B lymphomagenesis is an uncontrolled expansion of immature precursors that fail to complete their differentiation program. This failure could be at least partly explained by inappropriate expression of several oncogenic transcription factors, such as Pax5 and Myc. Both Pax5 and c-Myc are implicated in the pathogenesis of non-Hodgkin lymphomas. To address their role in lymphomagenesis, we analyzed B-cell lymphomas derived from p53-null bone marrow progenitors infected in vivo by a Myc-encoding retrovirus. All Myc-induced lymphomas invariably maintained expression of Pax5, which is thought to be incompatible with terminal differentiation. However, upon culturing in vitro, several cell lines spontaneously down-regulated Pax5 and its target genes CD19, N-Myc, and MB1. Unexpectedly, other B-cell markers (eg, CD45R) were also down-regulated, and markers of myeloid lineage (CD11b and F4/80 antigen) were acquired instead. Moreover, cells assumed the morphology reminiscent of myeloid cells. A pool of F4/80-positive cells as well as several single-cell clones were obtained and reinjected into syngeneic mice. Remarkably, pooled cells rapidly re-expressed Pax5 and formed tumors of relatively mature lymphoid phenotype, with surface immunoglobulins being abundantly expressed. Approximately half of tumorigenic single-cell clones also abandoned myeloid differentiation and gave rise to B lymphomas. However, when secondary lymphoma cells were returned to in vitro conditions, they once again switched to myeloid differentiation. This process could be curbed via enforced expression of retrovirally encoded Pax5. Our data demonstrate that some Myc target cells are bipotent B-lymphoid/myeloid progenitors with the astonishing capacity to undergo successive rounds of lineage switching.
Introduction
Trillions of highly specialized cells in the body of a multicellular organism are derived from a single totipotent cell—the fertilized egg. The descendants of this cell form the blastocyst and the inner cell mass, the latter being composed of pluripotent stem cells still capable of adopting any cell fate. However, with each successive differentiation step, the choice of fates becomes more limited. For instance, hematopoietic stem cells give rise to lymphoid and myeloid progenitors but not stromal tissues. Furthermore, lymphoid stem cells give rise to B and T lymphocytes and natural killer cells but not to macrophages, granulocytes, or other cells of myeloid lineage. Such lineage commitment relies on timely activation of appropriate transcription factors and silencing of inappropriate ones. In B-cell differentiation, key transcription factors are PU.1, E2A, EBF, and Pax5 (also known as BSAP; reviewed in Kee and Murre1). These factors play a dual role in commitment to the B-lymphoid lineage.
One of their functions is to ensure expression of genes required for B-cell maturation. For instance, E2A and EBF govern production of immunoglobulin (Ig) light chains and recombinases responsible for Ig gene rearrangements.2 The other function of these transcription factors is to preclude expression of genes specific for alternative cell fates. Failure to do so could have unwanted consequences. For example, ectopic expression of Notch on the surface of bone marrow (BM) progenitors causes a switch from B- to T-cell differentiation.3 Furthermore, the receptor for granulocyte-macrophage colony-stimulating factor (GM-CSF) causes preferential proliferation of myeloid precursors, potentially at the expense of B-cell precursors. Thus, for B-lymphoid differentiation, both Notch and GM-CSF receptor need to be silenced. Which transcription factor precludes expression of Notch in B-cell progenitors is not clear, but expression of GM-CSF receptor is known to be inhibited by Pax5.4,5 Consequently, in Pax5-null mice, pro-B lymphocytes are generated but do not remain committed to B-cell lineage.6 Under certain circumstances, they can even differentiate into functional T cells.7 Pax5 also plays a role in maintaining lineage identity: its forced inactivation in previously committed pro-B cells via homologous recombination results in the capacity to differentiate into macrophages in vitro and to reconstitute T-cell development in vivo.8
While the choice between pathways is obviously driven by transcription factors, how these transcription factors themselves are regulated is not completely understood.9,10 One possibility is that their regulation is extrinsic, or instructive, whereby the cell reacts to environmental and positional cues. The other, not necessarily mutually exclusive scenario, involves an intrinsic mechanism: each cell makes its choice in a random, stochastic manner. Busslinger et al have proposed that Pax5 activation occurs in such an inefficient manner to ensure that the progenitor cell retains other differentiation options.11 Moreover, since gene expression during differentiation is based largely on epigenetic mechanisms, there is always a potential for reversal.12 Thus, some cells, despite their seemingly committed status, might be able to redifferentiate into a different lineage, in particular during hematopoiesis.
Neoplastic cells have been very useful for the studies on “lineage promiscuity”13 as their differentiation programs are seldom completed. As early as 1957, a B-lymphoma cell line was established that upon culturing in vitro morphed into macrophagelike cells.14 Upon reinjection into animals, these cells were tumorigenic and gave rise to myeloid tumors. Similar cell lines were described in subsequent years: 70Z/3,15 Raf+Myc-induced neoplasms,16 and several others (referenced in Borrello and Phipps17). Interestingly, the conversion of macrophages into B cells has not been documented. Moreover, the propensity of B cells, but not T cells, to convert into macrophages was unexpected, in light of the prevailing view that B and T lymphocytes share a common nonmyeloid progenitor. It also implied the existence of bipotential B-macrophage progenitors. Indeed, such progenitors have been identified in hematopoietic malignancies,18 in fetal liver,19 and more recently in adult bone marrow.20 These and other findings have established that lineage switching is an integral part of at least some differentiation programs.
Still, the role of lineage infidelity in cell fate specification is poorly understood. Intriguing questions are (1) whether the reversal of cell fate is itself reversible, allowing myeloidlike ex–B cells to regain their lymphoid phenotype; and (2) how many transcription factors would need to oscillate to make such recurrent lineage infidelity possible. To answer these questions, we drew upon our recently developed model for B-cell lymphoma based on the infection of p53-null bone marrow progenitors with a retrovirus encoding the Myc oncoprotein.21 Using this approach, we have generated neoplastic lines that exclusively expressed B-cell markers when passaged in vivo. However, several of them, obtained from independently derived tumors, spontaneously acquired myeloid markers upon culturing in vitro. We thus set out to determine what transcription factors are involved in the switch between lymphoid and myeloid differentiation and whether this process could occur repeatedly in the same cell.
Materials and methods
Cell lines, tumors, and animals
Generation of murine Myc/p53-null B lymphomas syngeneic with C57BL6/J mice has been described earlier.21 For in vivo passaging, tumor tissues were dispersed, and 5 × 106cells were injected subcutaneously into C57BL6/J mice (Jackson Laboratory, Bar Harbor, ME). For in vitro culturing, cells were placed on a monolayer of gamma-irradiated S17 feeder cells and maintained in RPMI 1640 supplemented with 10% fetal bovine serum and interleukin-7 (IL-7; 0.1 ng/mL, R&D Systems, Minneapolis, MN), as described in Cumano et al.22 Single-cell clones of myeloid phenotype that attached tightly to S17 cells were maintained without a feeder layer, in the medium lacking IL-7 but supplemented with lipopolysaccharide (10 μg/mL, Sigma-Aldrich, St Louis, MO).
Flow cytometric analyses and cell sorting
The following monoclonal antibodies for flow cytometry were obtained from BD PharMingen (San Diego, CA): fluorescein isothiocyanate (FITC)– or phycoerythrin (PE)–conjugated anti-CD45R (B220, clone RA3-6B2), anti-CD19, anti-CD43 (clone S7), anti–surface IgM (sIgM; clone R6-60.2), anti-sIgD, anti-CD11b (Mac1, clone M1/70), and anti-Thy1.2 (clone 53-2.1). FITC-conjugated anti-F4/80, anti-CD4, and anti-CD8 antibodies were provided by Dr Christopher Hunter (University of Pennsylvania). Cultured or tumor cells were resuspended in phosphate-buffered saline supplemented with 0.1% bovine serum albumin (Sigma-Aldrich) and appropriate antibodies. Staining reactions were carried out on ice for 30 minutes. Cells were assayed using a FACS calibur flow cytometer (Becton Dickinson, Mountain View, CA) and the results were analyzed using the CELLQuest software (Becton Dickinson). For preparative purposes, cells were sorted using a FACSTAR+ (Becton Dickinson) or a MoFlo (Cytomation, Fort Collins, CO) cell sorter. Sorted cells were either pooled or placed individually in wells of a 96-well plate. GFP-positive cells were sorted without prior labeling.
Histologic and cytochemical staining
All tumor tissues were fixed in 10% neutral buffered-formalin (Fisher Scientific, Fair Lawn, NJ) and embedded in paraffin. Then, 5-micrometer sections were stained with hematoxylin and eosin (H&E). Cultured cells were spun onto slides, air-dried for 5 minutes, and stained with Hema-Quick II (Wright-Giemsa stain solution, Biochemical Sciences, Swedesboro, NJ) according to the manufacturer's recommendation.
Polymerase chain reaction (PCR) analyses
RNA isolation was performed using Tri-reagent (Sigma-Aldrich). Reverse transcription (RT) reactions were performed using SuperScript First-Strand Synthesis System for RT-PCR (Gibco BRL, Rockville, MD). PCRs were performed under the following conditions: denaturation at 95°C for 45 seconds, annealing at indicated temperatures for 45 seconds, and extension at 72°C for 60 seconds. All reactions were carried out for 35 cycles, with 5 minutes initial denaturing and 7 minutes final extension. “S” and “A” refer to sense and antisense primers, respectively. PCR analysis for VDJ-recombination was performed on genomic DNA, not cDNA (Table 1).
PCR primers for gene expression analyses
| Genes . | Primer sequences . | Annealing temperature, °C . | Reference/ source . |
|---|---|---|---|
| VJ558-JH4 | S: CAGGTCCAACTGCAGCAG | 67 | Li et al23 |
| A: CTCTCAGCCGGCTCCCTCAGGG | |||
| PU.1 | S: CGGATGACTTGGTTACTTACG | 62 | Lin and |
| A: GTAGGAAACCTGGTGACTGAG | Grosschedl24 | ||
| E12 | S: CGCACTGACCACGAGCTTCAC | 63 | Li et al25 |
| A: GGTCGTCCTCGTCCTCGTCTG | |||
| E47 | S: CGCACTGACCACGAGCTTCAC | 62 | Li et al25 |
| A: TCCAGGGACAGCACCTCATCTG | |||
| EBF | S: GCCAACAGCGAAAAGACC | 62 | J.G.M.'s lab |
| A: GCTTGGAGTTATTGTGGAC | |||
| Pax5 | S: CTACAGGCTCCGTGACGCAG | 61 | J.G.M.'s lab |
| A: GTCTCGGCCTGTGACAATAGG | |||
| N-myc | S: CGGTGGCTGCTCCTGCTCGTG | 64 | Nagy et al26 |
| A: TGTCCTGCCCCCTACCCAGAT | |||
| MB1 | S: GCCAGGGGGTCTAGAAGC | 66 | Li et al23 |
| A: TCACTTGGCACCCAGTACAA |
| Genes . | Primer sequences . | Annealing temperature, °C . | Reference/ source . |
|---|---|---|---|
| VJ558-JH4 | S: CAGGTCCAACTGCAGCAG | 67 | Li et al23 |
| A: CTCTCAGCCGGCTCCCTCAGGG | |||
| PU.1 | S: CGGATGACTTGGTTACTTACG | 62 | Lin and |
| A: GTAGGAAACCTGGTGACTGAG | Grosschedl24 | ||
| E12 | S: CGCACTGACCACGAGCTTCAC | 63 | Li et al25 |
| A: GGTCGTCCTCGTCCTCGTCTG | |||
| E47 | S: CGCACTGACCACGAGCTTCAC | 62 | Li et al25 |
| A: TCCAGGGACAGCACCTCATCTG | |||
| EBF | S: GCCAACAGCGAAAAGACC | 62 | J.G.M.'s lab |
| A: GCTTGGAGTTATTGTGGAC | |||
| Pax5 | S: CTACAGGCTCCGTGACGCAG | 61 | J.G.M.'s lab |
| A: GTCTCGGCCTGTGACAATAGG | |||
| N-myc | S: CGGTGGCTGCTCCTGCTCGTG | 64 | Nagy et al26 |
| A: TGTCCTGCCCCCTACCCAGAT | |||
| MB1 | S: GCCAGGGGGTCTAGAAGC | 66 | Li et al23 |
| A: TCACTTGGCACCCAGTACAA |
S indicates sense; A, antisense.
Generation of and infection by a murine Pax5-encoding retrovirus
Full-length Pax5 cDNA flanked by EcoRI sites27 was subcloned into the MIGR1 bicistronic retrovirus.28 Subconfluent BOSC packaging cells were transfected with 10 μg of the Pax5 retroviral DNA using Lipofectamine Plus (Gibco BRL). At 3 days after transfection, the conditioned medium was harvested, supplemented with polybrene (10 mg/mL, Sigma-Aldrich), and mixed with Myc5 secondary tumor cells. Infected cells were cultured in vitro and subjected to flow cytometric analyses.
Results
In vitro conversion to the myeloid phenotype of Myc-induced B lymphomas
To determine whether any of the Myc/p53-null B lymphomas described in our earlier paper21 had been derived from B-macrophage precursors, cells from primary tumors were cultured in vitro on the monolayer of gamma-irradiated S17 cells in the presence of IL-7.22 In the first experiment, none of 7 tumor explants tested were immediately capable of growing in vitro: all cultures went through crises in which the majority of cells died. From such crises, 2 apparently immortal cell lines have emerged, corresponding to tumors Myc3 and Myc5.
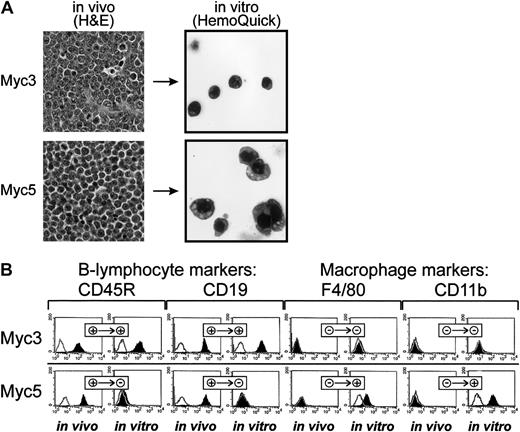
On gross and histolopathologic examinations, both tumors exhibited typical lymphoma characteristics: soft texture, scant stroma, and the presence of relatively monomorphic neoplastic cells with round or ovoid basophilic nuclei with clumped chromatin at the periphery and thin rims of eosinophilic cytoplasm (Figure 1A, top left and bottom left panels). However, upon culturing in vitro, profound changes between Myc3 and Myc5 have emerged. While Myc3 cells remained round and strictly nonadherent, Myc5 cells were somewhat irregularly shaped and often adhered to the S17 feeder layer (data not shown). To further contrast their morphologies, cells were spun onto the surface of glass slides and stained with the Wright-Giemsa dye. While cultured Myc3 cells resembled lymphocytes (top right panel), most Myc5 cells were much larger, with irregularly shaped nuclei, and often resembled granulocytes or monocytes (bottom right panel, top right and bottom right corners, respectively). These differences were suggestive of Myc3 retaining its B-cell differentiation and Myc5 converting to a myeloid lineage.
Myc5 B-lymphoma cells acquire myeloid phenotype in vitro.
(A) Histolopathologic (left) and cytologic (right) analyses of Myc3 and Myc5 lymphomas maintained in vivo and in vitro, respectively. On the left, sections of tumor tissues were stained with H&E (original magnification, × 40). On the right, cultured cells were spun onto slides and stained with Wright-Giemsa (HemoQuick, original magnification, × 60). (B) Flow cytometric analyses of surface markers on primary (in vivo) and cultured (in vitro) Myc3 and Myc5 cells. Cells stained with specified antibodies are indicated by black histograms; control unstained cells, with open lines. + and − refer to positive and negative staining, respectively.
Myc5 B-lymphoma cells acquire myeloid phenotype in vitro.
(A) Histolopathologic (left) and cytologic (right) analyses of Myc3 and Myc5 lymphomas maintained in vivo and in vitro, respectively. On the left, sections of tumor tissues were stained with H&E (original magnification, × 40). On the right, cultured cells were spun onto slides and stained with Wright-Giemsa (HemoQuick, original magnification, × 60). (B) Flow cytometric analyses of surface markers on primary (in vivo) and cultured (in vitro) Myc3 and Myc5 cells. Cells stained with specified antibodies are indicated by black histograms; control unstained cells, with open lines. + and − refer to positive and negative staining, respectively.
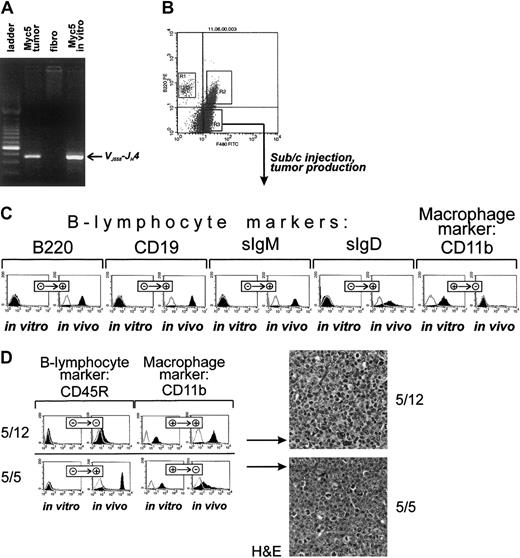
To confirm that this was indeed the case, cultured cells were stained with fluorescently labeled antibodies against B-lymphoid (CD45R and CD19) and myeloid (F4/80 and CD11b) markers. As evidenced by data presented in Figure 1B, top row, Myc3 cells were positive for the former and negative for the latter, regardless of growth conditions (in vivo as tumors or in vitro as cultured cells). In contrast, Myc5 cells (bottom row) expressed lymphoid markers in vivo but almost exclusively myeloid markers in vitro. No detectable CD19 and very little CD45R expression was seen using flow cytometric analysis. To rule out contamination with host macrophages or monocytes, we isolated genomic DNA from “converted” Myc5 cells and performed VDJ-recombination analysis as described in Li et al.23 Myc5 cells from both the primary tumor and cell culture (but not control murine fibroblasts) clearly contained the rearrangement, which was indicative of their B-cell origin (Figure 2A). We thus concluded that under our cell-culture conditions Myc5 cells spontaneously acquire a myeloid phenotype. In subsequent experiments, we have identified 2 additional Myc-induced tumors (MycA and MycB) whose cells adopted myeloid Myc5-like phenotype when cultured in vitro (data not shown). This suggested that lineage plasticity is a common attribute of Myc-transformed BM cells, not a peculiar trait of a single-cell clone.
Cultured myeloid Myc5 cells reacquire B-lymphoma markers in vivo.
(A) PCR analysis confirming the presence of VDJ-rearrangement in cultured Myc5 cells (far right lane). In preceding lanes, DNAs from primary Myc5 tumor and murine fibroblasts were used as positive and negative controls, respectively. Migration of the rearranged fragment is indicated by the arrow. (B) FACS of cultured Myc5 cells into B220-positive (R1), F4/80-positive (R3), and doubly positive (R2) populations. As indicated by the arrow, the R3 population was expanded and used for tumor production. (C) Flow cytometric analyses of surface markers on sorted Myc5 cells from the R3 fraction that were expanded in culture (in vitro) and subsequently injected into animals and allowed to form secondary tumors (in vivo). Cells stained with specified antibodies are indicated by black histograms; control unstained cells, with open lines. + and − refer to positive and negative staining, respectively. (D) The left panel depicts the same analyses as in panel C but performed on single cell subclones 5/12 and 5/5 from the R3 fraction. The right panel refers to H&E staining of sections of tumors derived from clones 5/12 and 5/5. Original magnification × 40.
Cultured myeloid Myc5 cells reacquire B-lymphoma markers in vivo.
(A) PCR analysis confirming the presence of VDJ-rearrangement in cultured Myc5 cells (far right lane). In preceding lanes, DNAs from primary Myc5 tumor and murine fibroblasts were used as positive and negative controls, respectively. Migration of the rearranged fragment is indicated by the arrow. (B) FACS of cultured Myc5 cells into B220-positive (R1), F4/80-positive (R3), and doubly positive (R2) populations. As indicated by the arrow, the R3 population was expanded and used for tumor production. (C) Flow cytometric analyses of surface markers on sorted Myc5 cells from the R3 fraction that were expanded in culture (in vitro) and subsequently injected into animals and allowed to form secondary tumors (in vivo). Cells stained with specified antibodies are indicated by black histograms; control unstained cells, with open lines. + and − refer to positive and negative staining, respectively. (D) The left panel depicts the same analyses as in panel C but performed on single cell subclones 5/12 and 5/5 from the R3 fraction. The right panel refers to H&E staining of sections of tumors derived from clones 5/12 and 5/5. Original magnification × 40.
The switch between B-lymphoid and myeloid phenotypes is reversible
To determine whether adoption of the myeloid fate is a terminal decision or could be reversed, we first obtained a pure population of CD45R-negative, F4/80-positive cells using fluorescent-activated cell sorting (FACS) (Figure 2B, fraction R3). These cells were separated from CD45R singly positive cells which have retained the B-cell phenotype (fraction R1) and from doubly positive cells undergoing transition to the myeloid phenotype (fraction R2). Cells from R3 fraction were expanded as described in “Materials and methods,” analyzed using flow cytometry, and injected subcutaneously into syngeneic mice. Injected cells readily formed tumors after a short latent period (2-3 weeks). However, staining with lineage-specific antibodies revealed that, unlike parental R3 cells, tumor cells no longer expressed myeloid markers (eg, CD11b, Figure 2C) but were once again strongly positive for B-cell markers (eg, CD45R and CD19). Interestingly, these secondary B lymphomas were also positive for sIgM and sIgD and thus possessed a more mature phenotype than primary Myc5 neoplasms (Figure 2C).
To rule out the possibility that secondary tumors were derived from rare CD45R-positive cells present in the R3 fraction, we obtained single-cell clones with confirmed myeloid phenotype. Expansion of such clones took several weeks, and during this period further myeloid differentiation occurred, as evidenced by their yet more adherent phenotype. Consequently, some of the clones were no longer tumorigenic. Those that were fell into 2 groups. Approximately half of clones (exemplified by clone 5/12) formed slow-growing tumors that have not acquired B-cell markers (Figure 2D, top left). Consistent with this, on histopathologic examination neoplastic cells exhibited nuclear and cellular polymorphism, with a large number of cells possessing lobated nuclei suggestive of granulocyte differentiation (Figure 2D, top right). However, other clones (exemplified by clone 5/5) were growing faster and formed tumors composed of cells that were strongly CD45R-positive and only weakly CD11b-positive (Figure 2C, bottom left). This suggested that they were undergoing the process of differentiation back to B-cell lineage. Indeed, H&E staining has revealed that cells with lobated nuclei were interspersed among larger cells with round hyperchromatic nuclei, distinct nucleoli, and a small amount of eosinophilic cytoplasm (Figure 2C, bottom right). The latter type was reminiscent of B cells comprising the original Myc5 tumor. Therefore, we concluded that even Myc5 single-cell clones of apparently myeloid phenotype were capable of resuming their original differentiation program.
EBF and Pax5 genes are silenced upon the switch to the myeloid phenotype
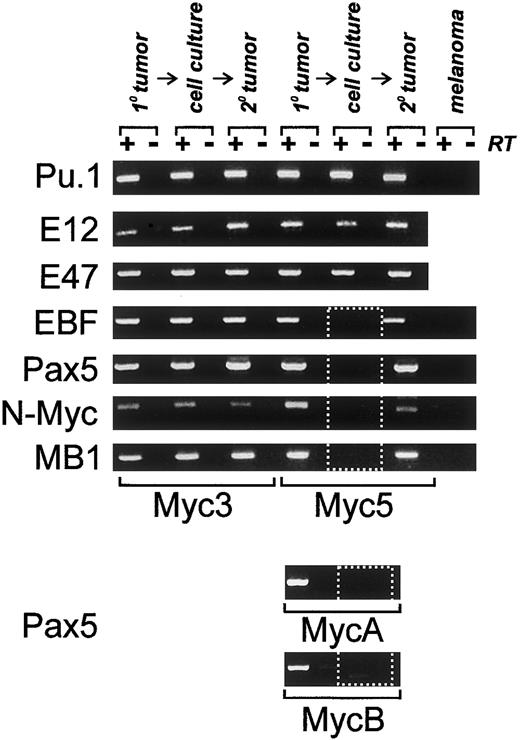
To determine what transcription factors are responsible for lineage infidelity in Myc5 cells, we utilized an RT-PCR approach. Oligonucleotide primers specific for genes encoding B-cell–specific transcription factors were generated. They were used to probe expression of PU.1, E12, E47, EBF, and Pax5. These factors are thought to appear in that order in differentiating B cells.1Predictably, all of them were expressed in control Myc3 cells, whether they had been obtained from the primary tumor, in vitro cultures, or a secondary tumor (Figure 3). In contrast, in Myc5 cells only the first 3 factors were expressed under all conditions, but EBF and Pax5 were expressed only in tumors but not cultured cells (Figure 3, dotted rectangle). Consistent with this finding, expression of several Pax5 target genes, such as N-Myc and MB1, was also undetectable in vitro. To extend the correlation between Pax5 silencing and the myeloid phenotype, we have also analyzed 2 other Myc5-like tumors, MycA and MycB. As evidenced by data in the bottom 2 panels, MycA and MycB tumors were positive for Pax5 expression, but the corresponding cell lines were not. We thus hypothesized that lineage infidelity was attributable to the silencing of EBF and Pax5 and could be prevented by enforced expression of one of these factors.
Expression of B-specific transcription factors and their targets is altered in some Myc-tumor cell lines.
All panels depict RT-PCR analyses of mRNAs encoding proteins specified on the left. Primary (1°) tumors, cultured cells, or secondary tumors derived from cultured cells (2°) were used as sources of RNA. +/−RT refers to reactions carried out in the presence and absence of reverse transcriptase. B16.F10 murine melanoma cells were used as a negative control. Dotted box indicates lack of expression of EBF,Pax5, and Pax5 target genes. Expression of the Pax5 protein in primary tumors was also confirmed using Western blotting (data not shown).
Expression of B-specific transcription factors and their targets is altered in some Myc-tumor cell lines.
All panels depict RT-PCR analyses of mRNAs encoding proteins specified on the left. Primary (1°) tumors, cultured cells, or secondary tumors derived from cultured cells (2°) were used as sources of RNA. +/−RT refers to reactions carried out in the presence and absence of reverse transcriptase. B16.F10 murine melanoma cells were used as a negative control. Dotted box indicates lack of expression of EBF,Pax5, and Pax5 target genes. Expression of the Pax5 protein in primary tumors was also confirmed using Western blotting (data not shown).
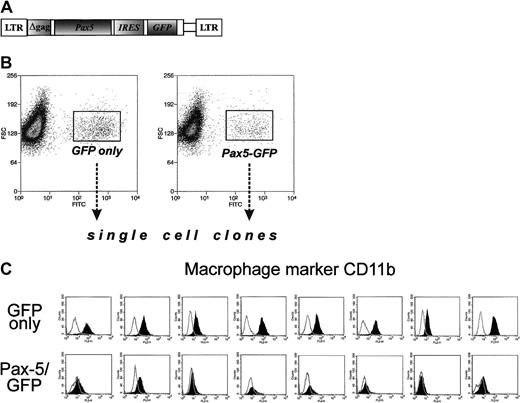
Enforced expression of Pax5 prevents acquisition of the myeloid-specific marker CD11b
To assess the role of Pax5 in lineage infidelity, we have generated a Pax5-encoding retrovirus. Pax5 cDNA was inserted into the bicistronic MIGR1 retrovirus carrying the GFP gene and the long terminal repeats from murine stem cell retrovirus28(Figure 4A). The retroviral construct was transiently transfected into packaging BOSC23 cells, and the supernatant was used to infect Myc5 cells from a secondary tumor. Upon infection, cells were subjected to FACS, and single “green” cells were sorted, placed into individual wells, and expanded. Control clones expressing “empty” vector were also obtained. Both types of clones were then stained with antibodies against B-cell marker CD19 and myeloid marker CD11b. None of the clones analyzed were positive for CD19, confirming the earlier observation that Pax5 alone is not sufficient for CD19 expression5 and differentiation into bona fide B cells. However, GFP and Pax5/GFP clones differed drastically in the pattern of CD11b expression. All 8 GFP clones were positive for this marker, indicating that they once again had reverted to the myeloid phenotype (Figure 4C, top row). In contrast, all 8 GFP/Pax5 clones were CD11b-negative (bottom row) and thus were not undergoing conversion to macrophagelike cells. This finding suggested that while enforced expression of Pax5 alone is not sufficient to maintain the B-cell phenotype, it is sufficient to block the switch from B-cell to myeloid differentiation.
Ectopic expression of Pax5 prevents acquisition of myeloid markers by Myc5 B-lymphoma cells.
(A) The diagram of the Pax5-encoding retrovirus. The GFP-encoding MIGR1 retroviral vector was used as a backbone. (B) FACS of Pax5- or control retrovirus-infected Myc5 cells from a secondary tumor. Sorted populations are indicated by rectangles and arrows. (C) Flow cytometric analyses of the myeloid CD11b surface marker on Myc5 cells infected by the control (top panels) and Pax5-encoding (bottom panels) retroviruses.
Ectopic expression of Pax5 prevents acquisition of myeloid markers by Myc5 B-lymphoma cells.
(A) The diagram of the Pax5-encoding retrovirus. The GFP-encoding MIGR1 retroviral vector was used as a backbone. (B) FACS of Pax5- or control retrovirus-infected Myc5 cells from a secondary tumor. Sorted populations are indicated by rectangles and arrows. (C) Flow cytometric analyses of the myeloid CD11b surface marker on Myc5 cells infected by the control (top panels) and Pax5-encoding (bottom panels) retroviruses.
Discussion
Here we demonstrate for the first time the existence of bone marrow precursors that upon neoplastic transformation “freeze” in a peculiar differentiation stage. The hallmark of this stage is the ability to retain the potential for both B-cell and myeloid differentiation even after the choice of fate is seemingly made. While the ability of some immature B lymphomas to acquire the myeloid phenotype has been previously reported,14-16 it was assumed that switching from lymphoid to myeloid lineage was irreversible. We have discovered that cells with overt myeloid phenotype could, upon return to in vivo conditions, retrace their steps and resume differentiation into relatively mature B cells expressing surface immunoglobulins, even though the original tumor was composed of prepro-B cells.21 Remarkably, even these sIgM- and sIgD-positive cells are not firmly committed to B-lymphoid lineage either as they readily reacquire the myeloid markers in culture.
It is also noteworthy that this astonishing capacity to undergo successive rounds of lineage switching apparently relies on a single pair of transcription factors, EBF and Pax5. We do not know at present whether regulations of EBF and Pax5 are achieved independently or whether gain and loss of Pax5 rely on regulation of EBF, its upstream activator.2 We have learned, however, that enforced expression of Pax5 is sufficient to prevent myeloid differentiation, although it is clearly not sufficient to sustain B-lymphoid differentiation. The latter finding corroborates earlier observations that concerted expression of several transcription factors is required for the B-cell phenotype.5,29,30 However, it appears that targeted inactivation of just one transcription factor (ie, Pax5) in neoplastic B cells would have a profound effect not only on their differentiation, but also on the rate of tumor growth. Thus, our results suggest that Pax5 might be a legitimate therapeutic target in lymphoplasmacytoid lymphomas with t(9;14)(p13;q32) translocations affecting the Pax5 gene. This hypothesis is in accord with the recent findings that even a brief inactivation of the Myc oncoprotein in pre-existing neoplasms results in sustained tumor regression.31 32 Whether or not this holds true for Pax5 could be verified in the future, as new gene targeting techniques are becoming available.
We thank Xin Yin and Gautam Rajpal for their help with performing RT-PCR assays and Dr Christopher Hunter (University of Pennsylvania) for the gift of several antibodies. We are indebted to various members of our laboratories for many stimulating discussions.
Prepublished online as Blood First Edition Paper, October 24, 2002; DOI 10.1182/blood-2002-06-1797.
Supported by National Cancer Institute grants CA 71881 and CA 97932 (A.T.-T.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Andrei Thomas-Tikhonenko, Department of Pathobiology, University of Pennsylvania, 3800 Spruce St, M/C 6051, Philadelphia, PA 19104-6051; e-mail:andreit@mail.vet.upenn.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal