Abstract
DNA viruses have evolved a number of mechanisms to inhibit the major cellular tumor-suppressor pathways. Viral oncogenes can override growth suppressive signals and extend the virus proliferative capacity. The Kaposi sarsoma–associated human herpesvirus 8 (KSHV) encodes a protein, cyclin K, that is similar to cellular cyclin D1 but behaves atypically. Cyclin K resists the actions of the p16 INK4a and p27Kip1 inhibitors and extends the range of cdk6 substrates, thereby inducing cell-cycle progression toward S phase. In this study, we show that cyclin K overrides growth suppressive signals through signal transducer and activator of transcription 3 (STAT3) inactivation. Cyclin K was found to associate with the activation domain of STAT3 to inhibit its DNA-binding and transcriptional activities. Overexpression of cyclin K and inhibition of STAT3 prevents the growth suppressive effect imposed by the interleukin 6–type cytokine, oncostatin M. Altogether, these results suggest that KSHV is able to override growth suppressive effects through multiple mechanisms, and they further indicate that cyclin K plays an important role in the oncogenic activity of these viruses.
Introduction
In 1994, partial viral DNA sequences were isolated from Kaposi sarcoma tumors, one of the highest neoplasms present in AIDS-afflicted patients. These sequences corresponded to a novel herpesvirus termed Kaposi sarcoma–associated herpesvirus (KSHV) or human herpesvirus-8 (HHV-8).1 Since that time, KSHV has been found in most epidemiologic forms of Kaposi sarcoma, implicating KSHV as one of the causative agents in the development of this neoplasm.2,3 KSHV is also implicated in Castelman disease, and the virus is also found in B lymphocytes, in which viral DNA is associated with abnormal lymphoproliferation such as primary effusion lymphoma.4 KSHV contains several viral oncogenes, and one important question to be resolved is what molecular basis governs the activation of cell transformation by this virus.2 Sequence analysis of the KSHV genome reveals at least 81 open reading frames that potentially code for viral gene products. Encoded by this genome with 53% amino acid sequence homology to human cyclin D1,4,5 viral cyclin K forms an active complex with cdk6 proteins that activates a wider range of substrates than the classic cyclin/cdk complexes.6,7 Notably, the cyclin K/cdk complex resists inhibition by cdk inhibitor proteins such as p21waf1 or p27Kip1,7,8 suggesting that cyclin K might not require p21waf1 for efficient assembly with the cdk subunit to form an active holoenzyme.9 Hence, cyclin K can activate more cell progression pathways while simultaneously failing to be regulated by cell-cycle inhibitory proteins. This unique dual capacity is most likely essential for KSHV oncogenicity.
Viral oncoproteins generally induce cell-cycle progression through a modulation of cellular gene expression and through a direct interaction with cell-cycle regulatory proteins to promote cell growth. Most DNA tumor viruses have evolved to inhibit major tumor-suppressor checkpoints.10 Human papillomavirus type 16 (HPV-16), adenovirus 5, and the simian virus 40 (SV40) produce viral proteins such as E6-E7, E1A, or large T antigens that induce cell transformation through interference with several targets such as p53 and retinoblastoma (Rb) proteins,10-12 the 2 main regulators of cell proliferation, apoptosis, and senescence. However, some of these viral proteins also deregulate various transcription factors such as signal transducer and activator of transcription 1 (STAT1)13 or coactivators such as p400, p300, and P/CAF.14-17 Binding to these transcriptional regulators enables the virus to target multiple functions of transcriptional activation and cell growth. Thus, viral progression through the cell cycle is controlled through a regulatory interplay between positive and negative regulators, leading to activation of growth-promoting genes and inhibition of tumor-suppressor genes. Through the binding to cdk proteins, Kaposi sarcoma–encoded cyclin K contributes to the transforming potential of KSHV. However, the use of cyclin K is probably a more widespread mechanism than initially recognized. Cyclin K expands the classical role of D-type cyclins since it has been shown to interfere with apoptotic or DNA replication pathways. Cyclin K has been found to interact with the replication protein Orc1 to promote DNA replication, whereas this effect was not observed with cyclin D1.18 Cyclin K can also phosphorylate Bcl-2 to inactivate its antiapoptotic functions and promote an efficient viral transmission.19
In this study, we expand the role of cyclin K, since we found that cyclin K overrides the growth suppressive effect of oncostatin M (OSM), a member of the interleukin-6 (IL-6) cytokine family that inhibits the replication of human tumor cells. Cyclin K interacts with STAT3 transcription factor and inhibits its DNA-binding and transcriptional activities, leading to the inhibition of OSM signaling. These results reveal an unexpected relationship between cyclin K and STAT3 proteins, and lead us to propose that cyclin K induces cell-cycle progression through the inhibition of growth suppressive pathways.
Materials and methods
Reagents, cell culture, and stable cell lines
Polyclonal STAT3 (C20), phospho-STAT3Tyr705, glutathione-S–transferase (GST; Z5), and phospho-STAT3Ser727 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal M2 antibody was obtained from Sigma (Saint-Quentin Fallavier, France). Rabbit serum–recognizing cyclin K was a generous gift of Dr S. Mittnacht. OSM receptor monoclonal antibodies were generated in the laboratory. Cell lines were grown in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS) and were stably transfected using the lipid reagent Fugene 6 (Roche Diagnostics, Meylan, France).
Plasmid constructs
Cyclin K–expressing vector (pcDNA3 derivative) was a gift of Dr D. Mann (Imperial College of Science, London, United Kingdom). The GST–cyclin K fusion construct was prepared in pGEX-2T using primers containing BamHI and EcoRI restriction sites. STAT3 fusion protein constructs have been previously described.20
TdT-mediated dUTP nick end labeling (TUNEL) analysis
Cell death was evaluated by the TUNEL technique according to the instructions of the manufacturer (Roche Diagnostics) and analyzed on a FACScan flow cytometer (Becton Dickinson, Le Pont de Claix, France).
Bioassay
Human melanoma A375 cells were plated at a density of 6 × 104 cells/mL in a 96-well plate in RPMI 1640 medium supplemented with 2% FCS. Incubation for 72 hours occurred in the presence of dilutions of the indicated cytokines. After a 72-hour incubation period, 0.5 μCi (0.0185 MBq) [3H]thymidine was added to each well for the last 4 hours of the culture and the incorporated radioactivity determined by scintillation counting.
Preparation of cell extracts and electrophoretic mobility shift assays (EMSAs)
Nuclear extracts, prepared as described before,20were preincubated (5 μg) for 5 minutes at room temperature in 25 mM NaCl, 10 mM Tris (tris(hydroxymethyl)aminomethane)–HCl, pH 7.5), 1 mM MgCl2, 5 mM EDTA (ethylenediaminetetraacetic acid, pH 8), 5% glycerol, and 1 mM DTT (dithiothreitol) with 1 μg poly dI-dC as a nonspecific competitor. A double-stranded nucleotide containing a STAT3-consensus binding site derived from the c-fos gene (sis-inducible element [Siem], 5′-CATTTCCCGTAAATCTTGTCG-3′) was end-labeled using the T4 kinase, and 10 pg of probe (20 000 cpm) was then added to the protein mixture for 15 minutes. Samples were then loaded on a 5% polyacrylamide gel (30:1) and separated by electrophoresis in 50 mM Tris, 0.38 M glycine, and 1 mM EDTA (pH 8.5). Gels were then dried and visualized by autoradiography. For the experiments presented in Figure 5, nuclear extracts (1 mg) were precleared with 40 μL strepdavidin-agarose (50% slurry in phosphate-buffered saline [PBS]) for one hour at 4°C. The double-stranded biotinylated oligonucleotide (40 pmol) containing the STAT3-consensus binding site was added and incubated for 1 hour at 4°C. Following one-hour incubation with streptavidin-agarose, the oligonucleotide-bound complexes were washed twice with buffer C and once with 20 mM Tris-HCl (pH 8) prior to addition of sample buffer. Protein detection by Western blot was performed as described in the next paragraph.
Immunoprecipitation and Western blot analysis
Immunoprecipitation reactions were performed in the presence of 1% nonidet P-40 (NP-40) with nuclear cell extracts (5 mg) precleared with 40 μL protein A–Sepharose (10% slurry in PBS) for 2 hours at 4°C. Cleared extracts were immunoprecipitated with 2 μg of the indicated antibodies overnight at 4°C followed by the addition of 40 μL protein A–Sepharose for one hour at 4°C. Immunoprecipitates were washed 3 times in buffer C and 1 time with 20 mM Tris-HCl (pH 8) prior to the addition of sample buffer. Following electrotransfer, membranes were analyzed by Western blot with the indicated antibodies diluted in Tris-buffered saline (TBS) buffer (10 mM Tris-HCl, pH 8; 150 mM NaCl) supplemented with bovine serum albumin (BSA; 2.5%) and milk powder (5%) for STAT3 (and GST), whereas only milk powder (5%) was added for cyclin K. Primary antibody incubation occurred for 2 hours for all Western blot procedures. Proteins were visualized using the enhanced chemiluminescence (ECL) system from Amersham Biosciences (Freiburg, Germany).
Fusion protein purification and pull-down experiments
Pull-down reactions were performed by incubating purified his-STAT3716–770 (100-200 ng) with 100 to 200 ng GST or GST–cyclin K coupled to glutathione beads in binding buffer (20 mM Tris-HCl, pH 7.5; 137 mM NaCl; 5 mM NaF; 5 mM Na3VO4; 1 mM DTT; and 1% Brij96 [Sigma]). After a 30-minute incubation at 4°C the beads were washed once with binding buffer, twice with binding buffer containing 0.5 M NaCl, and once with 20 mM Tris-HCl, pH 8. The inverse experiment was performed with his-STAT3716–770 coupled to nickel-agarose resin.
RNA extraction, Northern blot analysis, and luciferase assays
Northern blot analysis was performed essentially as described previously.21 RNA was extracted using the TRIzol reagent of which 5 μg total RNA was size fractionated on a denaturing 6% formaldehyde/1% agarose gel and transferred to Nitrocellulose (Amersham). Prehybridization occurred for 6 hours, after which hybridization was carried out overnight at 42°C in 5 mL 50-mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7), 0.75 M NaCl, 50% formamide, 3.5% sodium dodecyl sulfate (SDS), 5 × Denhardt, 2 mM EDTA, 0.1% SDS, 200 μg/mL salmon sperm DNA, and the appropriate radiolabeled probe. The full-length p21waf1 human cDNA was labeled with [α-32P]–deoxycytosine triphosphosphate using the random-priming labeling kit from Amersham (specific activity > 109 cpm/μg) and was used as a probe. Following hybridization, filters were washed 4 times in 0.1 × SSC and 0.1% SDS at room temperature for 20 minutes each. They were then exposed for 2 days to x-ray film with intensifying screens at −80°C. Luciferase assays were done as previously described.20
Gel filtration
For all gel filtration procedures, 0.25 mL of sample (5 μg bacterially expressed fusion proteins, 0.3-1 mg nuclear extract) was injected onto a Superose 12 gel filtration column (Amersham Biosciences). Analysis of endogenous proteins was carried out from nuclear extracts of A375 cells that were stimulated with OSM for 15 minutes. Samples were fractionated with a flow rate of 0.5 mL per minute; 0.3 mL fractions were collected and analyzed by Western blotting. Loading and elution of the system was carried out in PBS buffer. The column was calibrated using the following standards: blue dextran 2000 (void), apoferritine (443 kDa), amylase (200 kDa), alcohol dehydrogenase (150 kDa), albumine (66 kDa), carbonic anydrase (29 kDa), and cytochrome c (12.4 kDa).
Results
Cyclin K interacts with STAT3 and inhibits its transcriptional activity
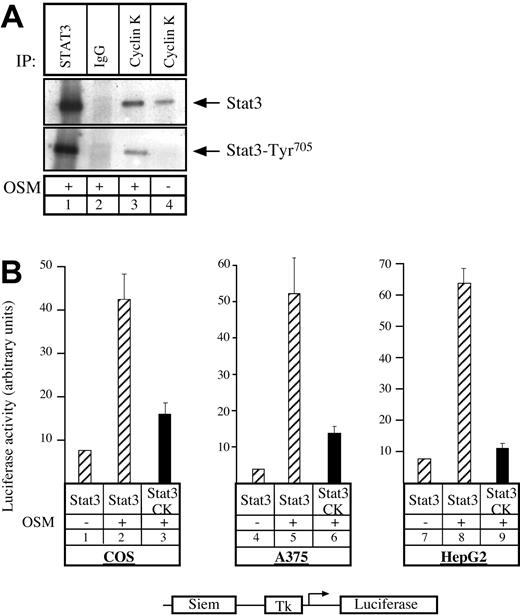
Having previously shown that the human homologue to cyclin K, cyclin D1, can interact with STAT3,20 we asked whether cyclin K would display the same biochemical characteristics. To test this, COS7 cells were transfected with vectors expressing STAT3 and cyclin K, serum starved, and then stimulated with OSM. Following stimulation, coimmunoprecipitations were performed using alternatively STAT3 polyclonal antibody, control immunoglobulin G (IgG) antibody, or M2 monoclonal antibody directed against flagged cyclin K (Figure 1A). Immunoblotting was performed with phospho-STAT3Tyr705 polyclonal antibodies (Figure 1A, bottom), followed by reblotting with STAT3 polyclonal antibodies (Figure 1A, top). Cyclin K and STAT3 were found to coimmunoprecipitate (Figure 1A, lane 3). This interaction was increased by OSM, however a weak interaction was detected between the 2 proteins in the absence of stimulation (Figure 1A, lane 4; Figure 6). We then hypothesized that cyclin K might affect the transcriptional activity of STAT3 proteins. To verify this, COS7 cells were cotransfected with a reporter construct containing 2 STAT3 consensus binding sites upstream of a thymidine kinase (tk) minimal promoter, together with vectors expressing STAT3 and cyclin K. Following transfection, cells were serum starved and stimulated with OSM, and luciferase activity was measured after 6 hours on cytoplasmic extracts. Inclusion of a STAT3-expressing vector in the transfection mix led to a 4- to 5-fold increase in reporter gene expression following cell stimulation (Figure 1B, lanes 1-2). Activation by STAT3 was inhibited in the presence of a cyclin K expression vector (Figure 1B, compare lanes 2 and 3). Further transitory reporter assays were performed in a panel of different cell lines, and expression of cyclin K inhibited the transcriptional activity of STAT3 in all cell types analyzed (Figure 1B, lanes 4-9).
Cyclin K interacts with and inhibits the transcriptional activity of STAT3.
(A) COS cells cotransfected with STAT3 and flagged cyclin K (CK)–expressing vectors were serum starved for 24 hours and stimulated for 30 minutes with OSM (10 ng/mL; lanes 1-3) or left untreated (lane 4). Nuclear extracts were immunoprecipitated with the appropriate antibodies, and the resulting precipitate was examined by Western blot probed with polyclonal antibodies directed against STAT3 proteins (top) or its Tyr705 phosphorylated form (bottom). (B) COS, A375, and HepG2 cells were cotransfected with a luciferase reporter gene containing 2 copies of a STAT3 consensus binding site linked to a minimal thymidine kinase promoter (Tk) together with vectors expressing STAT3 in the presence or absence of a vector encoding for cyclin K. Following transfection, cells were serum starved for 15 hours and left untreated or stimulated for 6 hours with OSM (10 ng/mL). Error bars represent SD.
Cyclin K interacts with and inhibits the transcriptional activity of STAT3.
(A) COS cells cotransfected with STAT3 and flagged cyclin K (CK)–expressing vectors were serum starved for 24 hours and stimulated for 30 minutes with OSM (10 ng/mL; lanes 1-3) or left untreated (lane 4). Nuclear extracts were immunoprecipitated with the appropriate antibodies, and the resulting precipitate was examined by Western blot probed with polyclonal antibodies directed against STAT3 proteins (top) or its Tyr705 phosphorylated form (bottom). (B) COS, A375, and HepG2 cells were cotransfected with a luciferase reporter gene containing 2 copies of a STAT3 consensus binding site linked to a minimal thymidine kinase promoter (Tk) together with vectors expressing STAT3 in the presence or absence of a vector encoding for cyclin K. Following transfection, cells were serum starved for 15 hours and left untreated or stimulated for 6 hours with OSM (10 ng/mL). Error bars represent SD.
Thus, we concluded from these results that cyclin K interacts with STAT3 and inhibits its transcriptional activity
Cyclin K interacts with the activation domain of STAT3
To further characterize the interaction between cyclin K and STAT3, we used a chimeric Gal4-STAT3 fusion protein corresponding to the activation domain of STAT3. Using a Gal4-dependent luciferase reporter gene, we found that cyclin K was able to inhibit transactivation by Gal4-STAT3 in COS cells (Figure2A, compare lanes 2 and 3) and in a panel of different cell lines (Figure 2A, lanes 6, 9). Importantly, cyclin K had no effect on the transcriptional activity of control Gal4 fusion proteins such as Gal4-VP16 (Figure 2A, compare lanes 11 and 12), indicating that the effect of cyclin K is specific. We then compared the relative inhibitory efficiencies of equivalent amounts of cyclin D1 and cyclin K on STAT3 activity. Using a Gal4-dependent luciferase reporter gene, we found that the cyclin-mediated inhibitory effects on Gal4-STAT3 transactivation increased with increasing amounts of cyclins (Figure 2B, compare lanes 3, 5, and 7 with 4, 6, and 8), but that cyclin D1 and cyclin K inhibited STAT3 transcriptional activity to the same extent.
Cyclin K interacts with the activation domain of STAT3.
(A) COS, A375, or HepG2 cells were cotransfected with vectors expressing a Gal4 luciferase reporter gene and the Gal4-STAT3 expression vector, in the presence or absence of the cyclin K expression vector (lanes 1-9). Following transfection, cells were serum starved and stimulated as described for Figure 1 for 4 hours. Identical experiments were performed in parallel in HepG2 cells using a Gal4-VP16 plasmid (lanes 10-12). (B) A375 cells were transfected as described in panel A with increasing amounts of cyclins (150 ng, lanes 3-4; 250 ng, lanes 5-6; 500 ng, lanes 7-8) in the presence of the Gal4-STAT3 expression vector. (C) Representation of the carboxy-terminal his-STAT3716–770 and full-length GST–cyclin K fusion protein used in pull-down experiments. SH2 indicates Src homology domain. Dotted lines illustrate the correspondence of fusion protein to the C terminal part of the protein below. (D) Purified GST–cyclin K fusion proteins (50-100 ng) were analyzed for binding to histidine or his-STAT3716–770fusion proteins (50-100 ng) corresponding to the activation domain of STAT3 immobilized on nickel-agarose beads (lanes 1-3). In parallel (lanes 4-6), his-STAT3716–770 (50-100 ng) was tested for binding to GST or full-length GST–cyclin K (50-100 ng) immobilized on sepharose beads. The same experiments were then repeated using equivalent amounts (3 pM) of purified GST–cyclin D or GST–cyclin K, with his or his-STAT3716–770 immobilized on nickel-agarose beads (lanes 7-13). Note that the Western blot experiment in panel D, lanes 7-13, was performed with an antibody directed against GST, and that lanes 12 to 13 correspond to a 10% input.
Cyclin K interacts with the activation domain of STAT3.
(A) COS, A375, or HepG2 cells were cotransfected with vectors expressing a Gal4 luciferase reporter gene and the Gal4-STAT3 expression vector, in the presence or absence of the cyclin K expression vector (lanes 1-9). Following transfection, cells were serum starved and stimulated as described for Figure 1 for 4 hours. Identical experiments were performed in parallel in HepG2 cells using a Gal4-VP16 plasmid (lanes 10-12). (B) A375 cells were transfected as described in panel A with increasing amounts of cyclins (150 ng, lanes 3-4; 250 ng, lanes 5-6; 500 ng, lanes 7-8) in the presence of the Gal4-STAT3 expression vector. (C) Representation of the carboxy-terminal his-STAT3716–770 and full-length GST–cyclin K fusion protein used in pull-down experiments. SH2 indicates Src homology domain. Dotted lines illustrate the correspondence of fusion protein to the C terminal part of the protein below. (D) Purified GST–cyclin K fusion proteins (50-100 ng) were analyzed for binding to histidine or his-STAT3716–770fusion proteins (50-100 ng) corresponding to the activation domain of STAT3 immobilized on nickel-agarose beads (lanes 1-3). In parallel (lanes 4-6), his-STAT3716–770 (50-100 ng) was tested for binding to GST or full-length GST–cyclin K (50-100 ng) immobilized on sepharose beads. The same experiments were then repeated using equivalent amounts (3 pM) of purified GST–cyclin D or GST–cyclin K, with his or his-STAT3716–770 immobilized on nickel-agarose beads (lanes 7-13). Note that the Western blot experiment in panel D, lanes 7-13, was performed with an antibody directed against GST, and that lanes 12 to 13 correspond to a 10% input.
Altogether, these results suggested that cyclin K interacts at least with the carboxy-terminal activation domain of STAT3. To verify this, in vitro pull-down experiments were performed using bacterially produced 6 × histidine-tagged STAT3 containing the 716 to 770 amino acids corresponding to the activation domain of STAT3 (his-STAT3716–770) and GST–cyclin K (Figure2C). We found that the GST–cyclin K fusion protein was retained by his-tagged STAT3716–770 and was not on the histidine beads (Figure 2D, lanes 1-3). The result was confirmed when the purified proteins were analyzed in the reverse sense: his-STAT3716–770 was retained by a GST–cyclin K fusion protein, whereas it was not significantly detected on the GST beads (Figure 2D, lanes 4-6). We also compared the relative binding affinity of his-STAT3716–770 with GST–cyclin K or with GST–cyclin D1 using equivalent amounts of purified cyclins (3 pM, Figure 2D, lanes 12-13). Under these conditions, we found that GST–cyclin K was significantly retained by his-tagged STAT3716–770 beads, whereas the binding of GST–cyclin D1 was relatively weak (Figure 2D, lanes 9-10). This result suggested to us that cyclin K might use a different mechanism compared with cyclin D1 to bind to the activation domain of STAT3 or that its affinity for the transcription factor is higher.
We concluded from these results that cyclin K interacts with STAT3 through its carboxy-terminal activation domain. These experiments also suggest that the interaction between cyclin K and STAT3 is probably direct, although we cannot rule out the possibility that cyclin K functions via another partner copurified from the bacteria.
Absence of OSM-induced growth arrest in A375 cells in the presence of cyclin K
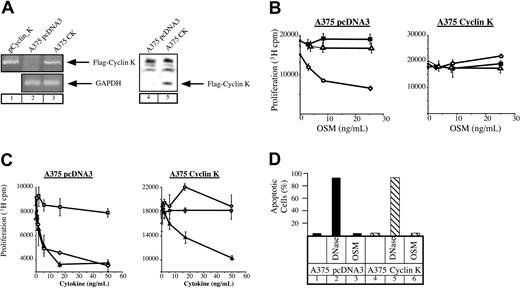
OSM mediates growth inhibition of different cell lines such as M1 myeloid leukemia cells, some breast and lung cancer cells, as well as melanocytes or primary melanomas. Since this effect relies on the activation of STAT3 (Kortylewski et al22 and Figure 4A), we hypothesized that cyclin K should override the growth inhibitory effect of OSM through STAT3 inactivation. To verify this, stable transfectants of A375 melanoma cells were generated and overexpression of cyclin K was examined by reverse transcription–polymerase chain reaction (RT-PCR) and Western blot analysis. Clones represent a population of cells to eliminate the possibility of cell phenotype abnormalities. As shown in Figure 3A, cyclin K mRNA and protein were observed in A375 transfectants, whereas they were undetectable in control clones. The revealed immunoreactive band corresponded to the reported molecular weight (28-30 kDa) of cyclin K containing the FLAG tag.5 We then examined the proliferative state of cyclin K–expressing A375 cloned cells in the presence or absence of OSM. Following the addition of OSM, growth of control cells was inhibited after 3 days in culture. In contrast, A375 cyclin K cells displayed continuous growth in the presence of OSM (Figure 3B). In our hands, IL-6 had no effect on cell proliferation regardless of the particular clone. To confirm and extend these results, identical experiments were performed using the growth inhibitory factor TGF-β. As expected, the growth of cyclin K–expressing cells was inhibited to the same extent by TGF-β compared with control clones (Figure 3C). Hence, the proliferative state of cyclin K–expressing cells was not a general phenomenon to increase cell proliferation. The apoptotic state of the cyclin K–expressing cells was examined to further explore the possibilities of the observed cell survival in the presence of OSM. TUNEL analysis indicated that the effect of OSM on cell proliferation was not mediated through activation of cell death (Figure 3D).
Lack of growth inhibition by OSM in cyclin K–expressing A375 cells.
(A) Expression of cyclin K was verified by RT-PCR (lanes 1-3) and Western blot (using the M2 Flag monoclonal antibody, lanes 4-5) in A375 stable transfectants. (B-C) Parental or cyclin K–expressing A375 cells were plated in triplicate in 96-well dishes in the presence of 2% serum and a dilution series of the indicated cytokines. After 72 hours, 3H-thymidine was pulsed for 4 hours and the cells were assessed for 3H incorporation by scintillation counting. Panel B: ○, OSM; ▵, IL-6; ▪, medium alone. Panel C: ▴, TGF-β; ○, OSM; ■, blank. (D) TUNEL analysis was performed on serum-starved A375 cloned cells after no treatment (lanes 1, 4) or treatment with OSM (10 ng/mL) for 24 hours (lanes 3, 6). DNase was added to a sample of nontreated cells for 10 minutes as a positive control (lanes 2, 5).
Lack of growth inhibition by OSM in cyclin K–expressing A375 cells.
(A) Expression of cyclin K was verified by RT-PCR (lanes 1-3) and Western blot (using the M2 Flag monoclonal antibody, lanes 4-5) in A375 stable transfectants. (B-C) Parental or cyclin K–expressing A375 cells were plated in triplicate in 96-well dishes in the presence of 2% serum and a dilution series of the indicated cytokines. After 72 hours, 3H-thymidine was pulsed for 4 hours and the cells were assessed for 3H incorporation by scintillation counting. Panel B: ○, OSM; ▵, IL-6; ▪, medium alone. Panel C: ▴, TGF-β; ○, OSM; ■, blank. (D) TUNEL analysis was performed on serum-starved A375 cloned cells after no treatment (lanes 1, 4) or treatment with OSM (10 ng/mL) for 24 hours (lanes 3, 6). DNase was added to a sample of nontreated cells for 10 minutes as a positive control (lanes 2, 5).
Taken together, these results suggest that expression of cyclin K overrides the inhibitory effect of OSM on cell proliferation.
Inhibition of the transcriptional and DNA-binding activities of STAT3 by cyclin K in A375 cells
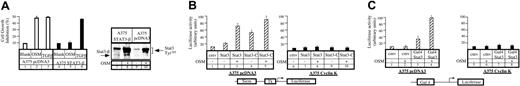
It has been previously shown that STAT3 activation plays an essential role in the growth inhibitory activities of oncostatin M.22 To confirm this result, we made use of a dominant-negative form of STAT3, STAT3β, a naturally occurring splice variant that lacks the C-terminal transactivation domain and functions in a dominant-negative manner to block STAT3-mediated gene activation.23-25 Stable transfectants of A375 cells were generated to evaluate the proliferative state of STAT3β-expressing cells in the presence or absence of OSM. Whereas STAT3β had no effect on cell proliferation in the absence of OSM, we found that STAT3β-expressing cells displayed continuous growth in the presence of OSM (Figure 4A, compare lanes 2 and 5). Overexpression of STAT3β was verified by Western blot analysis (Figure 4A, compare lanes 8 and 10). Importantly, STAT3β had no effect on the growth inhibitory effect of TGF-β (Figure 4A, compare lanes 3 and 6). Therefore, and as reported previously,22we concluded that STAT3 is a mediator of the growth arrest mediated by OSM in A375 cells.
STAT3 transcriptional activity in cyclin K–expressing cells.
(A) Parental or STAT3-β–expressing A375 cells were plated in triplicate in 96-well dishes in the presence of 2% serum and a dilution series of the indicated cytokines. Cell proliferation was evaluated as described for Figure 3. In parallel, overexpression of STAT3-β was verified by Western blot analysis using anti-STAT3 phosphorylated Tyr705 (A, lanes 7-10). (B) A375 control (lanes 1-5) or cyclin K–expressing cells (lanes 6-10) were cotransfected with a vector expressing a luciferase reporter gene containing 2 copies of a STAT3 consensus binding site linked to a minimal thymidine kinase promoter (Tk) together with vectors expressing STAT3 or its constitutive active form STAT3-C. Following transfection, cells were serum starved for 15 hours and left untreated or stimulated overnight with OSM (10 ng/mL). cmv indicates the corresponding parental expression vector. (C) A375 parental (lanes 1-4) or cyclin K–expressing cells (lanes 5-8) were cotransfected with the vector expressing a Gal4 luciferase reporter gene and Gal4 fusion protein linked to the STAT3 activation domain. Following transfection, cells were serum starved and stimulated as above for 4 hours.
STAT3 transcriptional activity in cyclin K–expressing cells.
(A) Parental or STAT3-β–expressing A375 cells were plated in triplicate in 96-well dishes in the presence of 2% serum and a dilution series of the indicated cytokines. Cell proliferation was evaluated as described for Figure 3. In parallel, overexpression of STAT3-β was verified by Western blot analysis using anti-STAT3 phosphorylated Tyr705 (A, lanes 7-10). (B) A375 control (lanes 1-5) or cyclin K–expressing cells (lanes 6-10) were cotransfected with a vector expressing a luciferase reporter gene containing 2 copies of a STAT3 consensus binding site linked to a minimal thymidine kinase promoter (Tk) together with vectors expressing STAT3 or its constitutive active form STAT3-C. Following transfection, cells were serum starved for 15 hours and left untreated or stimulated overnight with OSM (10 ng/mL). cmv indicates the corresponding parental expression vector. (C) A375 parental (lanes 1-4) or cyclin K–expressing cells (lanes 5-8) were cotransfected with the vector expressing a Gal4 luciferase reporter gene and Gal4 fusion protein linked to the STAT3 activation domain. Following transfection, cells were serum starved and stimulated as above for 4 hours.
In light of these results, our next aim was to characterize the effect of cyclin K on STAT3 activity in the A375 clones. Flow cytometry analysis indicated that comparable amounts of the OSM receptor was found on control and cyclin K–expressing clones (data not shown), indicating that OSM could confer an equal signal to STAT3 in each cell line. Accordingly, upon OSM stimulation, STAT3 translocated to the nucleus and was phosphorylated on Tyr705 and Ser727 to the same extent in each cell line. Hence, neither the nuclear localization nor the phosphorylation status of STAT3 was modified by cyclin K (data not shown; Figures5A, 6). We then evaluated the effect of cyclin K on STAT3 transcriptional activity following OSM stimulation. To this end, the stable cell lines were cotransfected with the Siem reporter construct, together with vectors expressing STAT3 or its constitutively active form STAT3-C.26 Following stimulation with OSM, STAT3 transcriptional activity was induced approximately 3- to 10-fold in control clones (Figure 4B, lanes 2-5). Markedly, cyclin K–expressing clones did not display any transcriptional activity of STAT3 or STAT3-C (Figure 4B, lanes 6-10). Moreover, using a Gal4-dependent luciferase reporter gene, OSM stimulation of control clones resulted in an increase in Gal4-STAT3–dependent transcriptional activity (Figure 4C, lanes 3-4), whereas in cyclin K–expressing cells, the transcriptional activity was not detected (Figure 4C, lanes 7-8).
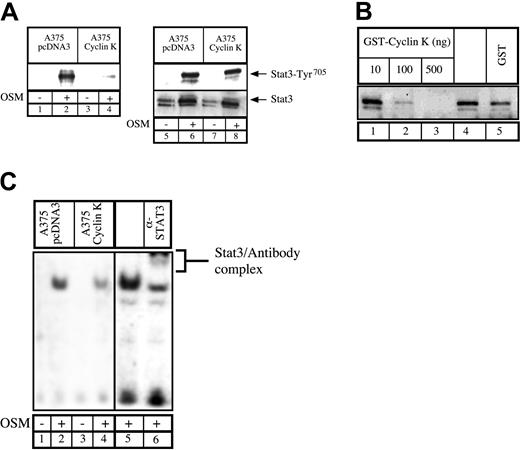
Inhibition of STAT3 DNA-binding activity by cyclin K.
(A) Parental or cyclin K–expressing A375 cells were serum starved for 48 hours and stimulated for 30 minutes with OSM. Nuclear extracts were incubated with a biotinylated double-stranded Siem oligonucleotide corresponding to a specific STAT3 DNA-binding site. Samples were analyzed by Western blot probed with polyclonal antibodies directed against STAT3 phosphorylated on Tyr705(lanes 1-4). In parallel, aliquots of the nuclear extracts were subjected to Western blot analysis using anti-STAT3 phosphorylated Tyr705 (lanes 5-8, top) prior to being stripped and reprobed with anti-STAT3 antibodies (lanes 5-8, bottom). (B) Normal A375 cells were serum starved for 48 hours and stimulated for 30 minutes with OSM (10 ng/mL). A titration of purified GST protein (500 ng, lane 5), GST–cyclin K (lanes 1-3), or buffer alone (lane 4) was incubated for 30 minutes with 30 μg nuclear extract prior to the addition of biotin-labeled double-stranded oligonucleotide probe. Samples were analyzed for DNA-binding activity by Western blot using polyclonal STAT3 phospho-Tyr705 antibodies. (C) DNA binding of STAT3 present in A375 parental or cyclin K–expressing cells was determined by EMSA. Supershift of STAT3 was performed (lane 6) with the addition of 2 μg polyclonal STAT3 antibodies.
Inhibition of STAT3 DNA-binding activity by cyclin K.
(A) Parental or cyclin K–expressing A375 cells were serum starved for 48 hours and stimulated for 30 minutes with OSM. Nuclear extracts were incubated with a biotinylated double-stranded Siem oligonucleotide corresponding to a specific STAT3 DNA-binding site. Samples were analyzed by Western blot probed with polyclonal antibodies directed against STAT3 phosphorylated on Tyr705(lanes 1-4). In parallel, aliquots of the nuclear extracts were subjected to Western blot analysis using anti-STAT3 phosphorylated Tyr705 (lanes 5-8, top) prior to being stripped and reprobed with anti-STAT3 antibodies (lanes 5-8, bottom). (B) Normal A375 cells were serum starved for 48 hours and stimulated for 30 minutes with OSM (10 ng/mL). A titration of purified GST protein (500 ng, lane 5), GST–cyclin K (lanes 1-3), or buffer alone (lane 4) was incubated for 30 minutes with 30 μg nuclear extract prior to the addition of biotin-labeled double-stranded oligonucleotide probe. Samples were analyzed for DNA-binding activity by Western blot using polyclonal STAT3 phospho-Tyr705 antibodies. (C) DNA binding of STAT3 present in A375 parental or cyclin K–expressing cells was determined by EMSA. Supershift of STAT3 was performed (lane 6) with the addition of 2 μg polyclonal STAT3 antibodies.
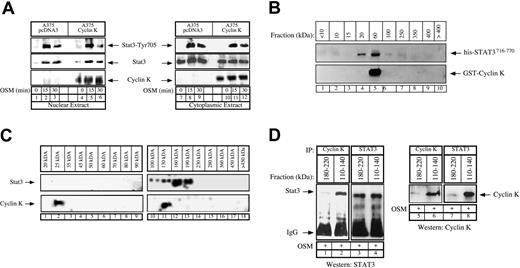
Stochiometry of the cyclin K–STAT3 complex.
(A) Control or cyclin K–expressing A375 cells were serum starved for 48 hours and stimulated with OSM (10 ng/mL) for the indicated times. Nuclear (lanes 1-6) or cytoplasmic (lanes 7-12) cell extracts were analyzed by Western blot analysis with polyclonal antibodies directed against STAT3 (middle panel), its Tyr705 phosphorylated forms (top panel), or cyclin K (bottom panel). (B) Purified GST–cyclin K and his-STAT3716–770 proteins (5 μg) were allowed to interact in vitro as described above, and aliquots of the binding reactions were then fractionated through Superose-12 filtration as described in “Materials and methods.” Eluate fractions were collected, and the presence of GST–cyclin K and his-STAT3716–770 was evaluated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting in each fraction. (C) A375 clones expressing cyclin K were stimulated for 15 minutes with OSM and nuclear extracts were prepared. Aliquots (300 μL) were subjected to Superose-12 filtration and STAT3 and cyclin K elutions were evaluated by SDS-PAGE and Western blotting in each fraction. Note that STAT3 (not cyclin K) was detected in some experiments in a complex with a size higher than 400 kDa but that this was not reproducible under the conditions used to detect the cyclin K association. (D) Following gel filtration of nuclear extracts, fractions corresponding to size 180 to 220 kDa (lanes 1, 3, 5, 7) and 110 to 140 kDa (lanes 2, 4, 6, 8) were analyzed by coimmunoprecipitation to detect the presence of a cyclin K–STAT3 complex. Fractions were immunoprecipitated with the indicated antibodies and analyzed by Western blot using antibodies directed against STAT3 proteins (lanes 1-4) or cyclin K (lanes 5-8).
Stochiometry of the cyclin K–STAT3 complex.
(A) Control or cyclin K–expressing A375 cells were serum starved for 48 hours and stimulated with OSM (10 ng/mL) for the indicated times. Nuclear (lanes 1-6) or cytoplasmic (lanes 7-12) cell extracts were analyzed by Western blot analysis with polyclonal antibodies directed against STAT3 (middle panel), its Tyr705 phosphorylated forms (top panel), or cyclin K (bottom panel). (B) Purified GST–cyclin K and his-STAT3716–770 proteins (5 μg) were allowed to interact in vitro as described above, and aliquots of the binding reactions were then fractionated through Superose-12 filtration as described in “Materials and methods.” Eluate fractions were collected, and the presence of GST–cyclin K and his-STAT3716–770 was evaluated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting in each fraction. (C) A375 clones expressing cyclin K were stimulated for 15 minutes with OSM and nuclear extracts were prepared. Aliquots (300 μL) were subjected to Superose-12 filtration and STAT3 and cyclin K elutions were evaluated by SDS-PAGE and Western blotting in each fraction. Note that STAT3 (not cyclin K) was detected in some experiments in a complex with a size higher than 400 kDa but that this was not reproducible under the conditions used to detect the cyclin K association. (D) Following gel filtration of nuclear extracts, fractions corresponding to size 180 to 220 kDa (lanes 1, 3, 5, 7) and 110 to 140 kDa (lanes 2, 4, 6, 8) were analyzed by coimmunoprecipitation to detect the presence of a cyclin K–STAT3 complex. Fractions were immunoprecipitated with the indicated antibodies and analyzed by Western blot using antibodies directed against STAT3 proteins (lanes 1-4) or cyclin K (lanes 5-8).
We concluded from these results that at least one mechanism by which cyclin K overrides the growth inhibitory effect of OSM on cell proliferation is through the inhibition of STAT3 transcriptional activity.
Inhibition of STAT3 DNA binding by cyclin K in A375 cells
To explain the effect of cyclin K on STAT3 transcriptional activity, we hypothesized that cyclin K might inhibit the DNA-binding activity of the transcription factor. Therefore, A375 cloned cells were analyzed for the ability of endogenous STAT3 to bind to DNA. Using first a biotinylated oligonucleotide containing the STAT3 consensus site, we found that endogeneous STAT3 associated with DNA upon OSM stimulation in control cells. By contrast, in cyclin K–expressing cells, the quantity of STAT3 bound to DNA was dramatically reduced (Figure 5A, lanes 1-4). Western blot analysis revealed equal quantity and phospho-Tyr705 state of nuclear STAT3 in the parental and cyclin K cells (Figure 5A, lanes 5-8). To verify this result, an in vitro experiment was performed in which OSM-stimulated A375 nuclear extracts were incubated with increasing concentrations of bacterially purified GST–cyclin K or GST alone. STAT3 DNA binding was then assessed using a biotinylated oligonucleotide containing the STAT3 consensus site. Increasing amounts of GST–cyclin K inhibited STAT3 DNA binding, whereas control GST had a minor effect (Figure 5B, compare lanes 4 with 1-3). Finally, the diminished binding of STAT3 to DNA was also seen in EMSA analysis using nuclear extracts from A375 cloned cells following stimulation with OSM. Following OSM stimulation, STAT3 DNA binding was observed in control cell lines, whereas this activity was significantly reduced in cyclin K–expressing cells (Figure 5C, compare lanes 1-2 with 3-4). The presence of STAT3 in the complex was verified by supershift with polyclonal STAT3 antibody using nuclear extract from control cells (Figure 5C, lanes 5-6).
Altogether, these results suggest that cyclin K inhibits STAT3 activation via multiple mechanisms. On one hand, cyclin K was found to inhibit Gal-STAT3, so we concluded that the inhibition of STAT3 was related to an inhibition of the transcriptional activity following binding of cyclin K to the activation domain. On the other hand, cyclin K was also able to block STAT3 activity through DNA-binding inhibition. Whether binding to the activation domain is related to DNA-binding inhibition remains to be determined (next paragraph).
Stochiometry of the cyclin K–STAT3 complex
As a first attempt to explain these inhibitory effects, we made the hypothesis that cyclin K might modify the nuclear and cytoplasmic localization of STAT3 in melanoma cells. To verify this, Western blot experiments were performed on cytoplasmic and nuclear extracts following OSM stimulation of control or cyclin K–expressing A375 clones. However, the overexpression of cyclin K did not modify the cellular localization, expression, or phosphorylation status of STAT3 (Figure 6A, compare lanes 1-3 with 4-6; lanes 7-9 with 11-12). Of note, OSM stimulation did not significantly affect cyclin K expression. Interestingly, cyclin K was detected in the nucleus but also in the cytoplasm of A375 cells (Figure 6A, lanes 10-12), suggesting that the protein might interact with nonphosphorylated forms of STAT3 as shown with pull-down experiments (Figure 2D) and in nonstimulated COS cells (Figure 1A, lane 4). Altogether, these results suggest that transcriptional inhibition of STAT3 by cyclin K is not mediated by a reduced nuclear expression of the transcription factor.
We then hypothesized that cyclin K might inhibit the DNA-binding activity by interfering with STAT3 dimerization, and we therefore began to characterize the stochiometry of the cyclin K–STAT3 complex. In vitro binding experiments were first performed using purified his-STAT3716–770 (19 kDa) and GST–cyclin K (50 kDa), and aliquots of the binding reactions were size-fractionated using Superose-12 chromatography. The presence of STAT3 and cyclin K in the eluted fractions was then assayed by SDS–polyacrylamide gel electrophoresis (PAGE) under reducing conditions and Western blotting. Under these conditions, his-STAT3716–770 was distributed in 2 size classes, 20 kDa and 60 kDa, whereas cyclin K was present only in the 60-kDa fraction (Figure 6B, lanes 4-5). This suggested that this fraction could contain free GST–cyclin K as well as a cyclin K–STAT3 complex with a 1:1 ratio (70 kDa). To extend these results, these gel filtration experiments were repeated using nuclear extracts of cyclin K–expressing cells that were stimulated with OSM for 15 minutes. The data in Figure 6C show that endogeneous nuclear STAT3 proteins were distributed in 2 broad size classes, 100 to 130 kDa and 160 to 190 kDa (Figure 6C, lanes 10-13). Very few monomeric forms of STAT3 proteins were observed in the 100-kDa region. Cyclin K was detected in 2 fractions, 25 kDa and 130 kDa (Figure 6C, lanes 2 and 11, respectively), suggesting again that a cyclin K–STAT3 complex with a 1:1 ratio might be present at 130 kDa. We then reasoned that this cyclin K–STAT3 complex with a 1:1 ratio should be detected only in the 110- to 140-kDa fractions, whereas a complex with a different composition should elute with a higher molecular weight. To verify this hypothesis, a series of elutions corresponding either to 110- to 140-kDa or to 180- to 220-kDa fractions were pooled and analyzed by coimmunoprecipitations using alternatively STAT3 or cyclin K polyclonal antibodies. Under these conditions, STAT3 and cyclin K were found to interact in the 110- to 140-kDa fractions but not in the 180- to 220-kDa elutions (Figure 6D, compare lane 1 with 2, 3 with 4). Although preliminary, these results suggest that cyclin K might interact with the monomeric form of STAT3 with a 1:1 ratio, and this could again explain the interactions observed either in pull-down experiments (Figure 2D) or in nonstimulated COS cells (Figure 1A, lane 4). Since STAT3 dimerization is necessary for the transcription factor to bind DNA, it will be important to determine whether the inhibitory effects of cyclin K are related to a reduced STAT3 dimer formation.
Cyclin K inhibits the induction of the p21waf1 gene by STAT3 proteins
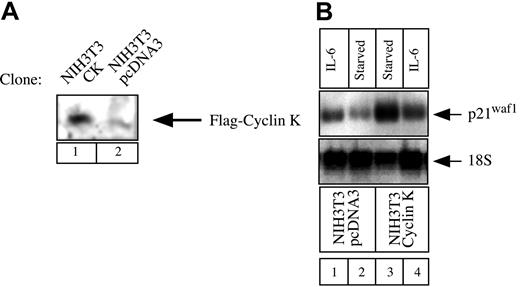
Our next aim was to determine whether cyclin K could function as a general inhibitor of IL-6–type cytokines and STAT3 activity under more physiologic conditions. STAT3 proteins can recognize a conserved response element in the promoter of the gene encoding the cell-cycle regulator p21waf1 and activate the induction of the p21waf1 mRNA.27,28 To determine the role of cyclin K on the regulation of the p21waf1 gene, we examined the regulation of the p21waf1 mRNA in NIH3T3 cells. NIH3T3 cells were chosen since we found that the p21waf1 gene was rapidly activated in this cell line by IL-6, which was not the case with OSM in A375 cells (data not shown and Coqueret et al21). To evaluate the effect of cyclin K, stable transfectants of NIH3T3 cells were generated, and overexpression of cyclin K was verified by Western Blot analysis (Figure 7A). Since cyclin K inhibited the transcriptional activity of STAT3 on luciferase reporter genes, we hypothesized that the expression of p21waf1 should be down-regulated in cyclin K–expressing clones. In accordance with previous results,27 29 Northern blot analysis revealed that IL-6 significantly induced p21waf1 mRNA in control NIH3T3 cells (Figure 7B, lanes 1-2, top panel). In contrast, this induction was not detected in cyclin K–expressing cells (Figure 7B, lanes 3-4, top panel). For an unknown reason, overexpression of cyclin K enhanced the steady-state level of p21waf1 mRNA. Equal loading was verified with control 18S ribosomal RNA staining.
Regulation of p21waf1 mRNA in cyclin K–expressing cell lines.
(A) NIH3T3 cells were stably transfected with an expression vector encoding cyclin K (lane 1) or the corresponding parental plasmid (lane 2). Following G418 selection, overexpression was verified using nuclear extracts analyzed by Western blot with the M2 Flag monoclonal antibody. (B) NIH3T3 cells were serum starved for 2 days and stimulated for 2 hours with IL-6 (10 ng/mL; lanes 1, 4) or left untreated (lanes 2-3). Total RNA was prepared and 10 μg RNA was subjected to Northern blot analysis using a mouse cDNA probe. The membrane was striped and reprobed with a 18S oligonucleotide (bottom panel).
Regulation of p21waf1 mRNA in cyclin K–expressing cell lines.
(A) NIH3T3 cells were stably transfected with an expression vector encoding cyclin K (lane 1) or the corresponding parental plasmid (lane 2). Following G418 selection, overexpression was verified using nuclear extracts analyzed by Western blot with the M2 Flag monoclonal antibody. (B) NIH3T3 cells were serum starved for 2 days and stimulated for 2 hours with IL-6 (10 ng/mL; lanes 1, 4) or left untreated (lanes 2-3). Total RNA was prepared and 10 μg RNA was subjected to Northern blot analysis using a mouse cDNA probe. The membrane was striped and reprobed with a 18S oligonucleotide (bottom panel).
Therefore, this result further confirms that cyclin K acts as an inhibitor of STAT3 activity, but also indicates that cyclin K functions under the physiologic conditions of the p21waf1 gene.
Discussion
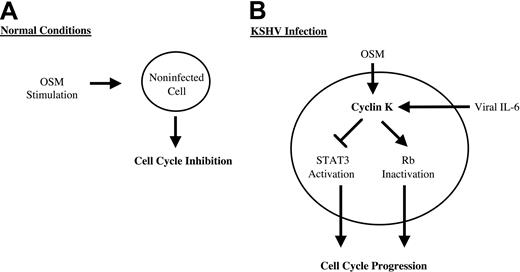
DNA tumor viruses such as KSHV have evolved a large array of strategies to deregulate the host signaling pathways and promote cell-cycle progression. Viral cyclins such as cyclin K are important to maintain infected cells in cycle since they can phosphorylate and inactivate key proteins that restrict progression into S phase. In addition to their cyclin D– and E–like kinase functions, they have distinct activities that directly act to protect from apoptosis or to stimulate the initiation of DNA replication. The results presented in this study extend these findings, further indicating that KSHV uses multiple mechanisms through the expression of cyclin K to steer cellular processes toward cellular growth. We found that cyclin K inactivated STAT3 and blocked the inhibitory effect of oncostatin M on cell proliferation. Primary cells are transformed when they are infected with KSHV particles, suggesting that the cyclin K oncogene could be implicated in the survival of primary cells in culture. Since overexpression of a dominant-negative form of STAT3 is sufficient to abrogate the inhibitory effect of OSM on cell growth,22 it is tempting to speculate that the elimination of OSM and STAT3 activities contributes to the tumorigenicity of KSHV. Interestingly, an acquired resistance to OSM is observed by KSHV-expressing cells, confirming that KSHV must produce viral proteins such as cyclin K to block OSM signaling.
A few possibilities exist regarding the molecular mechanisms by which STAT3 is inhibited by cyclin K. We showed that cyclin K could interact with and inhibit the STAT3 activation domain. Importantly, this domain was recently shown to be capable of recruiting the CBP and NcoA/SRC1a coactivators.28,30 Steric hindrance would in this case lead to transcriptional repression and block the interactions with the RNA polymerase II transcriptional machinery. Additionally, cyclin K blocked the DNA-binding activity of STAT3 while the phospho-Tyr705 state of STAT3 and its nuclear expression were not altered. Therefore, an alternative mechanism could be through physical hindrance of STAT3 DNA binding or through inhibition of dimer formation as suggested by the gel filtration experiments. It will be important to determine if the phosphorylation of STAT3 by cyclin K/cdk complexes could affect its ability to dimerize and thereby to bind DNA. Importantly, a large fraction of the 160 to 190 kDa STAT3-dimer fraction does not seem to interact with cyclin K. The reason for this is unknown, but it is tempting to speculate that the viral cyclin targets the formation or the activity of a high-molecular-weight STAT3 enhanceosome that we were not able to detect constantly under the conditions used in this study. If this hypothesis is correct, the 160 to 190 kDa STAT3 dimer would not be the primary target of the viral cyclin, probably because these proteins are not directly involved in transcriptional activation. Interestingly, the same effects have been reported with the adenovirus E1A protein.31 E1A represses IL-6 signaling through inhibition of STAT3 DNA binding, however the viral protein is also able to block the function of the STAT3 activation domain. These results further indicate that oncogenic proteins can inhibit STAT3 signaling through multiple pathways. Thus, STAT3 inactivation by E1A is probably an important step in the activation of cell transformation by the adenovirus, probably to override the antiproliferative effect of OSM.
Constitutive activation of STAT proteins has been demonstrated downstream of several oncogenes.32 An important correlation exists between activation of STAT3 and oncogenic transformation by Src.33-35 Other oncoproteins such as v-Abl, Lck, v-Fps, BCR, or v-Abl can also activate STAT transcription factors. Therefore, aberrant STAT activation contributes to the process of virus-induced cell transformation. In contrast, the SV40 large T antigen and v-Ras and v-Raf oncoproteins do not activate STAT3,36 whereas E1A and cyclin K inactivate the transcription factor. The reasons for these differences are currently under investigation; since STAT3 induces either differentiation or growth of a variety of cells, one might speculate that the oncogene effect will vary depending on the generation of growth-enhancing signals or growth arrest– or differentiation-inducing signals.37-39
We also found that cyclin K functioned as an inhibitor of STAT3 under physiologic conditions since cyclin K inhibited the induction of p21waf1 mRNA by IL-6. Surprisingly, the expression of this inhibitor was also significantly elevated in cells expressing cyclin K. Interestingly, it has been shown recently that the inactivation of STAT3 signaling induced the expression of p21waf138 in pro-B cells. Moreover, we have previously found that p21waf1 binds to the activation domain of STAT3 to inhibit its transcriptional activity.40 This could suggest that the inhibitory effect of cyclin K might be indirectly linked to an enhanced expression of p21waf1.
Altogether these results point to novel biologic functions for cyclin K to steer cellular processes toward cellular growth (Figure8). These results also suggest that the inactivation of STAT3 could be an important step in viral progression through the cell cycle. Since some of these effects are shared with its cellular homologue cyclin D1,20,41-45 we speculate that cyclin K will regulate the activity of other transcription factors previously shown to be regulated by cyclin D1, such as the estrogen receptor, v-myb, DPM1, acyclin D–binding Myb-like protein, and MyoD.42,43,45 46 This raises an interesting hypothesis that the regulation of these transcription factors could be an important step in the activation of cell transformation and cell-cycle progression by KSHV.
We thank Dr D. Mann, Dr J. F. Bromberg, and Dr J. E. Darnell for the gift of expression vectors and Dr S. Mittnacht for the gift of antibodies.
Prepublished online as Blood First Edition Paper, January 16, 2003; DOI 10.1182/blood-2002-07-1994.
Supported by a fellowship (A.L.) and a grant from the Agence Nationale de Recherches sur le SIDA and the Cancer and Solidarity Foundation; a fellowship (B.B.) from the Ministere de la Recherche et de la Technologie; and a fellowship (F.B.) from the Ligue Nationale Pour la Recherche Sur le Cancer.
A.L. and B.B. contributed equally to this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Olivier Coqueret, INSERM U564, 4 rue Larrey, CHU Angers, 49033 Angers Cedex, France; e-mail:olivier.coqueret@univ-angers.fr.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal