Intercellular adhesion molecule-4 (ICAM-4), a newly characterized adhesion molecule, is expressed early in human erythropoiesis and functions as a ligand for binding α4β1 and αVintegrin-expressing cells. Within the bone marrow, erythroblasts surround central macrophages forming erythroblastic islands. Evidence suggests that these islands are highly specialized subcompartments where cell adhesion events, in concert with cytokines, play critical roles in regulating erythropoiesis and apoptosis. Since erythroblasts express α4β1 and ICAM-4 and macrophages exhibit αV, ICAM-4 is an attractive candidate for mediating cellular interactions within erythroblastic islands. To determine whether ICAM-4 binding properties are conserved across species, we first cloned and sequenced the murine homologue. The translated amino acid sequence showed 68% overall identity with human ICAM-4. Using recombinant murine ICAM-4 extracellular domains, we discovered that hematopoietic α4β1- expressing HEL cells and nonhematopoietic αV-expressing FLY cells adhered to mouse ICAM-4. Cell adhesion studies showed that FLY and HEL cells bound to mouse and human proteins with similar avidity. These data strongly suggest conservation of integrin-binding properties across species. Importantly, we characterized a novel second splice cDNA that would be predicted to encode an ICAM-4 isoform, lacking the membrane-spanning domain. Erythroblasts express both isoforms of ICAM-4. COS-7 cells transfected with green flourescent protein constructs of prototypic or novel ICAM-4 cDNA showed different cellular localization patterns. Moreover, analysis of tissue culture medium revealed that the novel ICAM-4 cDNA encodes a secreted protein. We postulate that secretion of this newly described isoform, ICAM-4S, may modulate binding of membrane-associated ICAM-4 and could thus play a critical regulatory role in erythroblast molecular attachments.
Introduction
During terminal erythroid differentiation a diverse array of cell adhesion proteins is expressed on the erythroblast surface.1-9 These molecules mediate erythroblast interactions with both stromal cells and extracellular matrix fibronectin and laminin (reviewed in Hanspal10). Within the bone marrow microenvironment, developing erythroblasts surround a central macrophage forming substructures termed erythroblastic islands (or blood islands).11-14 Increasing evidence suggests that these islands are highly specialized bone marrow subcompartments where cell-cell adhesion events, in concert with cytokines, play critical roles in erythropoiesis and regulation of apoptosis.
Intercellular adhesion molecule-4 (ICAM-4; also known as Landsteiner Wiener [LW] blood group glycoprotein) is a newly characterized adhesion molecule expressed on early human erythroblasts concurrently with glycophorin A and Rh glycoprotein (RhGP).9 Located on chromosome 19, the ICAM-4gene contains 3 exons encoding a polypeptide that has a single membrane-spanning domain, 6 cysteine residues, and 4 potential N-glycosylation sites,15,16 2 of which are glycosylated in expressed human protein (Spring et al17; and T.J.M., unpublished observations, August 2002). Predicted membrane topology reveals 2 extracellular immunoglobulinlike domains (an N-terminal I1 set and a membrane proximal C2 set domain) that show very strong sequence homology (overall identity approximately 30%) with other members of the ICAM protein superfamily.15,18Very recently it has been shown that ICAM-4 binds α4β1 integrin on human hematopoietic cell lines and also αV-family integrins on α4β1-negative, nonhematopoietic cell lines.17 Since erythroblasts express α4β1 and macrophages exhibit αV, ICAM-4 is an attractive candidate for mediating cell-cell interactions within erythroblastic islands. There is also evidence that increased expression of ICAM-4 on sickle red blood cells may contribute to their abnormal adhesiveness.19 20 It therefore seems reasonable to postulate that this newly characterized integrin counterreceptor may function not only in cell-cell interactions during normal erythropoiesis, but it may also contribute to the pathophysiology of sickle cell disease by mediating adhesion of sickle cells to αV-expressing vascular endothelial cells.
To determine whether the integrin-binding properties of ICAM-4 are conserved across species, we cloned and sequenced the mouse gene encoding the murine homologue to human ICAM-4. The translated amino acid sequence showed 68% overall identity with human ICAM-4. Using recombinant murine ICAM-4 extracellular domains, we discovered that mouse ICAM-4 promotes adhesion of the same human hematopoietic α4β1-expressing HEL cells and the same nonhematopoietic αV-expressing FLY cells as human ICAM-4. Further, the HEL and FLY cells appear to adhere with similar avidity to mouse and human ICAM-4. These data strongly suggest conservation of the integrin-binding properties of ICAM-4 across species, thus strengthening the postulate that ICAM-4 has a key functional role during erythropoiesis. Importantly, during the course of these studies we characterized a second cDNA, which would be predicted to encode an isoform of ICAM-4 (ICAM-4S) lacking a membrane-spanning, hydrophobic sequence. COS-7 cells transfected with green flourescent protein (GFP) constructs of the prototypic ICAM-4 cDNA and the novel cDNA showed different cellular localization patterns for the 2 proteins. Moreover, analysis of tissue-culture medium revealed that the novel ICAM-4 cDNA encodes a secreted protein. We postulate that secretion of this newly described isoform may modulate binding interactions of membrane-associated ICAM-4 and could thus play a critical regulatory role in erythroblast adhesion.
Materials and methods
cDNA sequencing
Mouse expressed sequence tag (EST) clones (accession numbers AA050210, W98798, and W62001) that were closely related to human ICAM-4 were obtained from IMAGE Consortium (Lawrence Livermore National Laboratory, Livermore, CA; info@image.llnl.gov). Sequencing primers were sense and antisense vector primers, as well as sense and antisense mouse cDNA primers (5′-TCAATCTCGACGGGCTA-3′ and 5′-CAAGGGGGCCTGCAGAA-3′, respectively). DNA sequencing was performed using dye-labeled terminator chemistry on Applied Biosystems 373 or 373A automated DNA sequencers (Perkin Elmer, Foster City, CA, and Perkin Elmer, Warrington, United Kingdom, respectively).
Genomic cloning
The putative organization of the murine gene was deduced by aligning sequence of the human gene with sequences of murine EST clones. The mouse genome database (htgs) was screened with the putative ICAM-4 exon 1 sequence and a bacterial artificial chromosome (BAC) clone was identified (clone RP 23-37F10; Whitehead Institute/MIT Center for Genome Research, Cambridge, MA). Using a primer set (forward primer: 5′-TGCTCCCGTCGCTT-3′; reverse primer: 5′-GGCAGAGACTGAGGAGGAAG-3′), the BAC clone was found to contain the entire ICAM-4 gene, as determined by sequencing of polymerase chain reaction (PCR) products and by subcloning of BAC DNA. Subcloning of BAC DNA in a bluescript vector enabled sequencing of the entire murine ICAM-4 gene.
Chromosomal localization
We mapped the gene encoding mouse ICAM-4 using the Jackson Laboratory interspecific backcross [(C57BL/6JEi X SPRET/Ei) F1 X SPRET/Ei] panel.21 A 466-bp fragment was amplified using the ICAM-4 specific sequences 5′-TTCTTGGTGGTGAGCCTGAGAAGAG-3′ within exon 2 as forward primer, and 5′-CAAGTACCTGGCTGTGCAGATTAG-3′ within exon 3 as reverse primer. A BanI restriction fragment length polymorphism (RFLP) (C57BL/6JEi, 298- and 168-bp fragments; SPRET/Ei, 466-bp fragment) was used to follow the segregation of alleles in the 94 backcross progeny from the BSS panel on ethidium bromide–stained 2% agarose gels.
PCR
A mouse spleen cDNA library was screened by reverse transcription (RT)/PCR using primers binding to the 3′ end of exon 1 and the 3′ end of exon 3 of ICAM-4 (forward primer: 5′-CAGCTACTGGATGTGAGGC-3′; reverse primer: 5′-ACCAGGGTTGCGATGGAGGT-3′). mRNA isolated from proerythrobasts obtained from Friend virus–infected mice (FVA cells) was analyzed by RT/PCR for ICAM-4 and actin expression using forward primer 5′-CAGCTACTGGATGTGAGGC-3′ and reverse primer 5′-ACCAGGGTTGCGATGGAGGT-3′, and forward primer 5′-GTGACGAGGCCCAGAGCAAGAG-3′ and reverse primer 5′-GTGACGAGGCCCAGAGCAAGAG-3′, respectively. The entire coding sequence of ICAM-4 gene was amplified from BAC clone RP23-37F10 using primers binding to the 5′ end of exon 1 and the 3′ end of exon 3 (forward primer: 5′-TGCTCCCGTCGCTT-3′; reverse primer: 5′-GGCAGAGACTGAGGAGGAAG-3′).
Transfection and cell culture
ICAM-4 and ICAM-4S were cloned into a pEGFP-C3 vector downstream of GFP using BglII and SacII cloning sites. For transfection, COS-7 cells were grown in Dulbecco modified Eagle medium (DMEM; Invitrogen, Grand Island, NY) plus 10% fetal bovine serum, 1% penicillin-streptomycin for 24 hours, then seeded at 200 000/well onto coverslips. Plasmids were transiently transfected into COS-7 cells as described22 with minor modifications. In brief, 1.5 μg of plasmid cDNA was mixed with 5 μL LipofectAMINE2000 and 200 μL of Opti-MEM I (Invitrogen) at RT for 20 minutes and then added to the cells. Cells were cultured in DMEM plus 10% fetal bovine serum for 24 to 36 hours. Cells counts were obtained using a hemocytometer and viability determined by trypan blue staining.
Western blotting
At 24 hours after transfection, culture supernatants were collected and COS-7 cells were directly lysed in the plates with 200 μL of sodium dodecyl sulfate (SDS)–sample buffer after 3 washes with phosphate-buffered saline (PBS). Culture supernatants were centrifuged at 1500g for 10 minutes and transferred to new tubes. Equal volumes of 20% trichloroacetic acid (TCA) were added to the supernatants and after precipitation on ice for 30 minutes they were spun down in a microfuge at 4°C for 15 minutes. The resulting pellets were washed with 300 μL of cold acetone for 5 minutes at 4°C, dried, resuspended in 50μL of SDS-sample buffer, and boiled for 3 minutes. SDS–polyacrylamide gel electrophoresis (PAGE) of samples was performed on 7% acrylamide gels. The proteins were transferred onto nitrocellulose membrane using a semidry electroblotter (Integrated Separation Systems, Natick, MA). After blocking for one hour in PBS containing 5% nonfat dry milk, blots were washed in PBS, 0.1% Tween-20 then probed for one hour with mouse anti-GFP antibody (Roche, Indianapolis, IN) in PBS, 0.1% Tween-20. After several washes, blots were incubated with anti–mouse immunoglobulin G (IgG) coupled to horseradish peroxidase (Amersham, Arlington Heights, IL) diluted at 1/50 000, washed, and developed using the Renaissance chemiluminescence detection kit (NEN Life Science Products, Boston, MA).
Immunofluorescence microscopy
Transfected cells were washed 3 times in PBS, fixed with 4% paraformaldehyde in PBS at RT for 30 minutes, and rinsed in PBS. GFP-expressing cells were visualized at 480 nm excitation and 508 nm emission using a Zeiss Axiovert 135 microscope with a 63 × 1.25 oil immersion objective (Carl Zeiss MicroImaging, Thornwood, NY).
Mouse erythroblast RNA
Erythroblasts were obtained from mice infected with an anemia-inducing strain of Friend erythroleukemia virus as previously described.23 24 Cells were harvested from spleens of infected mice, separated by velocity sedimentation at unit gravity, and cells sedimenting at 6 mm/h or greater collected and cultured with 2 U/mL recombinant erythropoietin. Cultured cells were taken at various time points for RNA extraction. Cells at the 0 hour of culture were mainly proerythroblasts, which then differentiate over approximately 48 hours into late-stage erythroblasts and enucleated reticulocytes. Total RNA was isolated from cell pellets using RNeasy columns (Qiagen, Valencia, CA), and analyzed by RT/PCR.
Cells and reagents for adhesion assays
The majority of cell lines used in the study were obtained from the European Culture Collection (Wiltshire, United Kingdom) and were maintained in Iscoves modified Eagle medium (IMEM)/10% fetal bovine serum. The fibroblast line, FLYRD18 (FLY), a subclone of the HT1080 cell line, was a gift from Dr C. Porter, Hammersmith Hospital (London, United Kingdom). All reagents used were from Sigma (Dorset, United Kingdom) unless otherwise specified.
Function-blocking monoclonal antibodies to integrin subunits were anti-β1 clone13 (a gift from Dr K. Yamada, National Institutes of Health, Bethesda, MD); anti-β3clone PM6/13 (Harlan Sera-Lab, Loughborough, United Kingdom); anti-α2 clone JA218 (a gift from Prof M. Humphries, University of Manchester, United Kingdom); anti-α4 clones HP2/1 (Serotec, Oxford, United Kingdom) and Max68P (a gift from Dr T. Shock, Celltech plc, United Kingdom); anti-α5 clone SNAKA55 (a gift from Prof M. Humphries); anti-α6 clone NKI-GoH3 (Serotec); anti-αV clones 69.9.5 (Immunotech, High Wycombe, United Kingdom) and CLB-706 (Chemicon International, Harrow, United Kingdom); and anti-αVβ5clone P1F6 (Chemicon International). Mouse IgG1 and rat IgG control antibodies were from Sigma. Antihuman ICAM-4, clone BS56, was a gift from Dr H. Sonneborn (Biotest, Dreieich, Germany).
CAM Fc chimeric fusion (CAMFc) proteins
The CAMFc fusion proteins used in the study comprised the 2 extracellular domains of ICAM-4 or neural cell adhesion molecule (NCAM) and the hinge region and Fc domains of human IgG1.25 Human ICAM-4–Fc fusion protein (hICAM4Fc) cDNA was produced as described.17 A cDNA clone encoding the extracellular domains of NCAM in pIg was a gift from Dr D. Simmons (SmithKline Beecham, Harlow, Essex, United Kingdom). Murine ICAM-4–Fc fusion protein (mICAM4Fc) cDNA encoding the predicted leader sequence and the 2 predicted, extracellular IgSF domains (mICAM-4 amino acid residues 1 to 225) was amplified by PCR using sense primer (TTCCCAAGCTTTGTGCCATGGAGTCTGCCC), antisense primer (GTTTATGATCAACTTACCTGTTGCCTCACCGAGGACTGTCAACAT), and mICAM-4 cDNA template. The PCR product was digested with HindIII +BclI and ligated into HindIII +BamHI-cut pIg vector as described.25 Clones were verified by sequence analysis. CAMFc proteins were expressed in COS-7 cells as described25 and extracted from culture supernatant on protein A–Sepharose. Protein concentrations were determined by enzyme-linked immunosorbent assay. Antibodies used were goat anti–human IgG as the capture reagent, and peroxidase-conjugated goat F(ab)′2 anti–human IgG as the reveal reagent (both reagents are Fc-specific and absorbed for cross-reactivity to bovine IgG; Jackson ImmunoResearch Laboratories, West Grove, PA). AEVZ5.1, a human monoclonal IgG1 (International Blood Group Reference Laboratory, Bristol, United Kingdom), was used to obtain a standard curve (its protein concentration was previously determined by optical density280[OD280]).
Adhesion assays
Immulon-4 96-well plates (Dynex Technologies, Billingshurst, United Kingdom) were coated with 1 μg/well goat antihuman-Fc Ig (Sigma) overnight at 4°C. They were then washed 3 times with PBS and incubated overnight at 4°C with chimeric proteins at specified concentrations in PBS. The following day they were washed 3 times with PBS, then blocked with PBS containing 0.4% bovine serum albumin (BSA; Fraction V, Sigma) for 2 hours at RT. HEL cells were washed once in assay buffer (IMEM, 2mM EGTA (ethyleneglycoltetraacetic acid), 10 μg/mL human IgG [BPL, Elstree, Herts, United Kingdom]). FLY cells were lifted in PBS containing 2 mM EDTA (ethylenediaminetetraacetic acid) and 0.1% (wt/vol) BSA and washed once in IMEM containing 0.1% (wt/vol) BSA. Cells were resuspended at 107/mL in assay buffer and labeled with 10 μg/mL 2′, 7′-bis(2-carboxyethyl)–56-carboxyfluorescein acetoxymethyl ester (Sigma) for 15 minutes at 37°C. HEL cells were then activated by washing 3 times in assay buffer containing 2 mM Mn2+. After fluorescent labeling, FLY cells were washed 3 times in assay buffer then activated by incubation for 15 minutes at 37°C in assay buffer containing 80 μM phorbol ester (PMA, Sigma), followed by 2 washes in assay buffer containing 2 mM Mn2+. In experiments with functionally active antibodies the cells were then preincubated on ice for 15 minutes in assay buffer containing 2 mM Mn2+ and 10 μg/mL antibodies. The activated cells, with or without antibodies as appropriate, were then added to the CAMFc-coated plates to give 5 × 104/well in 100 μL and incubated for 30 minutes at 37°C. The plates were read on a fluorescence microplate reader (excitation 485 nm, emission 530 nm, Bio-Tek Instruments, Winooski, VT) prior to a series of standardized washes in assay buffer, and were read after each wash. The percentage of the input cells bound was calculated. Each data point represents the mean of 3 or more replicates. All assays were performed on at least 3 independent occasions, and representative experiments are shown.
Results
Cloning and characterization of mouse ICAM-4
Three mouse EST clones more closely related to human ICAM-4 than to any other member of the human immunoglobulin superfamily were identified by scanning the GenBank database with human ICAM-4 protein sequence. The 3 clones contained overlapping DNA sequences. ClonesAA050210 and W98798 were sequenced and comprised the protein coding region and the 3′ noncoding region of murine ICAM-4. CloneW62001 was partly sequenced to determine the 5′ untranslated region. The putative organization of the murine gene was deduced by aligning sequences of the human gene with sequences of murine EST clones. Surprisingly, murine EST clones contained intron 2 sequence in addition to exons 1, 2, and 3 (GenBank accession number AF296283). To determine whether ICAM-4 mRNA lacking intron 2 was also present, we screened a mouse spleen cDNA library by PCR using primers binding to the 3′ end of exon 1 and the 3′ end of exon 3. Two products of 594 bp and 456 bp were amplified (Figure 1). Sequencing revealed that the larger one contained intron 2 while the smaller one lacked intron 2 (GenBank accession number AF296282). We therefore concluded that there are 2 ICAM-4 mRNA species in mouse. Since the 139-bp intron 2 has a stop codon at 111, we predicted that exon 3 sequence would not be translated from mRNA containing intron 2.
PCR analysis of ICAM-4 in spleen cDNA library and BAC and EST clones.
Mouse spleen cDNA library and EST clones W98798 and AA050210 were screened using primers binding to the 3′ end of exon 1 and the 3′ end of exon 3. The coding sequence of murine ICAM-4 gene was amplified from BAC clone RP23-37F10 using primers binding to the 3′ end of exon 1 and the 3′ end of exon 3.
PCR analysis of ICAM-4 in spleen cDNA library and BAC and EST clones.
Mouse spleen cDNA library and EST clones W98798 and AA050210 were screened using primers binding to the 3′ end of exon 1 and the 3′ end of exon 3. The coding sequence of murine ICAM-4 gene was amplified from BAC clone RP23-37F10 using primers binding to the 3′ end of exon 1 and the 3′ end of exon 3.
To clone the murine ICAM-4 gene, the high-throughput sequence database of National Center for Biotechnology Information (NCBI) was screened with the putative ICAM-4 exon 1 sequence and a BAC clone (no. RP23-37F10) was identified. To verify that clone RP23-37F10 contained the coding region ofICAM-4, we amplified the entire coding sequence using primers binding to the 5′ end of exon 1 and the 3′ end of exon 3. Sequencing of PCR products confirmed that the BAC clone included the entire ICAM-4 gene (GenBank accession number AF296282). Murine ICAM-4 was 1.1 kb and contained 3 exons, ranging in size from 113 to 370 bp (Figure2). Characteristic 5′ splice donor gt and 3′ splice acceptor ag motifs were present in all of the intron/exon boundary sequences (Table 1). The 3 exonic sequences derived from our genomic sequence were completely homologous with the sequence of ICAM-4 cDNA derived from EST clones. Binding motifs for erythroid transcription factors GATA 1 and SP 1 were identified 73 nucelotide (nt) and 38 nt upstream of the initiation codon, respectively. Organization and size of the murine gene was very similar to that of its human counterpart, which also contains 3 exons distributed over 2.65 kb.16
mRNA splice variants of mouse ICAM-4gene.
Depiction of the 2 mRNA species of ICAM-4. Vertical bars represent the 3 exons with the size of each exon shown above the bar. Sizes of the first and third exons indicate the number of bases in coding sequence only. Intron 1, containing 113 bases, is spliced out in both isoforms, while intron 2 is excluded in ICAM-4 and included in ICAM-4S mRNA.
mRNA splice variants of mouse ICAM-4gene.
Depiction of the 2 mRNA species of ICAM-4. Vertical bars represent the 3 exons with the size of each exon shown above the bar. Sizes of the first and third exons indicate the number of bases in coding sequence only. Intron 1, containing 113 bases, is spliced out in both isoforms, while intron 2 is excluded in ICAM-4 and included in ICAM-4S mRNA.
Organization of the mouse ICAM-4 gene
| Exon . | Nucleotide number* . | Splice acceptor site . | Splice donor site . |
|---|---|---|---|
| 1 | 1-370 | NA | TTACA/gtgagggagaccggg |
| 2 | 484-789 | atctctcttgccttag/AACGG | CCTCG/gtgaggcatcctgta |
| 3 | 929-1042 | tcactcctgcccacag/CTTTA | NA |
| Exon . | Nucleotide number* . | Splice acceptor site . | Splice donor site . |
|---|---|---|---|
| 1 | 1-370 | NA | TTACA/gtgagggagaccggg |
| 2 | 484-789 | atctctcttgccttag/AACGG | CCTCG/gtgaggcatcctgta |
| 3 | 929-1042 | tcactcctgcccacag/CTTTA | NA |
NA indicates not applicable.
+1 taken as the first nucleotide of the initiation codon in the cDNA sequence.
Chromosomal localization of mouse ICAM-4
ICAM-4 was nonrecombinant with D9Hun3, placing the gene 8 centimorgan (cM) distal to the centromere on mouse chromosome 9, a region that shows extensive conserved synteny with human 19p13.3, where the human ICAM-4 gene is located.26 Notably, ICAM-1 andICAM-5 map to the same region of mouse chromosome 9.26 Our data have been added to Mouse Genome Database under accession number J:78208 and can be accessed through the website (http://www.jax.org). No obvious potential candidate mouse mutations map to the region containing ICAM-4 on chromosome 9.26
Amino acid sequence and functional comparisons of mouse and human ICAM-4
When the translated amino acid sequence of ICAM-4 cDNA from exons 1 to 3 was aligned with the sequence of human ICAM-4, using the “BLAST 2 sequences” program (NCBI Entrez), it revealed 68% identity overall. Of note, critical cysteine residues as well as other key residues within each strand of the predicted extracellular IgSF domains are conserved, strongly suggesting that the disulfide-bonded IgSF domains will be similarly folded in murine and human polypeptides. Moreover, the unusual “LRT” motif in domain 1, which replaces the so-called “LETSL” integrin-binding motif that is common to other ICAM family members, is conserved. In addition, 2 of 4 potential glycosylation sites (N59CS and N181VT) are conserved, and there is strong evidence that these sites are glycosylated in the expressed, human protein (Spring et al17; and T.J.M., unpublished observations, August 2002). These data suggest that ICAM-4 counterreceptors in mice and humans may be similar.
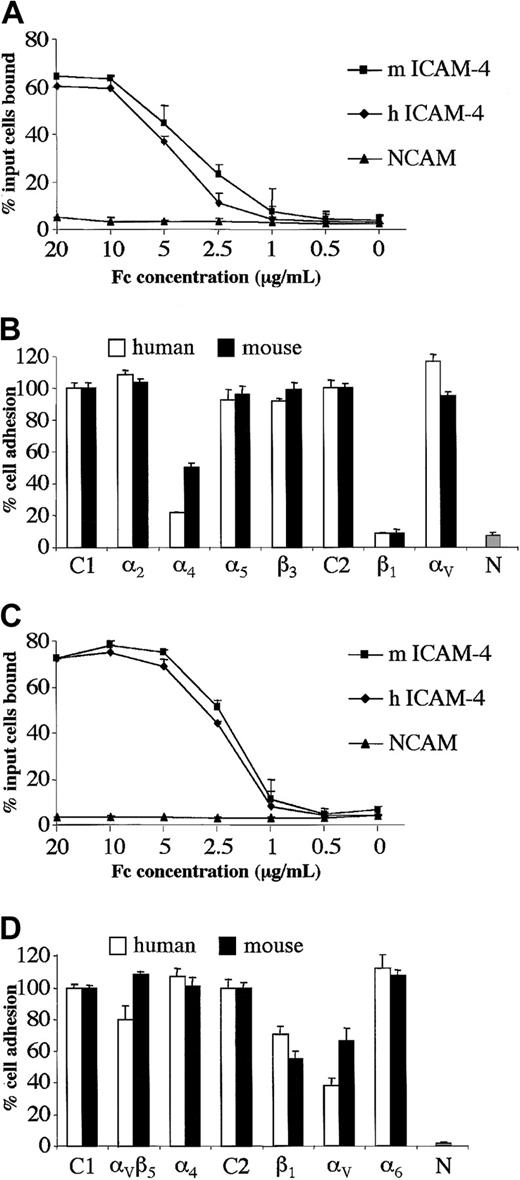
To begin to determine whether ICAM-4 ligand-binding properties are conserved across species, we expressed the predicted extracellular IgSF domains of mouse ICAM-4 as an Fc fusion protein and performed cell-based adhesion assays. Binding of hematopoietic α4β1-expressing HEL cells to mouse ICAM-4 was tested, since this cell line had been utilized to characterize human ICAM-4 binding interactions.17 HEL cells adhered to mouse ICAM-4–Fc. Further, dilution analysis experiments strongly suggest that HEL cells adhere to mouse or human ICAM-4–Fc proteins with near-identical avidity (Figure 3A). Since we have previously shown that HEL cell binding to human ICAM-4 is mediated by α4β1 integrin,17we tested whether α4β1 is also the counterreceptor on HEL cells for murine ICAM-4. Blocking β1 antibody abrogated adhesion of HEL cells to both human and murine ICAM-4, while blocking α4 antibodies inhibited binding to murine ICAM-4 and human ICAM-4 50% and 80%, respectively (Figure 3B). HEL cells also express integrin subunits β3, α2, α5, and αV.17 However, blocking antibodies to these 4 integrin subunits had no effect on binding of HEL cells to either human or murine ICAM-4 (Figure 3B).
Human hematopoietic and nonhematopoietic cell lines adhere to mouse or human ICAM-4 with similar avidity.
Adhesion to mouse and human ICAM-4 is mediated by α4β1 integrin on hematopoietic cells and by αV integrins on nonhematopoietic cells. (A) Adhesion of Mn2+-activated HEL cells to mouse (m) or human (h) ICAM-4–Fc or NCAM-Fc control protein. The results are the percentage of total input cells bound ±SD (n = 3). Dilution analysis was performed and the concentrations of protein applied to microplate wells are indicated as abscissa. (B) Adhesion of Mn2+-activated HEL cells to mouse or human ICAM-4–Fc or NCAM-Fc control protein in the presence of function-blocking antibodies against integrin α or β subunits. The fusion proteins were applied at a concentration of 10 μg/mL. Results are shown as the percentage of total input cells bound relative to binding to control antibody ±SD (n = 6). Adhesion in the presence of mouse IgG1 (C1) or rat IgG (C2) control antibodies is shown as 100% (actual values obtained were in the range of 42%-50% or 55%-60% adhesion of input cells to human or mouse ICAM-4–Fc, respectively). Percent adhesion in the presence of mouse antibodies against α2, α4, α5 or β3, or rat antibodies against β1 or αV subunits is shown. N indicates percent adhesion to NCAM-Fc control protein in the absence of antibody. (C) Adhesion of PMA+Mn2+-activated FLY cells to mouse (m) or human (h) ICAM-4–Fc or NCAM-Fc control protein. The results are the percentage of total input cells bound ±SD (n = 3). Concentrations of protein applied to microplate wells are indicated as abscissa. (D) Adhesion of PMA+Mn2+-activated FLY cells to mouse or human ICAM-4–Fc or NCAM-Fc control protein in the presence of function-blocking antibodies against integrin α or β subunits. Fusion proteins were applied at a concentration of 10 μg/mL. Results are shown as the percentage of total input cells bound relative to binding to control antibody ±SD (n = 6). Adhesion in the presence of mouse IgG1 (C1) or rat IgG (C2) control antibodies is shown as 100% (actual values obtained were in the range of 65%-75% or 76%-80% adhesion of input cells to human or mouse ICAM-4–Fc, respectively). Percent adhesion in the presence of mouse antibodies against αVβ5 integrin complex, α4subunit or rat antibodies against β1, αV, or α6 subunits is shown. N indicates percent adhesion to NCAM-Fc control protein in the absence of antibody.
Human hematopoietic and nonhematopoietic cell lines adhere to mouse or human ICAM-4 with similar avidity.
Adhesion to mouse and human ICAM-4 is mediated by α4β1 integrin on hematopoietic cells and by αV integrins on nonhematopoietic cells. (A) Adhesion of Mn2+-activated HEL cells to mouse (m) or human (h) ICAM-4–Fc or NCAM-Fc control protein. The results are the percentage of total input cells bound ±SD (n = 3). Dilution analysis was performed and the concentrations of protein applied to microplate wells are indicated as abscissa. (B) Adhesion of Mn2+-activated HEL cells to mouse or human ICAM-4–Fc or NCAM-Fc control protein in the presence of function-blocking antibodies against integrin α or β subunits. The fusion proteins were applied at a concentration of 10 μg/mL. Results are shown as the percentage of total input cells bound relative to binding to control antibody ±SD (n = 6). Adhesion in the presence of mouse IgG1 (C1) or rat IgG (C2) control antibodies is shown as 100% (actual values obtained were in the range of 42%-50% or 55%-60% adhesion of input cells to human or mouse ICAM-4–Fc, respectively). Percent adhesion in the presence of mouse antibodies against α2, α4, α5 or β3, or rat antibodies against β1 or αV subunits is shown. N indicates percent adhesion to NCAM-Fc control protein in the absence of antibody. (C) Adhesion of PMA+Mn2+-activated FLY cells to mouse (m) or human (h) ICAM-4–Fc or NCAM-Fc control protein. The results are the percentage of total input cells bound ±SD (n = 3). Concentrations of protein applied to microplate wells are indicated as abscissa. (D) Adhesion of PMA+Mn2+-activated FLY cells to mouse or human ICAM-4–Fc or NCAM-Fc control protein in the presence of function-blocking antibodies against integrin α or β subunits. Fusion proteins were applied at a concentration of 10 μg/mL. Results are shown as the percentage of total input cells bound relative to binding to control antibody ±SD (n = 6). Adhesion in the presence of mouse IgG1 (C1) or rat IgG (C2) control antibodies is shown as 100% (actual values obtained were in the range of 65%-75% or 76%-80% adhesion of input cells to human or mouse ICAM-4–Fc, respectively). Percent adhesion in the presence of mouse antibodies against αVβ5 integrin complex, α4subunit or rat antibodies against β1, αV, or α6 subunits is shown. N indicates percent adhesion to NCAM-Fc control protein in the absence of antibody.
Human ICAM-4 is unusual in that it has a second counterreceptor, αV, that is widely expressed on nonhematopoietic cells.17 Because erythroblasts interact with macrophages within the bone marrow microenvironment and macrophages exhibit αV integrins on their surface, we were interested in testing adherence of αV-expressing cells to mouse ICAM-4. We discovered that nonhematopoietic αV-expressing FLY cells adhered to mouse ICAM-4–Fc. Moreover, dilution analysis suggested that FLY cells adhered to mouse or human proteins with very similar avidity (Figure 3C). Binding to both human and murine ICAM-4 was partially inhibited by blocking αV antibodies (Figure3D). FLY cells do not express α4β1 but do express several other integrins of the β1 and β3 families.17 Blocking antibodies to the α subunits of these integrins did not inhibit binding of FLY cells to murine ICAM-4 (Figure 3D and data not shown), consistent with data presented for human ICAM-4.17 Our findings strongly imply conservation of the integrin-binding properties of ICAM-4 across species, thus strengthening the postulate that ICAM-4 could play a key functional role during erythropoiesis.
Amino acid sequence comparisons of mouse ICAM-4 isoforms
When the translated amino acid sequences of the 2 mouse ICAM-4 mRNAs (one with and one without intron 2) were analyzed we found that both isoforms are 261 amino acids in length (Figure4). Amino acid residues 1 to 224, which constitute the extracellular domain, are identical. The polypeptide translated from cDNA containing intron 2 terminates at a stop codon TAG (nt 787-789) within intron 2, and therefore does not contain the transmembrane and cytoplasmic domain sequences encoded by exon 3. Interestingly, however, in the polypeptide translated from cDNA containing intron 2, much of the hydrophobic sequence in the putative transmembrane domain of the prototypical ICAM-4 is replaced by hydrophilic residues encoded by intron 2. We, therefore, speculated that this ICAM-4 isoform might be secreted, and will henceforth refer to it as ICAM-4S.
Alignment of ICAM-4 and ICAM-4S amino acid sequences.
Hydrophobic residues in ICAM-4 are starred; hydrophilic residues in ICAM-4S are underlined. The junction between the extracellular and transmembrane domains of ICAM-4 is amino acid residue 251.
Alignment of ICAM-4 and ICAM-4S amino acid sequences.
Hydrophobic residues in ICAM-4 are starred; hydrophilic residues in ICAM-4S are underlined. The junction between the extracellular and transmembrane domains of ICAM-4 is amino acid residue 251.
Erythroblasts express 2 isoforms of ICAM-4
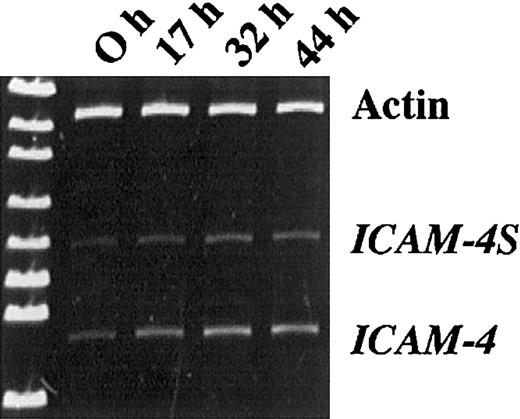
To determine whether ICAM-4S mRNA is expressed in differentiating erythroblasts, we analyzed mRNA isolated from proerythrobasts obtained from Friend virus–infected mice (FVA cells). This carefully characterized model system of terminal erythroid differentiation closely mimics in vivo erythropoiesis.23 27-32 Using primers binding to exon 1 and exon 3, products of 594 bp and 456 bp were amplified by PCR (Figure 5). These products correspond to the predicted sizes of the 2 ICAM-4 isoforms, thereby demonstrating that erythroblasts do, indeed, express both isoforms. To explore whether a developmentally regulated change in isoform expression occurs during terminal differentiation, we compared RT/PCR products amplified from mRNA of FVA cells cultured for 0, 17, 32, and 44 hours. Cells were 98% proerythroblasts at the 0 hour initiation of culture and well hemoglobinized at 44 hours, with many enucleating forms. Actin, ICAM-4, and ICAM-4S were amplified at each time point so that densitometry measurements of ICAM-4 and ICAM-4S could be normalized to actin. The band corresponding to ICAM-4 increased in intensity between the 0- and 32-hour time points and subsequently decreased approximately 6% to 10%. In contrast, ICAM-4S continued to increase over 44 hours, strongly suggesting that ICAM-4S expression is continuously up-regulated late in erythroid differentiation.
Erythroblasts express 2 isoforms of ICAM-4.
RT/PCR analysis of mRNA isolated from erythroblasts obtained from Friend virus–infected mice and further differentiated in culture. Using primers binding to exon 1 and exon 3, products of 594 bp and 456 bp were amplified. At the 0 hour initiation of culture, cells were 98% proerythroblasts. After 44 hours in culture, cells were well hemoglobinized. Actin was amplified at each time point so that densitometry measurements of ICAM-4 and ICAM-4S could be normalized to actin. Normalization ratios for ICAM-4/actin were 0.189, 0.326, 0.513, and 0.484 for 0, 17, 32, and 44 hours, respectively. Normalization ratios for ICAM-4S/actin were 0.103, 0.133, 0.231, and 0.247 for 0, 17, 32, and 44 hours, respectively.
Erythroblasts express 2 isoforms of ICAM-4.
RT/PCR analysis of mRNA isolated from erythroblasts obtained from Friend virus–infected mice and further differentiated in culture. Using primers binding to exon 1 and exon 3, products of 594 bp and 456 bp were amplified. At the 0 hour initiation of culture, cells were 98% proerythroblasts. After 44 hours in culture, cells were well hemoglobinized. Actin was amplified at each time point so that densitometry measurements of ICAM-4 and ICAM-4S could be normalized to actin. Normalization ratios for ICAM-4/actin were 0.189, 0.326, 0.513, and 0.484 for 0, 17, 32, and 44 hours, respectively. Normalization ratios for ICAM-4S/actin were 0.103, 0.133, 0.231, and 0.247 for 0, 17, 32, and 44 hours, respectively.
ICAM-4S is secreted
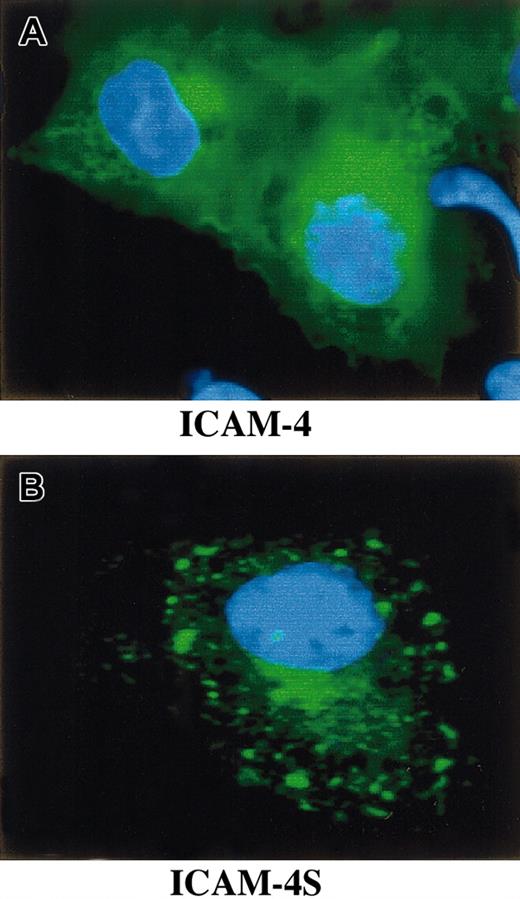
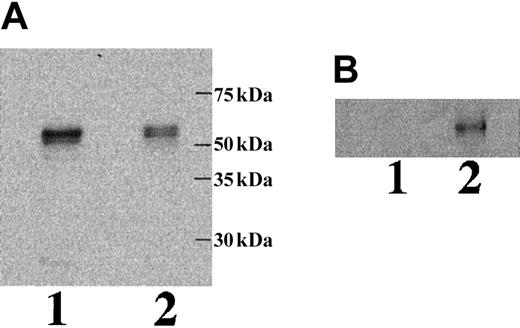
To study whether mRNA containing intron 2 is effectively translated and, if so, whether the translated polypeptide is a secreted isoform of ICAM-4, we performed a series of transfection experiments. COS-7 cells were transfected with a GFP ICAM-4 fusion construct, a GFP ICAM-4S fusion construct, or vector alone. Immunofluorescence microscopy revealed that cells expressing ICAM-4 had diffuse cytoplasmic and plasma membrane staining (Figure6). In contrast, cells expressing ICAM-4S had bright focal areas of staining distributed throughout the cytoplasm but no plasma membrane staining, indicating that ICAM-4S was not stably assembled on the plasma membrane. Western blot analysis of whole cell lysates probed with anti-GFP showed a band of approximately 58 kDa in both ICAM-4– and ICAM-4S–transfected samples (Figure7A). We next asked whether transfected COS-7 cells secrete ICAM-4S. To address this question, cell culture supernatants were collected and their proteins TCA-precipitated and analyzed by Western blotting. To ensure that the source of ICAM-4S in the supernatant was not disrupted nonviable cells, percent cell viability was determined by trypan blue exclusion. No differences in cell viability were noted between ICAM-4 and ICAM-4S cultures. In both cultures, 90% of cells were viable. To control for variation in amounts of GFP fusion protein expressed per culture, supernatant gel loads were normalized to the amount of GFP fusion protein in a particular culture. This was accomplished by determining the amounts of GFP fusion proteins by densitometry of Western blots of equivalent numbers of ICAM-4– and ICAM-4S–transfected cell lysates probed with anti-GFP. Having established these parameters, we detected a band in supernatant from cells transfected with the ICAM-4S construct that was not present in supernatant from ICAM-4–expressing cells (Figure 7B). We conclude from these data that mRNA containing intron 2 is effectively translated and that the resulting polypeptide is a secreted isoform of ICAM-4.
Expression of GFP ICAM-4 and ICAM-4S fusion constructs in transfected COS-7 cells.
COS-7 cells were transfected with a GFP ICAM-4 or a GFP ICAM-4S fusion construct, fixed with 4% paraformaldehyde, and visualized using a Zeiss Axiovert 135 microscope with a 63 × 1.25 oil immersion objective. ICAM-4 (green) localized to the plasma membrane and cytoplasm (A). In contrast, ICAM-4S (green) was predominantly present in bright foci within the cytoplasm (B). Blue is DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride) stain of nucleus.
Expression of GFP ICAM-4 and ICAM-4S fusion constructs in transfected COS-7 cells.
COS-7 cells were transfected with a GFP ICAM-4 or a GFP ICAM-4S fusion construct, fixed with 4% paraformaldehyde, and visualized using a Zeiss Axiovert 135 microscope with a 63 × 1.25 oil immersion objective. ICAM-4 (green) localized to the plasma membrane and cytoplasm (A). In contrast, ICAM-4S (green) was predominantly present in bright foci within the cytoplasm (B). Blue is DAPI (4′,6-diamidine-2′-phenylindole dihydrochloride) stain of nucleus.
Western blot analysis of ICAM-4 in transfected COS-7 cells and culture supernatant.
COS-7 cells were transfected with a GFP ICAM-4 or a GFP ICAM-4S fusion construct. Whole cell lysates (A) and TCA-precipitated proteins from tissue culture supernatants (B) were analyzed by immunoblots of 7% acrylamide gels probed with anti-GFP. ICAM-4 (lane 1); ICAM-4S (lane 2).
Western blot analysis of ICAM-4 in transfected COS-7 cells and culture supernatant.
COS-7 cells were transfected with a GFP ICAM-4 or a GFP ICAM-4S fusion construct. Whole cell lysates (A) and TCA-precipitated proteins from tissue culture supernatants (B) were analyzed by immunoblots of 7% acrylamide gels probed with anti-GFP. ICAM-4 (lane 1); ICAM-4S (lane 2).
Discussion
A major finding of the current study is that there is a secreted isoform of ICAM-4. In several EST clones and a mouse spleen cDNA library we discovered a distinct mRNA species containing intron 2 sequence. This mRNA would be predicted to encode an ICAM-4 isoform lacking a hydrophobic transmembrane spanning domain. Consistent with this prediction, immunofluorescence microscopy of COS-7 cells transfected with GFP constructs of ICAM-4 cDNAs with or without intron 2 sequence showed that the fusion protein translated from cDNA lacking intron 2 localized to plasma membranes of transfected cells, while the isoform expressed from the construct containing intron 2 was detected as discrete cytoplasmic foci. Moreover, the isoform expressed from the construct containing intron 2 was secreted into the tissue-culture medium of transfected cells. On this basis we have termed the secreted form of ICAM-4, ICAM-4S. Interestingly, Bailly et al isolated a similar clone from a human bone marrow cDNA library, although they have not reported whether the encoded human protein is secreted.15 We postulate that secretion of ICAM-4S may modulate binding interactions of ICAM-4 and could thus play a critical regulatory role in erythroblast adhesion.
Our observations of the developmental time frame of ICAM-4S mRNA expression suggest that this isoform's regulatory role occurs late in erythropoiesis. We found an up-regulation of ICAM-4S mRNA late in the terminal differentiation of mouse erythroblasts. We postulate that secreted ICAM-4S competes with cellular ICAM-4 for specific binding sites, resulting in decreased adhesive interactions between membrane ICAM-4 and its binding partners. This molecular mechanism, in conjunction with down-regulation of α4β1 in late erythroblasts, could function in releasing erythroblasts from their anchorage to the erythroblastic island. Precedent for a soluble protein repressing the function of its transmembrane form exists, for example the soluble extracellular region of the ligand for Notch receptors.33
Our current data indicate striking similarities between mouse and human ICAM-4. The murine ICAM-4 gene contains 3 exons distributed over 1.1 kb. Hence the organization and size of the murine gene closely resembles its 2.65 kb human counterpart, which also contains 3 exons.15,16 Alignment of human- and murine-translated amino acid sequences reveals 68% identity overall. There is noteworthy conservation of critical cysteine residues as well as other key residues within each strand of the predicted extracellular IgSF domains, supporting similar folding of the disulfide-bonded IgSF domains in human and murine polypeptides. Finally, murine ICAM-4 is located on chromosome 9 in a region with highly conserved synteny with human 19p13.3, where the human ICAM-4 gene is located.26These observed characteristics of mouse ICAM-4 gene and protein structure suggest that its functional properties may be analogous to human ICAM-4.
Indeed, our findings indicate conservation of integrin-binding properties of ICAM-4 across species. Specifically, we observed that mouse ICAM-4–Fc promoted adhesion of HEL and FLY cells comparable to human ICAM-4–Fc. Further, data obtained with blocking antibodies indicate that α4β1 and αV are counterreceptors for both human and mouse ICAM-4, on HEL and FLY cells, respectively. We noted that the effect of blocking antibodies was similar with mouse and human protein but not in each instance identical. Although antibody to α4 inhibited HEL cell adhesion to both mouse and human ICAM-4, the blocking was somewhat less pronounced with mouse protein. Likewise, there were differences in the degrees of inhibition of FLY cell binding observed with antibodies to β1 and αV. These results may reflect the fact that there is 68% identity between the 2 ICAMs, but much higher identity between the respective integrins: −88% for β1, 83% for α4, 89% for αV, and 88% for β5. Incomplete inhibition may also result from the antibody specificities since they have all been defined by inhibition with other more well-characterized ligands. Although the binding sites for different ligands may be overlapping on integrins, they may have distinct properties. Thus certain antibodies may inhibit binding of one ligand and not of another, or inhibit one interaction more effectively than another. Thus partial or no inhibition using certain integrin antibodies may not be a surprising finding. It is also possible that the observed incomplete blockade of cell binding by antibodies indicates an interaction of ICAM-4 with a second, as yet unidentified, receptor on HEL and FLY cells.
Prior studies document the presence of ICAM-4 on the surface of early human erythroblasts.9 Our current data showing ICAM-4 mRNA expression in FVA proerythroblasts strongly suggest that ICAM-4 undergoes similar developmental regulation in mice and humans. The timing of ICAM-4 expression on the surface of erythroblasts makes it an attractive candidate for mediating cell-cell adhesion within erythroblastic islands. Although increasing evidence suggests that developmentally important interactions occur between cells in this marrow subcompartment, only a few cell-binding partners have been identified. A heparin-dependent binding protein, Emp (erythroblast macrophage protein), expressed on both erythroblasts and central macrophages, appears to mediate erythroblast-erythroblast and erythroblast-macrophage associations via homophilic binding.13,34 Disruption of this specific interaction markedly inhibits erythroid terminal maturation and enucleation13 and promotes apoptosis.34 A second adhesive interaction between components of the erythroblastic island involves α4β1 integrin expressed on erythroblasts, and its counterreceptor, VCAM-1, expressed by central macrophages.35 Antibodies to either VCAM-1 or α4β1 interfere with binding of erythroblasts to central macrophages and disrupt the island structure.35 In addition, it was recently reported that Fas and Fas ligand can regulate apoptosis within the erythroblastic island.36 Fas ligand–bearing mature orthochromatic erythroblasts demonstrate a Fas-based cytotoxicity against Fas-expressing immature erythroblasts. Since high levels of erythropoietin abolish this cytotoxicity, these data suggest the presence of a negative regulatory feedback between mature and immature erythroblasts. Bone marrow macrophages may also modulate erythroblast apoptosis via a Fas-independent mechanism. They produce RCAS1 (receptor binding cancer antigen expressed in SiSo cells), the soluble form of which induces mitochondrial membrane permeabilization and activation of caspases-8 and -3 in RCAS1 receptor-bearing immature erythroblasts.37 In aggregate, these findings lend support to the concept of erythroblastic islands as highly specialized bone marrow subcompartments, where adhesion events, in concert with cytokines, play critical roles in regulating erythropoiesis. As a binding partner of both α4β1 and αV integrins, ICAM-4 could play a multifunctional role within the erythroblastic island. ICAM-4–α4β1 associations may mediate adhesion between adjacent erythroblasts, while ICAM-4–αVinteractions may effect binding of erythroblasts to the central macrophage. We speculate that the various molecular attachments among cellular components of erythroblastic islands are in a dynamic state during development and that the signaling pathways they stimulate promote differentiation and enucleation. The secreted isoform ICAM-4S may be a key regulator of these molecular attachments.
We would like to thank Drs Jan-Fang Cheng and Thiebaut-Noel Willig (Lawrence Berkeley National Laboratory, Berkeley, CA) for invaluable advice on identification and characterization of the BAC clone–containing ICAM-4 gene. We are very grateful to Mr Peter Martin for DNA sequencing and to Ms Gail Mosley and Mr Kevin Peet for their expert assistance in manuscript preparation.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-08-2529.
Supported in part by National Institutes of Health grants DK56267, DK26263, DK32094, and HL31579, and by the Director, Office of Health and Environment Research Division, US Department of Energy, under contract DE-AC03-76SF00098.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Joel Anne Chasis, Lawrence Berkeley National Laboratory, Building 74, 1 Cyclotron Rd, Berkeley, CA 94720; e-mail:jachasis@lbl.gov.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal