Abstract
Signal transducers and activators of transcription (STATs) have been reported to play a critical role in the differentiation of several myeloid cell lines, although the importance of STATs in the differentiation of primary human hematopoietic cells remains to be established. Terminal eosinophil differentiation is induced by interleukin-5 (IL-5), which has also been demonstrated to activate STAT5. We have investigated whether STAT5 plays a critical role during eosinophil differentiation using umbilical cord blood–derived CD34+ cells. In this ex vivo system, STAT5 expression and activation are high early during differentiation, and STAT5 protein expression is down-regulated during the final stages of eosinophil differentiation. Retroviral transductions were performed to ectopically express wild-type and dominant-negative STAT5a (STAT5aΔ750) in CD34+ cells. Transduction of cells with STAT5a resulted in enhanced proliferation compared with cells transduced with empty vector alone. Interestingly, ectopic expression of STAT5a also resulted in accelerated differentiation. In contrast, ectopic expression of STAT5aΔ750 resulted in a block in differentiation, whereas proliferation was also severely inhibited. Similar results were obtained with dominant-negative STAT5b. Forced expression of STAT5a enhanced expression of the STAT5 target genes Bcl-2 andp21WAF/Cip1, suggesting they may be important in STAT5a-mediated eosinophil differentiation. These results demonstrate that STAT5 plays a critical role in eosinophil differentiation of primary human hematopoietic cells.
Introduction
Eosinophils play an important role in immunity against helminth infection. Parasite infection results in enhanced eosinophil numbers and in migration toward and degranulation at the site of parasite invasion. In vitro studies demonstrated that eosinophils are capable of adhering to parasites in the presence of different antibody isotypes (immunoglobulin E [IgE], IgG, IgA)1,2 and complement components (C3b).3 At the site of contact, eosinophils degranulate, resulting in damage to the tegumental membranes and eventually in the death of parasites.4 The toxic effect of eosinophilic granule proteins, such as eosinophilic cationic protein (ECP), is complemented by the generation of toxic oxygen metabolites.5 Besides their role in immunity against parasitic infection, eosinophils are thought to play an important role in the pathogenesis of allergic diseases, among them asthma and dermatitis. Eosinophils are recruited to the site of allergic inflammation by chemoattractants, such as C5a and platelet activating factor (PAF).6,7 Release of granular contents at the inflammatory locus can result in long-term tissue damage. For example, airway epithelium can be damaged during airway inflammation.8 9
Eosinophils are derived from pluripotent hematopoietic stem cells (HSCs) in the bone marrow. These progenitors are defined as precursors for all lineages of mature blood cells, and they are capable of self-renewal. Such cells can be divided into long-term repopulating hematopoietic stem cells (LT-HSCs) and short-term repopulating hematopoietic stem cells (ST-HSCs). ST-HSCs can differentiate to multipotent progenitor cells, which are capable of differentiation toward a subset of the hematopoietic lineage. These multipotent stem cells include the common lymphoid precursor10 and the common myeloid precursor.11 Common myeloid progenitor cells, known as the granulocyte/erythrocyte/macrophage/megakaryocyte colony-forming unit (CFU-GEMM), can differentiate toward the erythroid, megakaryocytic, and myelomonocytic lineage. It has been demonstrated that genes specific for erythroid, myeloid, or megakaryocytic lineages are transcribed in the common myeloid progenitors before commitment to a single lineage. Genes specific for differentiation toward other lineages are down-regulated on commitment to a single lineage.12 Myeloid differentiation is regulated by a variety of cytokines, including erythropoietin (EPO), granulocyte–colony-stimulating factor (G-CSF), thrombopoietin (TPO), interleukin-3 (IL-3), granulocyte macrophage–colony-stimulating factor (GM-CSF), macrophage–colony-stimulating factor (M-CSF), and IL-5. IL-3 and GM-CSF are cytokines that regulate proliferation and survival during myeloid differentiation of various lineages, whereas EPO, TPO, G-CSF, M-CSF, and IL-5 are required for the final maturation of erythrocytes,13 megakaryocytes, platelets,14,15 neutrophils, monocytes,16 and eosinophils,17 18 respectively.
Hematopoietic cytokines can activate several signal transduction pathways, including the Ras/Raf/Erk,19-23phosphatidylinositol 3 kinase (PI3K),24-26 and Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway.27,28 The JAK/STAT signal transduction pathway is thought to play an important role in myeloid differentiation,29 and the signaling paradigm mediating this pathway has been elucidated in detail.30 In short, on ligand binding, cytokine receptor dimerization results in the activation of members of the receptor-associated JAK family through cross-phosphorylation. Subsequently, tyrosine phosphorylation of the intracellular domains of the dimerized receptor occurs, enabling STAT transcription factors to associate with the receptor. STATs are subsequently phosphorylated by the receptor-associated JAKs. Homodimers or heterodimers are formed, and, after translocation to the nucleus, transcription can be regulated through STAT-specific DNA-binding sites.31-34
It has been demonstrated that the stimulation of cells with IL-5, which is required for terminal eosinophil differentiation, results in the activation of STAT5.35 36 In this study, we have investigated the role of STAT5 during IL-5–mediated eosinophil differentiation. Our data demonstrate that STAT5 is expressed throughout the eosinophil differentiation program of CD34+progenitor cells. STAT5 expression and activation are high during early differentiation, but STAT5 expression is down-regulated during terminal eosinophil differentiation. To determine whether STAT5 plays a critical role in the differentiation process, wild-type STAT5a, dominant-negative STAT5a, and dominant-negative STAT5b were ectopically expressed in CD34+ cells by retroviral transduction. Our experiments demonstrated that STAT5a and STAT5b play an important role in proliferation and maturation during eosinophil differentiation. Ectopic expression of STAT5a was found to induce the expression of Bcl-2, an antiapoptotic protein, and p21WAF/Cip1, a cyclin-dependent kinase inhibitor, in differentiating eosinophils. These data demonstrate that STAT5 plays a critical role in regulating the differentiation of primary human hematopoietic cells toward eosinophils.
Materials and methods
Isolation and culture of human CD34+ cells
CD34+ cells were isolated as previously described.35 In short, mononuclear cells were isolated from umbilical cord blood by density-gradient centrifugation over a Ficoll-Paque solution (density, 1.077 g/mL). Magnetic-activated cell sorter (MACS) immunomagnetic cell separation (Miltenyi Biotech, Auburn, CA) using a hapten-conjugated antibody against CD34, which was coupled to beads, was used to isolate CD34+ cells. CD34+ cells were cultured in Iscove modified Dulbecco medium (Gibco, Paisley, United Kingdom) supplemented with 10% fetal calf serum (FCS), 50 μM β-mercaptoethanol, 10 U/mL penicillin, 10 μg/mL streptomycin, and 2 mM glutamine at a density of 0.3 × 106 cells/mL. Cells were differentiated toward eosinophils with the addition of stem cell factor (SCF) (50 ng/mL), FLT-3 ligand (50 ng/mL), GM-CSF (0.1 nM), IL-3 (0.1 nM), and IL-5 (0.1 nM). Every 3 or 4 days, cells were counted and fresh medium was added to a density of 0.5 × 106 cells/mL. After 3 days of differentiation, only IL-3 and IL-5 were added to the cells.
Neutrophil differentiation was induced with the addition of SCF (50 ng/mL), FLT-3 (50 ng/mL) ligand, GM-CSF (0.1 nM), IL-3 (0.1 nM), and G-CSF (30 ng/mL). After 6 days of culture, only G-CSF was added to the cells.
Viral transduction of CD34+ cells
We used a bicistronic retroviral DNA construct consisting of the gene of interest and an internal ribosomal entry site (IRES) followed by the gene encoding for enhanced green fluorescent protein (eGFP) (LZRS-eGFP). LZRS-eGFP retrovirus was produced by stable transfection of the retroviral packaging cell line, Phoenix-ampho,37 by calcium-phosphate coprecipitation. Cells were plated in 6-cm dishes 24 hours before transfection. Five minutes before transfection, 25 μM chloroquine diphosphate was added to the cells. Ten micrograms DNA was used per transfection. Medium was refreshed 16 hours after transfection. After an additional 24 hours, cells were split into 75-cm2 culture flasks (Greiner, Frickenhausen, Germany), and 2 μg/mL puromycin was added to the cells. After 2 weeks of selection, cells were plated into 25-cm2 flasks (Greiner) and were grown to a confluence of 90%. Subsequently, cells were grown in a minimal amount of medium (2.5 mL) for 24 hours. Viral supernatants were collected and filtered through a 0.45-μm filter.
CD34+ cells were transduced in 24-well dishes precoated with 20 μg/cm2 recombinant human fibronectin fragment CH-296 (RetroNectin; Takara, Otsu, Japan) for 2 hours and with 2% bovine serum albumin (BSA) for 30 minutes. Transduction was performed by the addition of 0.5 mL viral supernatant to 0.5 mL medium containing 0.5 × 106 cells. Twenty-four hours after transduction, 0.7 mL medium was removed from the cells, and 0.5 fresh virus supernatant was added together with 0.5 mL fresh medium and cytokines (IL-3 and IL-5). The percentage of eGFP-positive living cells was determined every 3 or 4 days by fluorescence-activated cell sorter (FACS) analysis (FACS Vantage; Becton Dickinson, San Jose, CA).
Histochemical staining of eosinophil and neutrophil precursors
The percentage of eosinophil differentiation was determined by histochemical staining of the cells. The percentage of cells differentiating toward eosinophils was determined by Luxol Fast-Blue (LFB) staining (Avocado Research Chem, Heysham, United Kingdom), a dye that specifically stains eosinophil granules. Cytospins were prepared from 5 × 104 differentiated eosinophils. Slides were dried on silica gel for 24 hours before fixing them in dry acetone for 10 minutes. Slides were stained with 0.15% wt/vol LFB (Avocado Research) in urea-saturated ethanol for 2 hours.
May-Grünwald-Giemsa staining was used to analyze differentiating eosinophils and neutrophils. Cytospins were prepared from 5 × 104 differentiating eosinophils and were fixed in methanol for 3 minutes. After fixation, cytospins were stained in a 50% eosin methylene blue solution (Sigma-Aldrich GmbH, Seelze, Germany) for 20 minutes and rinsed in water for 5 seconds, and the nuclei were counterstained with 10% Giemsa solution (Merck kGaA, Darmstadt, Germany) for 15 minutes. During eosinophil differentiation, different stages of maturation can be observed. Cells differentiate from blast cells to promyelocyte type 1, promyelocyte type 2, myelocyte, metamyelocyte, and, finally, mature eosinophils with segmented nuclei. These stages can be distinguished by cell size, ratio of cytoplasm to nucleus, presence of azurophilic granules, appearance of eosinophilic granules, and nucleus shape. Juvenile eosinophils were characterized as cells belonging to the stages of promyelocyte 2, myelocyte, metamyelocyte, and mature eosinophils. These cells were all observed to contain eosinophilic granules.
Electrophoretic mobility shift assay
Nuclear extracts were prepared from differentiating eosinophils as described previously.38 Oligonucleotides were labeled by filling in the cohesive ends with [α-32P] dCTP using Klenow fragment of DNA polymerase 1. Nuclear extracts were incubated in a final volume of 20 μL, containing 10 mM HEPES (N-2-hydroxylethypiperazine-N'-2-ethanesulfonic acid), pH 7.8, 50 mM KCl, 1 mM EDTA (ethylenediaminetetraacetic acid), 5 mM MgCl2, 10% (vol/vol) glycerol, 5 mM dithiothreitol, 2 μg poly(dI-dC), 20 μg BSA, and 1 ng 32P-labeled oligonucleotide for 20 minutes at room temperature. Subsequently, samples were subjected to electrophoresis for 3 hours on a 5% nondenaturing polyacrylamide gel. Supershift analysis was performed by preincubating 10 μg nuclear extract with 3 μg antibody against STAT5, STAT3 (Transduction Laboratories, Lexington, KY), and STAT1 (Transduction Laboratories, Lexington, KY) for 2 hours on ice before adding the binding buffer and the 32P-labeled oligonucleotide.
Western blot analysis
Western blot analysis was performed using standard techniques. In short, for the detection of STAT5 expression and phosphorylation, differentiating eosinophils were lysed in Laemmli buffer (0.12 M Tris HCl, pH 6.8, 4% sodium dodecyl sulfate [SDS], 20% glycerol, 0.05 μg/μL bromophenol blue, and 35 mM β-mercaptoethanol) and were boiled for 5 minutes. Equal amounts of total lysate were analyzed by 8% SDS–polyacrylamide gel electrophoresis (SDS-PAGE). Proteins were transferred to Immobilon-P and incubated with blocking buffer (Tris-buffered saline/Tween 20) containing 5% low-fat milk or BSA for 16 hours at 4°C before incubation with either an N-terminal STAT5 antibody (Santa Cruz Technology, Santa Cruz, CA), an antiphospho-STAT5 (Y695) antibody (Cell Signaling Technology, Beverly, CA), an antibody against β-actin (Santa Cruz Technology), an antibody against Bcl-2 (Santa Cruz Technology), or an antibody against p21WAF/Cip1(Santa Cruz Technology) for 2 hours in the same buffer. Subsequently, blots were incubated with peroxidase-conjugated secondary antibodies for 1 hour. Enhanced chemiluminescence (ECL) was used as a detection method according to the manufacturer's protocol (Amersham Pharmacia, Amersham, United Kingdom).
Measurement of apoptosis
CD34+ cells were transduced with a retroviral vector (SFCMM-3) expressing a truncated nerve growth factor receptor (NGFR) as a surface marker. During eosinophil differentiation, the percentage of Annexin V–positive cells was determined. Cells were incubated for 20 minutes on ice with a primary mouse antibody against NGFR. After washing the cells with phosphate-buffered saline (PBS), the cells were simultaneously incubated with a phycoerythrin (PE)–conjugated goat-antimouse antibody and Annexin V–fluorescein isothiocyanate (FITC) in Annexin-binding buffer (Bender Medsystems, Austria) for 20 minutes on ice. Percentages of transduced and apoptotic cells were determined by FACS analysis. As a positive control, a cell-permeable tat-peptide consisting of the BH3 domain of the proapoptotic protein Bim (YGRKKRRQRRREIWIAQELRRIGDEFNAYY) was used. Cells were incubated for 1 hour with 40 μM peptide at 37°C.
Results
Characterization of ex vivo eosinophil differentiation
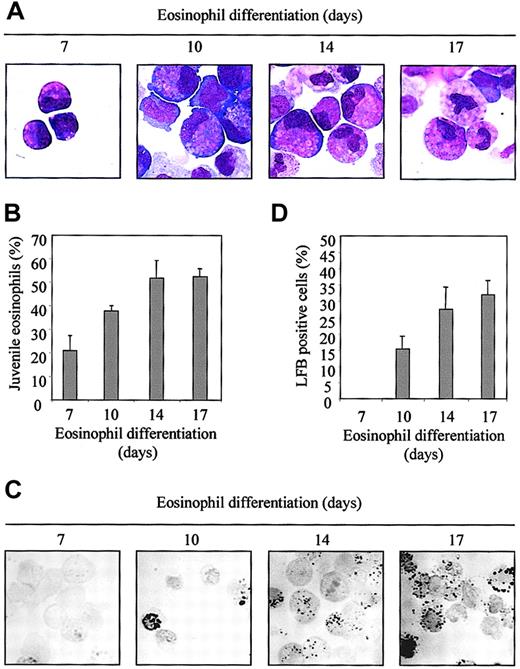
CD34+ progenitor cells, isolated from umbilical cord blood, were differentiated toward eosinophils in the presence of the cytokines IL-3 and IL-5, as described in “Materials and methods.” The morphology of differentiating eosinophils was analyzed by May-Grünwald-Giemsa staining (Figure1A). The percentage of juvenile eosinophils is shown in Figure 1B. After 7 days of differentiation, 20% of the cells were characterized as juvenile eosinophils, and most of the cells belonged to the promyelocyte 1 stage. After 17 days of culture, differentiation increased to 58% of juvenile eosinophils, of which most were mature myelocytes. During eosinophil differentiation, granules are formed. Final maturation of eosinophil differentiation was further investigated with LFB staining, which specifically stains eosinophil granules (Figure 1C). The percentage of LFB-positive cells increased from 15% after 10 days of differentiation to 35% after 17 days of differentiation (Figure 1D).
Eosinophil differentiation from CD34+ cells ex vivo.
Isolated CD34+ cells from cord blood were cultured in presence of SCF, FLT-3, GM-CSF, IL-3, and IL-5 for 3 days (as described in “Materials and methods”). After 3 days of differentiation cells were cultured in the presence of IL-3 and IL-5 only. After 7, 10, 14, and 17 days of differentiation, cytospins were made and (A) were stained with the May-Grünwald-Giemsa solution. (B) The percentage of juvenile eosinophils is expressed as an average ± SEM of 4 individual experiments on each day. (C) Cytospins were also stained with LFB for 2 hours after fixation with dry acetone. (D) The percentage of LFB-positive cells is expressed as an average ± SEM of 4 individual experiments on each day.
Eosinophil differentiation from CD34+ cells ex vivo.
Isolated CD34+ cells from cord blood were cultured in presence of SCF, FLT-3, GM-CSF, IL-3, and IL-5 for 3 days (as described in “Materials and methods”). After 3 days of differentiation cells were cultured in the presence of IL-3 and IL-5 only. After 7, 10, 14, and 17 days of differentiation, cytospins were made and (A) were stained with the May-Grünwald-Giemsa solution. (B) The percentage of juvenile eosinophils is expressed as an average ± SEM of 4 individual experiments on each day. (C) Cytospins were also stained with LFB for 2 hours after fixation with dry acetone. (D) The percentage of LFB-positive cells is expressed as an average ± SEM of 4 individual experiments on each day.
STAT5 expression and activation during eosinophil differentiation
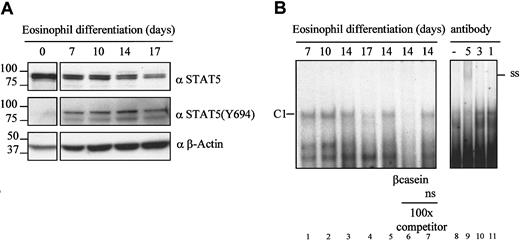
To investigate the regulation of STAT5a during eosinophil differentiation, protein lysates were prepared from CD34+cells immediately after isolation from umbilical cord blood and during subsequent eosinophil differentiation. Equal amounts of protein were separated by SDS-PAGE, and Western blotting was performed with an N-terminal antibody against STAT5 or an antibody against tyrosine-phosphorylated STAT5 (Figure2A). STAT5 was highly expressed in CD34+ cells as the nonphosphorylated, inactive form. However, STAT5 became phosphorylated on stimulation with IL-5. STAT5 expression and phosphorylation levels were high early during eosinophil differentiation, but protein expression was down-regulated after 14 to 17 days of differentiation. To demonstrate that STAT5 phosphorylation indeed correlates with STAT5 activation during eosinophil differentiation, nuclear extracts were prepared, and electrophoretic mobility shift assays (EMSAs) were performed using a β-casein element as a STAT5-binding site.39 Figure 2B shows that the specific slower migrating (C1) DNA-protein complex, corresponding to STAT5a (as demonstrated by supershift analysis with an antibody against STAT5),40 decreased after 17 days of differentiation. These experiments demonstrate that STAT5 is indeed expressed in differentiating eosinophils but is down-regulated during terminal maturation.
STAT5 expression and activation during eosinophil differentiation.
(A) Protein lysates were prepared from CD34+ cells, and cells differentiated toward eosinophils for 7, 10, 14, and 17 days. Western Blot analysis was performed with an N-terminal antibody against STAT5 (top panel), an antibody against tyrosine-phosphorylated STAT5 (middle panel), and as a control for equal loading with an antibody against β-actin (bottom panel). The experiment shown is representative of 3 additional experiments. (B) Nuclear extracts were prepared from differentiating eosinophils at days 7, 10, 14, and 17. Nuclear extracts were analyzed by EMSA using a 32P-labeled β-casein probe. Competition experiments were performed with an unlabeled β-casein probe in lane 6 and with a nonspecific probe in lane 7. Supershift analysis was performed with nuclear extracts prepared from differentiating eosinophils stimulated with IL-5 for 15 minutes, with antibodies against STAT5, STAT3, and STAT1, respectively (lanes 9-11). The experiment shown is representative of 3 individual experiments.
STAT5 expression and activation during eosinophil differentiation.
(A) Protein lysates were prepared from CD34+ cells, and cells differentiated toward eosinophils for 7, 10, 14, and 17 days. Western Blot analysis was performed with an N-terminal antibody against STAT5 (top panel), an antibody against tyrosine-phosphorylated STAT5 (middle panel), and as a control for equal loading with an antibody against β-actin (bottom panel). The experiment shown is representative of 3 additional experiments. (B) Nuclear extracts were prepared from differentiating eosinophils at days 7, 10, 14, and 17. Nuclear extracts were analyzed by EMSA using a 32P-labeled β-casein probe. Competition experiments were performed with an unlabeled β-casein probe in lane 6 and with a nonspecific probe in lane 7. Supershift analysis was performed with nuclear extracts prepared from differentiating eosinophils stimulated with IL-5 for 15 minutes, with antibodies against STAT5, STAT3, and STAT1, respectively (lanes 9-11). The experiment shown is representative of 3 individual experiments.
Retroviral transduction of CD34+ progenitor cells
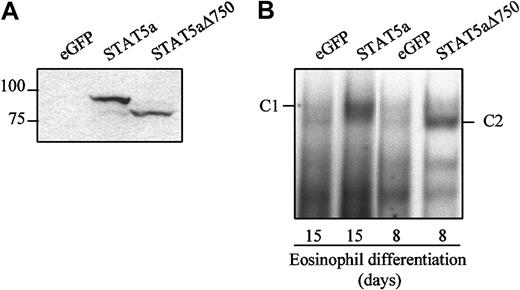
To investigate the role of STAT5a during eosinophil differentiation, a bicistronic retroviral DNA construct was used coexpressing eGFP. CD34+ cells, differentiated toward eosinophils for 2 days, were transduced with virus expressing either STAT5a, STAT5aΔ750, or empty vector. STAT5aΔ750 is a dominant-negative STAT5 mutant that can bind to DNA but is unable to activate transcription.41 Expression of STAT5a and STAT5aΔ750 in transduced cells was confirmed by Western blotting of protein lysates from transduced cells with a STAT5 antibody (Figure3A). EMSA was also performed to analyze the DNA binding of STAT5a and STAT5aΔ750 in transduced cells (Figure3B). After 7 or 14 days of differentiation, cells transduced with eGFP alone, STAT5aΔ750, or STAT5a were starved of cytokines for 16 hours before stimulation with IL-5 for 15 minutes. Nuclear extracts were analyzed using a β-casein element as a probe for STAT5 binding. Nuclear extracts prepared from transduced cells with STAT5a retrovirus showed increased DNA binding compared with cells transduced with eGFP. Stimulation of cells transduced with STAT5aΔ750 also resulted in increased DNA binding to the β-casein element. The DNA-protein complex C2 migrated faster than the DNA-protein complex C1 in cells transduced with STAT5a (Figure 3B), clearly demonstrating that retroviral transduction results in ectopic expression of STAT5a and its inactive mutant.
Expression and activation of STAT5a in transduced cells.
(A) Protein lysates were prepared from CD34+ cells transduced with eGFP, STAT5a, or STAT5aΔ750, and Western blot analysis was performed with an N-terminal antibody against STAT5. (B) Nuclear extracts were made of cells transduced with eGFP, STAT5a, or STAT5aΔ750 after either 15 or 8 days of eosinophil differentiation. Cells were starved of cytokines for 16 hours before stimulation with or without IL-5 for 15 minutes. Nuclear extracts were analyzed with EMSA using a 32P-labeled β-casein probe.
Expression and activation of STAT5a in transduced cells.
(A) Protein lysates were prepared from CD34+ cells transduced with eGFP, STAT5a, or STAT5aΔ750, and Western blot analysis was performed with an N-terminal antibody against STAT5. (B) Nuclear extracts were made of cells transduced with eGFP, STAT5a, or STAT5aΔ750 after either 15 or 8 days of eosinophil differentiation. Cells were starved of cytokines for 16 hours before stimulation with or without IL-5 for 15 minutes. Nuclear extracts were analyzed with EMSA using a 32P-labeled β-casein probe.
Regulation of proliferation during eosinophil differentiation by STAT5
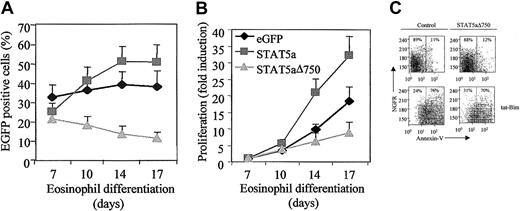
To determine whether STAT5 plays a critical role in regulating eosinophil differentiation, umbilical cord blood–derived cells were transduced with either eGFP, STAT5a, or STAT5aΔ750. The percentage of eGFP-positive cells represents the percentage of transduced cells and was determined by FACS analysis (Figure4A). Transduction efficiency of cells transduced with eGFP was approximately 32%, whereas transduction of cells with STAT5a or STAT5aΔ750 resulted in average transduction efficiency of approximately 25% and 21%, respectively. We observed an increase in the percentage of eGFP-positive cells after transduction with STAT5a (from 25% to 48%), indicating that STAT5a positively influences proliferation during eosinophil differentiation compared with cells transduced with eGFP alone. We also observed a decrease in eGFP-positive cells after transduction with STAT5aΔ750 compared with cells transduced with eGFP alone. This demonstrates that STAT5a is both necessary and sufficient for progenitor cell proliferation.
Regulation of proliferation during eosinophil differentiation by STAT5.
CD34+ cells were transduced with eGFP, STAT5a, or STAT5aΔ750. (A) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. Results are expressed as averages ± SEMs of 5 individual experiments. (B) Proliferation was determined by counting the eGFP-positive cells. Results are expressed as -fold increases in cell numbers compared with day 7 as averages ± SEMs of 5 individual experiments. (C) Cells were transduced with either empty vector or STAT5aΔ750. The percentage of apoptotic cells in NGFR-positive cells was determined with Annexin V–FITC by FACS analysis. As a positive control for apoptosis, cells were preincubated for 1 hour with a cell-permeable Bim tat-peptide.
Regulation of proliferation during eosinophil differentiation by STAT5.
CD34+ cells were transduced with eGFP, STAT5a, or STAT5aΔ750. (A) The percentages of eGFP-positive cells were determined by FACS analysis after 7, 10, 14, and 17 days of differentiation. Results are expressed as averages ± SEMs of 5 individual experiments. (B) Proliferation was determined by counting the eGFP-positive cells. Results are expressed as -fold increases in cell numbers compared with day 7 as averages ± SEMs of 5 individual experiments. (C) Cells were transduced with either empty vector or STAT5aΔ750. The percentage of apoptotic cells in NGFR-positive cells was determined with Annexin V–FITC by FACS analysis. As a positive control for apoptosis, cells were preincubated for 1 hour with a cell-permeable Bim tat-peptide.
To determine whether STAT5 was indeed critical in the regulation of proliferation during eosinophil differentiation, CD34+cells were transduced with eGFP, STAT5a, or STAT5aΔ750. Proliferation was determined by counting the total amount of eGFP-positive cells. Transduction of cells with eGFP resulted in an induction of proliferation of approximately 15-fold between day 7 and day 17 (Figure4B). Transduction of cells with STAT5a resulted in an increase in proliferation to approximately 27-fold, and this was mainly because of enhanced proliferation during the first 14 days of differentiation. In contrast, transduction of cells with STAT5aΔ750 resulted in a decrease of proliferation.
To investigate whether the inhibition of proliferation in cells ectopically expressing dominant-negative STAT5a resulted from enhanced apoptosis, Annexin V staining was performed in transduced cells. It has previously been described that eGFP tends to leak from apoptotic cells42; in our experiments, no eGFP-positive apoptotic or dead cells could be detected (results not shown). Therefore, an alternative retroviral vector expressing a truncated NGFR as a surface marker was used. The percentage of apoptotic cells during eosinophil differentiation is very low, approximately 10%. No difference could be observed in the percentage of Annexin V–positive cells between cells transduced with empty vector alone and cells transduced with STAT5aΔ750 (Figure 4C). A cell-permeable tat-peptide consisting of the BH3 domain of Bim was used to demonstrate that apoptosis could be measured in our CD34+ cells differentiating toward eosinophils (Figure 4C, lower panel). These data demonstrate that STAT5a is indeed required for optimal proliferation during eosinophil differentiation and does not play a critical role in regulating apoptosis.
Regulation of eosinophil differentiation by STAT5
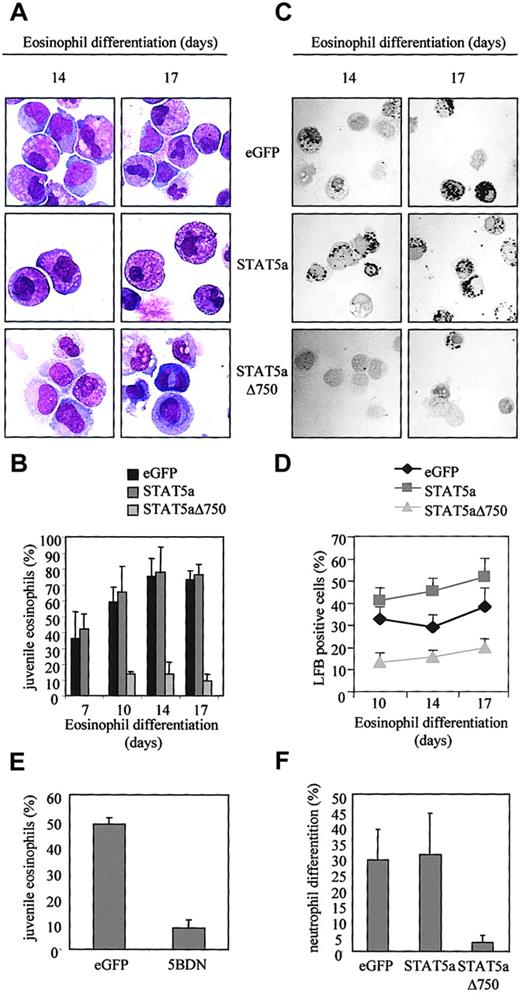
To investigate whether STAT5a also plays a role in eosinophil differentiation, cells were again transduced with eGFP, STAT5a, or STAT5aΔ750. After 7, 10, 14, and 17 days of differentiation, eGFP-positive cells were sorted by FACS from the nontransduced cells and cytospins prepared. The morphology of the differentiating eosinophils was analyzed by May-Grünwald-Giemsa staining as described in “Materials and methods” (Figure5A). The percentage of juvenile eosinophils is expressed in Figure 5B. Transduction of cells with eGFP or STAT5a resulted in approximately 70% juvenile eosinophils after 17 days of differentiation, of which most were mature myelocytes or metamyelocytes.
Regulation of eosinophil differentiation by STAT5.
CD34+ cells were transduced with eGFP, STAT5a, or STAT5aΔ750. After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. (A) Cytospins were stained with May-Grünwald-Giemsa solution. (B) Eosinophil differentiation was expressed as a percentage of juvenile eosinophils. Data are expressed as averages ± SEMs of 5 independent experiments. (C) Cytospins were fixed with dry acetone for 10 minutes before they were stained with LFB for 2 hours. (D) The percentage of LFB-positive cells was determined by microscopy, and data are expressed as averages ± SEMs of 5 individual experiments. (E) CD34+ cells were transduced with eGFP or dominant-negative STAT5b (5BDN). After 14 days of differentiation, cytospins were made of eGFP-positive cells and stained with May-Grünwald-Giemsa solution. The percentages of juvenile eosinophils are expressed as averages ± SEMs of 3 independent experiments. (F) CD34+ cells transduced with eGFP, STAT5a, or STAT5aΔ750 were differentiated toward neutrophils. After 17 days of differentiation, eGFP-positive cells were sorted, and cytospins were made and stained with May-Grünwald-Giemsa solution. Neutrophil differentiation is expressed as the percentages of cells consisting of banded or segmented nuclei. Data are expressed as averages ± SEMs of 3 independent experiments.
Regulation of eosinophil differentiation by STAT5.
CD34+ cells were transduced with eGFP, STAT5a, or STAT5aΔ750. After 14 and 17 days of differentiation, transduced cells were separated from the nontransduced cells, and cytospins were made. (A) Cytospins were stained with May-Grünwald-Giemsa solution. (B) Eosinophil differentiation was expressed as a percentage of juvenile eosinophils. Data are expressed as averages ± SEMs of 5 independent experiments. (C) Cytospins were fixed with dry acetone for 10 minutes before they were stained with LFB for 2 hours. (D) The percentage of LFB-positive cells was determined by microscopy, and data are expressed as averages ± SEMs of 5 individual experiments. (E) CD34+ cells were transduced with eGFP or dominant-negative STAT5b (5BDN). After 14 days of differentiation, cytospins were made of eGFP-positive cells and stained with May-Grünwald-Giemsa solution. The percentages of juvenile eosinophils are expressed as averages ± SEMs of 3 independent experiments. (F) CD34+ cells transduced with eGFP, STAT5a, or STAT5aΔ750 were differentiated toward neutrophils. After 17 days of differentiation, eGFP-positive cells were sorted, and cytospins were made and stained with May-Grünwald-Giemsa solution. Neutrophil differentiation is expressed as the percentages of cells consisting of banded or segmented nuclei. Data are expressed as averages ± SEMs of 3 independent experiments.
After the transduction of cells with STAT5a, approximately the same percentages of juvenile eosinophils and cells transduced with eGFP were observed. However, because cells transduced with STAT5a showed an increase in proliferation, there was an increase in total numbers of juvenile eosinophils. Transduction of cells with STAT5aΔ750 resulted, after 17 days of differentiation, in a very low percentage of juvenile eosinophils, approximately 7%, which were promyelocytes type 2. Most of the cells belonging to the eosinophil lineage were blocked in differentiation at the stage of promyelocyte type 1. Proliferation of cells transduced with STAT5aΔ750 was also inhibited, resulting in a decrease of approximately 95% in total numbers of juvenile eosinophils compared with cells transduced with eGFP.
Eosinophil differentiation was also analyzed by LFB staining of the granules (Figure 5C). After fixation, cells were stained with LFB for 2 hours. After 10 days of differentiation, approximately 33% of the cells transduced with eGFP were positive for LFB. This percentage increased to 38% after 21 days of differentiation (Figure 5D). Transduction of cells with STAT5a resulted in accelerated eosinophil differentiation. After only 10 days of differentiation, more than 40% of the cells were positive for LFB. Percentages of LFB-positive cells increased to 52% after 17 days of differentiation. In contrast, cells ectopically expressing STAT5aΔ750 were potently inhibited in eosinophil differentiation, as measured by LFB staining. The percentage of LFB-positive cells remained less than 20%, whereas proliferation was severely disturbed (Figure 5D). These data clearly demonstrate that STAT5a plays a critical role during eosinophil differentiation.
To determine whether STAT5a and STAT5b may have different functional activities in eosinophil differentiation, cells were transduced with eGFP or dominant-negative STAT5b (Figure 5E). Expression of dominant-negative STAT5b in CD34+ cells resulted in a block in eosinophil differentiation that was comparable to the inhibition in differentiation caused by STAT5aΔ750. Proliferation was also comparably inhibited by ectopic expression of dominant-negative STAT5b (data not shown), suggesting that the 2 STAT5 isoforms have similar, if not redundant, roles.
Although STAT5 is thought to play a role in the differentiation of neutrophils, this has only been demonstrated in the murine 32D cell line.43 To determine whether STAT5a also plays a role in neutrophil differentiation from human CD34+ cells, cells were transduced with eGFP or STAT5aΔ750 and were differentiated toward neutrophils in response to G-CSF (Figure 5F). The percentage of differentiating neutrophils was defined as the percentage of cells consisting of either banded or segmented nuclei. Transduction of cells with eGFP or STAT5a resulted in approximately 30% differentiated neutrophils, whereas transduction of cells with STAT5aΔ750 resulted in less than 5% differentiated neutrophils. Neutrophil differentiation was blocked at the stage of promyelocytes, similar to what was observed in eosinophil differentiation. Proliferation was also inhibited after transduction with STAT5aΔ750 (data not shown).
Regulation of STAT5 target genes during eosinophil differentiation
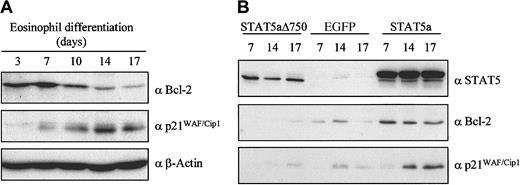
Our results demonstrate that STAT5 plays an important role during eosinophil differentiation. To confirm the functionality of STAT5, the expression of several STAT5 target genes was evaluated. Protein lysates were prepared from differentiating eosinophils, and these were separated by SDS-PAGE (Figure 6A). Western blotting was performed with antibodies against Bcl-2 and p21WAF/Cip1, both previously described STAT5 target genes.44 45 Bcl-2 expression was high early during eosinophil differentiation, whereas expression was down-regulated during final maturation. In contrast, p21WAF/Cip1expression was absent after 3 days of eosinophil differentiation and was up-regulated during final maturation.
Regulation of STAT5 target genes during eosinophil differentiation.
(A) Protein lysates were prepared from CD34+ cells differentiated toward eosinophils for 3, 7, 10, 14, and 17 days. Western blot analysis was performed with antibodies against Bcl-2 (top panel) and p21WAF/Cip1 (middle panel) and as a control for equal loading with an antibody against β-actin (bottom panel). The experiment shown is representative of 3 independent experiments. (B) CD34+ cells were transduced with eGFP, STAT5a, or STAT5aΔ750. After 7, 14, and 17 days of differentiation, eGFP-positive cells were sorted, and protein lysates were prepared and analyzed by Western blotting.
Regulation of STAT5 target genes during eosinophil differentiation.
(A) Protein lysates were prepared from CD34+ cells differentiated toward eosinophils for 3, 7, 10, 14, and 17 days. Western blot analysis was performed with antibodies against Bcl-2 (top panel) and p21WAF/Cip1 (middle panel) and as a control for equal loading with an antibody against β-actin (bottom panel). The experiment shown is representative of 3 independent experiments. (B) CD34+ cells were transduced with eGFP, STAT5a, or STAT5aΔ750. After 7, 14, and 17 days of differentiation, eGFP-positive cells were sorted, and protein lysates were prepared and analyzed by Western blotting.
Ectopic expression of STAT5a and STAT5aΔ750 perturbed the expression of Bcl-2 and p21WAF/Cip1 (Figure 6B). Forced expression of STAT5a in CD34+ cells resulted in the enhanced expression of Bcl-2, which was not down-regulated during final maturation. In addition, p21WAF/Cip1 expression was enhanced under these conditions. In agreement, ectopic expression of STAT5aΔ750 in CD34+ cells resulted in a decreased expression of Bcl-2 and p21WAF/Cip1. These data indicate that increased expression of Bcl-2 and p21WAF/Cip1 can be regulated by STAT5a, and this may play a role in the processes regulating eosinophil differentiation.
Discussion
In this study we have investigated the role of STAT5a during eosinophil differentiation. Our experiments indicate that, during eosinophil differentiation of CD34+ umbilical cord blood progenitors, STAT5a protein is differentially expressed and its activity is regulated. Ectopic expression of STAT5a results in an enhanced proliferation and an accelerated differentiation of umbilical cord blood–derived CD34+ cells. Furthermore, ectopic expression of STAT5aΔ750 results in an inhibition of proliferation and a block in differentiation at the promyelocytic stage.
Although previous studies have suggested that STAT transcription factors may play an important role during myeloid differentiation, these studies were performed in myeloid leukemic cell lines, not in primary hematopoietic cells.43 We used an ex vivo differentiation system that resulted in the differentiation of a high percentage of juvenile eosinophils (Figure 1). Several studies have used similar ex vivo differentiation protocols for eosinophil differentiation from CD34+ progenitor cells derived from cord blood or peripheral blood.46-48 Recently, Hashida et al49 demonstrated that 80% of the cord blood–derived CD34+ cells in their study developed into mature eosinophils after 17 days of differentiation. However, they only distinguished between blast cells and mature eosinophils without mentioning the other stages of eosinophil differentiation. The same percentages of cells that resembled eosinophils, after 21 days of differentiation, were described by Velazquez et al.50 Our differentiation protocol resulted in approximately the same percentages of differentiated and juvenile eosinophils as these previous studies. This ex vivo system of eosinophil differentiation resulted in a high number of juvenile eosinophils that were positive for LFB and eosinophil peroxidase (data not shown), and it is an important tool for analyzing the role of transcription factors during early stages of the differentiation process.
Retroviral transduction experiments were performed to ectopically express either STAT5a or STAT5aΔ750 in CD34+ cells. It is often thought that enhanced differentiation is accompanied by a decrease in proliferation. In contrast, our experiments demonstrated enhanced differentiation and increased proliferation in cells transduced with STAT5a compared with cells transduced with eGFP (Figures 4B, 5A-D).
A role for STAT5 in proliferation during myeloid differentiation has been suggested because of the observation that the expression of dominant-negative STAT5 results in an inhibition of proliferation in IL-3–dependent cell lines.43,51 Bone marrow–derived macrophages obtained from STAT5a(−/−) knockout mice also proliferate more slowly on GM-CSF stimulation.52Furthermore, it has been demonstrated that thymocytes derived from STAT5a(−/−) STAT5b(−/−) mice fail to proliferate in response to stimulation with IL-2.53Proliferation in cells transduced with STAT5a might be enhanced by a reduction of apoptosis. Bcl-x is an antiapoptotic protein that might play a role in survival during eosinophil differentiation.54 It is expressed in CD34+cells and is down-regulated during the final stages of neutrophil differentiation, whereas expression remains high during monocyte differentiation.55 It has also been demonstrated40 that A1, another antiapoptotic Bcl-2 family member, is down-regulated in STAT5a-deficient mice. We have demonstrated that STAT5 expression is high during early eosinophil differentiation, whereas STAT5 expression is down-regulated during final maturation. STAT5 might modulate survival during eosinophil differentiation through the transcriptional regulation of members of the Bcl-2 family. We have demonstrated that the expression of Bcl-2 and Bcl-xl (data not shown) was high during early eosinophil differentiation and was down-regulated during final maturation. Ectopic expression of STAT5a in CD34+ cells resulted in enhanced levels of Bcl-2 during eosinophil differentiation. Previously, it had been demonstrated that ectopic expression of dominant-negative STAT5 in ts21E26-transformed chicken myeloblasts results in increased apoptosis54. This was corroborated by the demonstration of enhanced apoptosis in bone marrow cells derived from STAT5a/b(−/−) knockout mice. However, we demonstrate that the ectopic expression of dominant-negative STAT5a in differentiating eosinophils results in decreased levels of Bcl-2, without affecting the levels of apoptosis. This suggests that although STAT5a might enhance survival during eosinophil differentiation, the absence of STAT5a and Bcl-2 is not sufficient in themselves to cause apoptosis. It is likely that the survival of differentiating eosinophils is regulated by several redundant signaling pathways. Proliferation during eosinophil differentiation might be enhanced by regulation of the cell-cycle regulators cyclin D156 and cyclin D2. Dominant-negative STAT5a has been demonstrated to inhibit the induction of cyclin D2 mRNA levels by IL-2 in a pro–B-cell line,57 and it inhibited IL-3–induced cyclin D1 promoter activity.58 Another STAT5 target gene implicated in playing a role in the regulation of proliferation is Pim-1. Northern blot analysis demonstrated that the constitutive activation of STAT5a in an IL-3–dependent cell line resulted in the up-regulation of cyclin D2, Bcl-x, and Pim-1. Furthermore, the ectopic expression of Pim-1 in Ba/F3 cells resulted in IL-3–independent proliferation. Importantly, it has also been demonstrated that Pim-1 levels are high in leukemic cell lines that express constitutively activated STAT5.59
Although little is known about STAT5 target genes regulating differentiation, the activation of p21WAF1/Cip1 60 and p27kip1 cyclin-dependent kinase inhibitors have been demonstrated to be involved in megakaryocytic differentiation.61 Ectopic expression of p21WAF/Cip1 and p27kip1 in a human megakaryoblastic leukemia cell line resulted in the induction of megakaryocyte differentiation. Ectopic expression of p21WAF/Cip1 also resulted in the morphologic differentiation of Ba/F3 cells.59 We have demonstrated that p21WAF/Cip1 expression levels are up-regulated during eosinophil differentiation. Ectopic expression of STAT5a also resulted in enhanced expression of p21WAF/Cip1, whereas p21WAF/Cip1 expression is reduced in cells transduced with dominant-negative STAT5a. This suggests that transcriptional activation of p21WAF/Cip1 is involved in STAT5-dependent eosinophil differentiation, by inducing a cell-cycle arrest that might be necessary for the induction of terminal differentiation.
In chickens, it has been demonstrated that CCAAT/enhancer binding protein (C/EBP)α and C/EBPβ62 play important roles in regulating eosinophil differentiation.63 C/EBPs have been demonstrated to synergistically activate transcription with STAT5 in some promoter studies.64 For example, C/EBP is required for full basal promoter activity of the oncostatin M promoter, an IL-6 subfamily member, whereas STAT5 is responsible for GM-CSF–induced promoter activity.65 Furthermore, it has been demonstrated that STAT5 induces transcriptional activation of β-casein,66,67 whereas C/EBP-binding sites are also required for hormonal transcription activation of β-casein.68 69 It is possible that the cooperation between STAT5 and C/EBPs is responsible for final maturation during eosinophil differentiation.
The 2 STAT5 gene products, STAT5a and STAT5b, are highly homologous and are thought to have overlapping functional roles.70 Recently, it has been demonstrated that STAT5b can be differentially regulated compared with STAT5a. Both isoforms are phosphorylated by Src kinases, whereas only STAT5b subsequently translocates to the nucleus in NIH 3T3 cells Furthermore, it has been demonstrated that STAT5b may have unique transcriptional functions. For example, STAT5a and STAT5b can regulate transcription of the estrogen receptor-α gene, whereas regulation of the estrogen receptor-β gene is exclusively mediated by STAT5b.71These results suggest that STAT5a and STAT5b are nonredundant and can have unique functions in some cell types. However, it has also been demonstrated that STAT5a and STAT5b have redundant functions in T cells, including IL-2 regulation of CD25 and natural killer (NK) cell function.72 Ectopic expression of dominant-negative STAT5a or dominant-negative STAT5b in differentiating eosinophils resulted in a similar block in differentiation and in comparably reduced levels of proliferation. This indicates that during eosinophil differentiation the 2 STAT5 isoforms have similar, if not redundant, functions.
STAT5 expression and phosphorylation are high during the early stages of eosinophil differentiation, whereas protein expression of STAT5 is down-regulated at later stages. This indicates that STAT5 might play a critical role, mainly during early eosinophil differentiation. Indeed, the transduction of cells with STAT5a results in an acceleration of differentiation but does not result in cells with more mature morphology. To support this hypothesis, it has been recently suggested that STAT5 plays a role in early hematopoiesis. In STAT5a(−/−) and STAT5b(−/−) knockout mice, high numbers of early erythroblasts were observed that could not progress in the differentiation process.73
This is the first study that clearly demonstrates that STAT5a plays a critical role in human eosinophil differentiation. The precise molecular mechanism underlying this process remains to be clarified. However, our data suggest that the transcriptional regulation of critical cell cycle and antiapoptotic genes is important in regulating differentiation fate.
We thank Dr G. Nolan for kindly providing us with the LZRS vector and the phoenix-ampho packaging cell line. We also would like to thank Dr H. Spits for providing us with the LZRS construct expressing dominant-negative STAT5b and M. Stijns for providing us with the SFCMM-3 vector and the antibody against NGFR.
Prepublished online as Blood First Edition Paper, August 15, 2002; DOI 10.1182/blood-2002-03-0740.
Supported by research grants from GlaxoSmithKline and by a grant from the Dutch Cancer Society (M.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Leo Koenderman, Department of Pulmonary Diseases, G03.550, University Medical Center, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands; e-mail:l.koenderman@hli.azu.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal