On the other hand, GATA-1 is required for terminal maturation and survival of erythroid cells beyond the proerythroblast stage23,24 and negatively regulates cell cycle.25-27 A complex regulatory network between GATA-1 and GATA-2 has been postulated, in which GATA-1 represses GATA-2 expression during erythroid differentiation.28 Taken together, sustained expression of GATA-2 may explain in part the high mitogenic activity of Gfi-1B–expressing erythroblasts. A database search revealed 2 potential Gfi-1B binding sites in intron 1 of the mouse GATA-2 gene, but no corresponding site in the human GATA-2 gene. The possibility that Gfi-1B directly regulates GATA-2 gene expression remains to be determined. On the other hand, it is also possible that Gfi-1B negatively regulates GATA-1 function, which results in derepression of GATA-2 and relief from cell cycle repression by GATA-1. Sustained expression of GATA-2 is indicative of a potential link between Gfi-1B and GATA family regulators.
Notably, erythroblasts expressing Gfi-1B failed to differentiate beyond the proerythroblast stage and showed massive apoptosis after 7 to 10 days of culture (Figure 3). They did not show up-regulation of Bcl-xL mRNA (Figure 7). Coexpression ofBcl-xL with Gfi-1B efficiently rescued erythroblasts from apoptosis, revealing an even higher mitogenic effect of Gfi-1B on erythroblasts (Figure 3A-C). In addition, expression ofBcl-xL allowed erythroblasts to further differentiate beyond the proerythroblast stage (Figure 3D). A database search revealed no potential Gfi-1B binding sites in the proximal promoters of human and murine Bcl-xl genes. Therefore, it is unlikely that Gfi-1B directly regulates Bcl-xl gene expression. It has been reported that GATA-1 and EPO cooperate to promote erythroid cell survival by regulating Bcl-xL expression.29Erythroblasts expressing Gfi-1B showed normal GATA-1 expression (Figure 7) and EPO-responsiveness in terms of growth and differentiation (Figures 2A and 4) but did not up-regulateBcl-xL mRNA. Taken together, there remains a possibility that Gfi-1B represses GATA-1 function as discussed above. An alternative interpretation is that the observed apoptosis occurs due to hyperproliferative signals triggered by Gfi-1B. p53 provides an “oncogene checkpoint” function that guards cells against hyperproliferative signals.30 When proliferative signals exceed a critical threshold, the alternative reading frame (ARF)–dependent checkpoint is activated, and ARF triggers a p53-dependent response that induces growth arrest and/or apoptosis. Massive apoptosis of erythroblasts could be associated with deregulated cell cycle progression induced by excessive Gfi-1B protein. However, ARF/INK4a was not up-regulated in erythroblasts expressing Gfi-1B. In addition,p21Cip1/Waf1, which is induced by p53, was not up-regulated either (data not shown). The mechanisms responsible for apoptosis and failure in Bcl-xLup-regulation remain to be determined.
Gfi-1 and Gfi-1B have been characterized as transcriptional repressors. The SNAG domain functions as a transcriptional repression domain and is responsible for both Gfi-1–mediated inhibition of G1arrest induced by IL-2 withdrawal in T cells3 and Gfi-1B–mediated inhibition of IL-6–induced G1 arrest and differentiation of M1 myeloid cells.2 Recently, the non–zinc finger domain of Gfi-1 has been demonstrated to directly interact with PIAS3, an inhibitory regulator of STAT3, and enhance STAT3-mediated transcriptional activation.9 In terms of erythroid expansion induced by Gfi-1B and Gfi-1, however, the SNAG and non–zinc finger domains were dispensable, whereas the zinc domain was essential and sufficient (Figure 5). These data contrast with previous findings that demonstrated essential roles of the SNAG and non–zinc finger domains in Gfi-1 and /or Gfi-1B functions.2,3,9Therefore, we analyzed the ability of Gfi-1B to modulate transcription in hematopoietic cells. In contrast with the previous data showing transcriptional repression by Gfi-1B, Gfi-1B acted as a transcriptional activator on an artificial promoter (Figure 6). Mapping the functional domains revealed that the zinc finger domain acts as an activation domain, whereas the SNAG domain serves as a repression domain, as expected, as does the non–zinc finger domain. Interestingly, the transactivation activity was significant in an erythroid cell line. On the other hand, we also confirmed that Gfi-1B behaves as a repressor of the SOCS-1 promoter, one of the well-characterized Gfi-1B target promoters. These data establish for the first time that the function of Gfi-1B to modulate transcription is dependent on both the promoter and cell type context. Although the zinc finger domain has been characterized only as a DNA binding domain,5 our findings suggest that the zinc finger domain itself could mediate or enhance mitogenic signals, possibly by activating transcription of as-not-yet-identified target genes. The fact that the zinc finger domain is highly conserved between Gfi-1 and Gfi-1B may explain the functional redundancy between Gfi-1 and Gfi-1B in erythroid expansion in vitro.
Another biologic phenotype induced by enforced expression ofGfi-1B, as well as Gfi-1, is suppression of myelopoiesis. In contrast with a drastic expansion of erythroblasts, development of granulocytes and macrophages was significantly inhibited. This function stands in contrast to the role of Gfi-1B in erythroid cells. Given the predominant expression of Gfi-1but not Gfi-1B in myeloid cells, our findings suggest distinct roles of Gfi-1 from those of Gfi-1B in myeloid development.
In summary, we demonstrated that Gfi-1B supports proliferation of immature erythroblasts through its zinc finger domain and regulates transcription during erythropoiesis. These findings strongly implicate involvement of Gfi-1B in normal erythropoiesis and support its potential role in hematopoietic stem cells and megakaryocytes as well (Figure 8).
Note added in proof. Saleque et al31 have recently reported that Gfi-1b–deficient embryos die with failure to produce definitive enucleated erythrocytes. The fetal liver of mutant mice contains erythroid and megakaryocytic precursors arrested in their development whereas myelopoiesis is normal.
We thank Dr Yoshihiro Shiina (Shiina Hospital, Ushiku, Ibaraki, Japan) and Dr Nikunj Somia (University of Minnesota) for providing us with human cord blood and 293gp cells, respectively; Dr Daniel G. Tenen for critical reading of the manuscript; Yohei Morita for technical assistance; and Miss Michie Itoh for excellent secretarial assistance.
Prepublished online as Blood First Edition Paper, June 14, 2002; DOI 10.1182/blood-2002-01-0182.
Supported in part by grants from the Ministry of Education, Culture, Sport, Science and Technology, and Core Research for Evolutional Science and Technology (CREST) of Japan Science and Technology Corporation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Atsushi Iwama and Hiromitsu Nakauchi, Laboratory of Stem Cell Therapy, Center for Experimental Medicine, The Institute of Medical Science, University of Tokyo, 4-6-1 Shirokanedai, Minato-ku, Tokyo, 108-8639, Japan; e-mail: aiwama@ims.u-tokyo.ac.jp, ornakauchi@ims.u-tokyo.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal