Abstract
The standard approach to assess hematopoietic stem cell (HSC) engraftment in experimental bone marrow transplantation models relies on detection of donor hematopoietic cells in host bone marrow following death; this approach provides data from only a single time point after transplantation for each animal. In vivo bioluminescence imaging was therefore explored as a method to gain a dynamic, longitudinal profile of human HSC engraftment in a living xenogeneic model. Luciferase expression using a lentiviral vector allowed detection of distinctly different patterns of engraftment kinetics from human CD34+ and CD34+CD38- populations in the marrow NOD/SCID/β2mnull mice. Imaging showed an early peak (day 13) of engraftment from CD34+ cells followed by a rapid decline in signal. Engraftment from the more primitive CD34+CD38- population was relatively delayed but by day 36 increased to significantly higher levels than those from CD34+ cells (P < .05). Signal intensity from CD34+CD38--engrafted mice continued to increase during more than 100 days of analysis. Flow cytometry analysis of bone marrow from mice after death demonstrated that levels of 1% donor cell engraftment could be readily detected by bioluminescence imaging; higher engraftment levels corresponded to higher image signal intensity. In vivo bioluminescence imaging provides a novel method to track the dynamics of engraftment of human HSC and progenitors in vivo. (Blood. 2003;102: 3478-3482)
Introduction
Key aspects of the biology of hematopoietic stem cells (HSCs) have been studied for many years using elegant murine models of bone marrow transplantation. More recently, immunodeficient murine models have been used successfully to study human HSCs.1 In all of these models, HSCs are infused into recipient animals, and donor engraftment is determined by analysis (usually flow cytometry) of bone marrow harvested after recipient animals are killed. This approach, although able to accurately measure the levels of engraftment and the contribution of donor HSCs to different hematopoietic lineages, has 2 main disadvantages. First, sampling limitations may occur by harvesting only certain parts of the marrow space. Thus, the anatomic distribution of donor engraftment is assumed to be equivalent within and between experimental animals. Second, engraftment is measured at a single time point (after death) and so the kinetics of engraftment within an individual animal are unknown.
Bioluminescence imaging is a technique that detects visible light released when the enzyme luciferase reacts with its substrate, luciferin.2 The development of a method to detect luciferase expression in vivo has facilitated the study of infectious disease,3 gene expression,4 and tumor growth and response to therapy in small animal models.5 Lymphocyte trafficking into joints and the central nervous system (CNS) has also been demonstrated in vivo.6 We now show that by accomplishing high efficiency, stable transduction, and expression of luciferase in HSCs, bioluminescence imaging can reveal novel, dynamic information on the pattern of human HSC and progenitor engraftment and proliferation within the marrow space of living hosts.
Materials and methods
Isolation of human HSCs
Umbilical cord blood (CB) was obtained from normal deliveries under protocols approved by the Committee on Clinic Investigations of Childrens Hospital Los Angeles institutional review board. CD34+ cells were isolated from mononuclear cells to 80% to 90% purity with a magnetic-activated cell sorting (MACS) CD34 enrichment column (Miltenyi Biotec, Oberlin, CA). CD34+CD38- cells were isolated using a fluorescence-activated cell sorter (FACSVantage; Becton Dickinson Immunocytometry Systems [BDIS], San Jose, CA) from CD34+-enriched populations following fluorescence-labeled antibody staining with CD34-fluorescein isothiocyanate (FITC; BDIS) and CD38-phycoerythrin (PE; leu-17; BDIS), as previously described.7
Lentiviral vector expressing the luciferase reporter gene
SMPU-R-MNCU3-LUC is a lentiviral vector based on HIV-1 that transduces the firefly luciferase gene. The backbone vector SMPU-R has deletions of the enhancers and promoters of the HIV-1 long terminal repeat (LTR; SIN), has minimal HIV-1 gag sequences, contains the cPPT/CTS sequence from HIV-1, has 3 copies of the UES polyadenylation enhancement element from SV40, and a minimal HIV-1 RRE (gift from Paula Cannon, Childrens Hospital Los Angeles).8 The vector has the U3 region from the MND retroviral vector as an internal promoter9 driving expression of the firefly luciferase gene from SP-LUC+ (Promega no. E178A; Promega, Madison, WI).
Transduction of HSCs and progenitors with luciferase vector
CD34+ and CD34+CD38- cells were transduced in fibronectin-coated plates with virus supernatant containing SMPU-R-MNCU3-LUC vector in the presence of interleukin 3 (IL-3), IL-6, and stem cell factor (SCF), as described.10 A second cycle of transduction was performed 8 hours later by removing old medium and adding new virus supernatant and medium. Twenty-four hours after the initial transduction, cells were thoroughly washed 3 times with phosphate-buffered saline (PBS) before transplantation or in vitro analysis.
Detection of luciferase expression in CD34+ cells by immunohistochemistry
An aliquot of transduced CD34+ cells was cultured for 3 days in IL-3, IL-6, and SCF, after which cells were harvested and prepared on cytospin slides. Slides were stained with monoclonal antiluciferase antibody (Novus, Littleton, CO) 1:100 for 1 hour, followed by donkey polyclonal antibody to mouse IgG-FITC (Novus) 1:100 for 30 minutes. The slides were mounted with Vectashield medium with DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratory, Burlingame, CA). Cultured nontransduced CD34+ cells were used as negative controls.
HSC and progenitor transplantation and bioluminescent imaging
Eight- to 10-week old NOD/SCID/β2mnull mice, obtained from Jackson Laboratories (Bar Harbor, ME), were sublethally irradiated with 300 cGy 2 hours prior to infusion of luciferase-transduced CD34+ cells (1 × 105 cells/mouse) or CD34+CD38- cells (4 × 104 cells/mouse) via tail vein.11 In vivo optical imaging was performed with a prototype IVIS 3-dimensional bioluminescence/fluorescence optical imaging system (Xenogen, Alameda, CA) at different time points.
Prior to imaging, each mouse was given an intravenous injection of luciferin (Promega) at a dose of 125 mg/kg, as described.12 General anesthesia was then induced with 5% isoflurane and the mouse was placed in the light-tight heated chamber; anesthesia was continued during the procedure with 2% isoflurane introduced via nose cone. The imaging system consists of a cooled, back-thinned charge-coupled device (CCD) camera to capture both a visible light photograph of the animal taken with light-emitting diodes and the luminescent image, and a rotating mirror and translatable animal stage that allow for images to be acquired over 360° if desired.
After acquiring photographic images of each mouse, anterior and posterior luminescent images were acquired with 1- to 3-minute exposure times. The resulting gray scale photographic and pseudocolor luminescent images were automatically superimposed by software so that identification of any optical signal with location on the mouse was facilitated. Optical images were displayed and analyzed with the Igor (WaveMetrics, Lake Oswego, OR) and IVIS Living Image (Xenogen) software packages. Regions were manually drawn around the bodies of the mice to assess signal intensity emitted. Optical signal was expressed as photon flux, in units of photons/s/cm2/steradian. Maximum photons/s/cm2/steradian, although sometimes reported in bioluminescent studies, were not used because this method is sensitive to noise and can provide incorrect results.
Flow cytometry detection of engraftment
At the end of each experiment, mice were killed by inhalation of a mixture of 75% CO2/25% O2 and marrow was harvested from different parts of the skeleton. Direct immunofluorescence staining was performed with antibody against human leukocyte common antigen CD45 (BDIS). Cells were analyzed by FACSCalibur (BDIS).
Statistical analysis
Data were presented as mean ± SD. The unpaired t test was used for comparisons between groups at each time point. P < .05 was considered significant.
Results
Expression of luciferase transgene in HSCs and progenitor cells
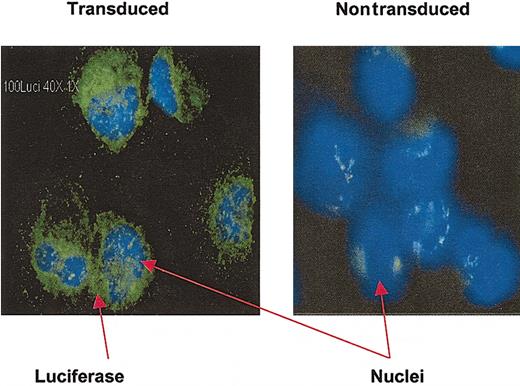
Luciferase expression in HSCs and progenitor cells was accomplished using a lentiviral (HIV-1)-based vector. The proportion of CD34+ cells expressing the luciferase transgene was assessed in vitro by analyzing an aliquot of transduced cells after 3 days of culture. Using immunohistochemical detection of luciferase protein, high-efficiency gene expression was noted (Figure 1; Table 1).
Detection of luciferase expression in CD34+ cells by immunohistochemistry. Cytospin slides prepared from transduced CD34+ cells after 3 days of culture were stained with monoclonal antiluciferase antibody. Luciferase-positive cells have green cytoplasm; nuclei stained with DAPI are blue. Nontransduced, cultured CD34+ cells were used as a negative control. Original magnification, × 40.
Detection of luciferase expression in CD34+ cells by immunohistochemistry. Cytospin slides prepared from transduced CD34+ cells after 3 days of culture were stained with monoclonal antiluciferase antibody. Luciferase-positive cells have green cytoplasm; nuclei stained with DAPI are blue. Nontransduced, cultured CD34+ cells were used as a negative control. Original magnification, × 40.
Immunohistochemical detection of luciferase in transduced and nontransduced CD34+ cells
. | No. of luciferase-positive cells . | Total no. of cells . | Luciferase-positive cells, % . |
|---|---|---|---|
| Nontransduced cells | 6 | 75 | 8 |
| Transduced CD34+ cells | 109 | 123 | 89 |
. | No. of luciferase-positive cells . | Total no. of cells . | Luciferase-positive cells, % . |
|---|---|---|---|
| Nontransduced cells | 6 | 75 | 8 |
| Transduced CD34+ cells | 109 | 123 | 89 |
Time course of signal increase and decay after luciferin administration
The time course of peak and decay of the bioluminescent signal after each administration of the luciferin substrate is an important consideration in quantitative and comparative studies. We therefore studied the signal kinetics using luciferase-expressing human CD34+ cells in our immunodeficient mouse model. Mice that received transplants of luciferase-expressing CD34+ cells 2 weeks earlier were given intravenous or intraperitoneal injections of luciferin. Images of the animals were acquired continuously beginning immediately after injection, each with a 3-minute exposure. Peak signals were detected within 5 minutes after intravenous injection and then decreased rapidly over the next few minutes (data not shown). These kinetics are unlike many previous animal models using luciferase in which time to both peak signal and decay are more delayed.12,13 The route of injection of luciferin (intravenous or intraperitoneal) produced no significant differences in signal kinetics. Therefore, to standardize the measurement of signal in this model, all images in subsequent experiments were acquired 2 minutes after intravenous injection of luciferin.
Bioluminescence imaging of engraftment patterns of human progenitor and HSC populations in immunodeficient mice
The pattern of short- and long-term human HSC and progenitor engraftment was assessed with longitudinal imaging studies in immunodeficient (NOD/SCID/β2mnull) mice. Mice were sublethally irradiated 2 hours before intravenous infusion with transduced human CD34+ cells or the more primitive CD34+CD38- subpopulation believed to contain HSCs (Figure 2).
Experimental schema for imaging of luciferase-expressing human progenitor cells in a xenogeneic transplantation model.
Experimental schema for imaging of luciferase-expressing human progenitor cells in a xenogeneic transplantation model.
Serial images were acquired over the course of 7 to 15 weeks after transplantation at intervals of 2 to 7 days, beginning 1 to 8 days after cell infusion. Luciferin was injected intravenously no more than 2 minutes before each imaging run. Mice that had not received human cells were used as negative controls to assess background luminescence; no detectable signal was apparent either before or after luciferin injection in the negative controls.
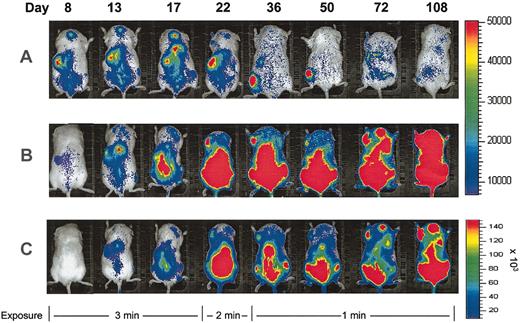
In mice that received CD34+ transplants, human cells could be detected by bioluminescence imaging as early as 24 hours after transplantation. Whole body signal measurements over the course of the experiment showed a sudden increase in signal 2 to 3 weeks after injection of CD34+ cells, after which time the signal rapidly decreased. Low signal levels were detected beyond 100 days after transplantation (Figure 3A).
Longitudinal imaging of human HSC and progenitor engraftment. Images are of representative mice that received transplants of luciferase-expressing human CD34+ (A) or CD34+CD38- cells (B-C) and were imaged serially. All successive images in each row were taken from the same experimental mouse after transplantation at days shown. Exposure time was reduced for all animals at later time points as shown to avoid image saturation. Identical scales are used for transplants of CD34+ and CD34+CD38- cells in panels A and B. In panel C, the same images shown in panel B are scaled to decrease signal intensity and more clearly demonstrate the anatomic distribution of engraftment.
Longitudinal imaging of human HSC and progenitor engraftment. Images are of representative mice that received transplants of luciferase-expressing human CD34+ (A) or CD34+CD38- cells (B-C) and were imaged serially. All successive images in each row were taken from the same experimental mouse after transplantation at days shown. Exposure time was reduced for all animals at later time points as shown to avoid image saturation. Identical scales are used for transplants of CD34+ and CD34+CD38- cells in panels A and B. In panel C, the same images shown in panel B are scaled to decrease signal intensity and more clearly demonstrate the anatomic distribution of engraftment.
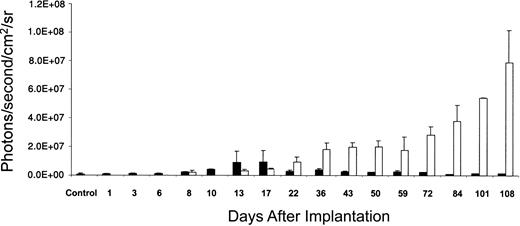
A different pattern of engraftment kinetics was seen consistently when CD34+CD38- cells were transplanted (Figure 3B-C). Signals were still detectable early, but were initially at lower levels than in the CD34+ transplants. Luminescence signals began to rise during the second week after transplantation and continued to increase in mice that received CD34+CD38- transplants throughout the period of study (108 days). Quantification of signal intensity from mice that received transplants of CD34+ cells (n = 5) and mice that received CD34+CD38- cells (n = 3) showed a highly reproducible pattern of engraftment kinetics for each population (Figure 4). From day 36, the signal in mice that received CD34+CD38- cells was significantly higher than that in mice that received CD34+ cells (P < .05).
Quantification of bioluminescent signals in mice that received transplants of luciferase-transduced human CD34+ and CD34+CD38- cells. Summary of data from 5 animals that received transplants of CD34+ cells and 3 animals receiving CD34+CD38- cells in 3 separate experiments showing quantification of photon flux from each entire animal plotted against the number of days after transplantation. Means and SDs are presented. From day 36, mean signal from animals that received CD34+CD38- transplants was significantly higher than mean signal from those that received CD34+ transplants (P < .05). Data from only one CD34+ animal are available on days 72 to 108. On days 1, 3, 6, and 10, mice that received transplants of CD34+CD38- cells were not imaged. Control is a mouse that did not receive a transplant, after intravenous injection of luciferin. ▪ indicates CD34+; □, CD34+CD38-. sr indicates steradian.
Quantification of bioluminescent signals in mice that received transplants of luciferase-transduced human CD34+ and CD34+CD38- cells. Summary of data from 5 animals that received transplants of CD34+ cells and 3 animals receiving CD34+CD38- cells in 3 separate experiments showing quantification of photon flux from each entire animal plotted against the number of days after transplantation. Means and SDs are presented. From day 36, mean signal from animals that received CD34+CD38- transplants was significantly higher than mean signal from those that received CD34+ transplants (P < .05). Data from only one CD34+ animal are available on days 72 to 108. On days 1, 3, 6, and 10, mice that received transplants of CD34+CD38- cells were not imaged. Control is a mouse that did not receive a transplant, after intravenous injection of luciferin. ▪ indicates CD34+; □, CD34+CD38-. sr indicates steradian.
Anatomic distribution of bioluminescent signals in the murine skeleton
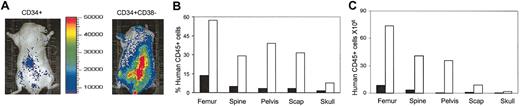
Throughout the course of imaging, the strongest signals appeared in the vertebral column, pelvis, and femur; lower signals were detected in the skull and humeroscapula (Figure 5A).
Anatomic distribution of human cell engraftment. Mice receiving transplants of CD34+ and CD34+CD38- cells were scanned 9 weeks after transplantation (A) and then killed. Bone marrow cells were harvested from each region of the skeleton and analyzed by FACS for percentage (B) and total number (C) of donor human CD45+ cells. Scap indicates scapula; ▪, CD34+; and □, CD34+CD38-.
Anatomic distribution of human cell engraftment. Mice receiving transplants of CD34+ and CD34+CD38- cells were scanned 9 weeks after transplantation (A) and then killed. Bone marrow cells were harvested from each region of the skeleton and analyzed by FACS for percentage (B) and total number (C) of donor human CD45+ cells. Scap indicates scapula; ▪, CD34+; and □, CD34+CD38-.
At the end of the experiments, mice were killed and the percentages and total numbers of human cells in each part of the skeleton were quantified by harvesting all the bone marrow from each area separately and measuring the presence of cells that express the pan-leukocyte human-specific marker CD45 (Figure 5B-C). Consistent with the pattern of distribution of the luminescence signal, the highest percentage of human CD45+ cells was found in marrow harvested from the spine, femur, and pelvis. Engraftment levels as low as 1.3% as detected by flow cytometry corresponded to readily detectable signals by bioluminescence imaging. The higher bioluminescent signal in the mice that received CD34+CD38- transplants corresponded to higher levels of engraftment based on FACS analysis. These results demonstrate the ability to detect low levels of engraftment of human HSCs and progenitors and to quantitate different levels of engraftment using bioluminescence imaging.
Discussion
These studies are the first to describe the use of in vivo bioluminescence imaging to detect marrow engraftment of HSCs. Compared with other imaging modalities such as positron emission tomography (PET), single photon emission computed tomography (SPECT), and magnetic resonance imaging (MRI), bioluminescence imaging offers higher sensitivity,14 ease of use, and high throughput for imaging of small animals. This high sensitivity holds even though the light emitted by the firefly luciferase-luciferin reaction has a peak wavelength at about 560 nm, because a significant portion of its light emission occurs above 600 nm, a wavelength where light more readily passes through tissue. Light-emitting reporters that emit at longer wavelengths (in the red and near-infrared range) may provide advantages in the future in terms of tissue penetration of their signal, but they have not yet been well studied in vivo.15
The limitations of light propagation through tissue currently restrict the application of bioluminescence imaging to small animals where the signal can easily penetrate at all depths. High-resolution MRI appears to be another useful way to track cells in vivo,16 especially in humans, and can provide the added component of 3-dimensional information. However, MRI requires high loading of cells with magnetic label and comparatively long imaging times, and is not as sensitive or quantifiable as bioluminescence imaging.14,17,18 Although PET and SPECT imaging provide 3-dimensional information and can also be applied both in small animals as well as in humans, they require the use of radioactive substances, are not as sensitive as bioluminescence imaging, and generally demand longer imaging times.14,18 In the case of PET, expensive cyclotron facilities are also required. The development of multiple reporter systems that combine optical and radionuclide probes will enable multimodality imaging and provide the dual benefits of high sensitivity and precise anatomic localization.
In the studies reported here, the bioluminescence model possessed the sensitivity to detect and quantitate different levels of chimerism of human donor cells within the marrow space, providing a dynamic profile of engraftment and proliferation in live recipient animals. The technique was able to demonstrate the biologic differences between the cell populations transplanted. The increase in luminescence signal at weeks 2 to 3 after transplantation with CD34+ cells is consistent with engraftment and expansion from short-term repopulating progenitors. The subsequent lower but stable signal is consistent with stable engraftment of a small fraction of long-term repopulating cells within the CD34+ population. These findings are compatible with studies by Glimm et al,19 who demonstrated that the NOD/SCID/β2mnull model supports 2 distinct populations of engrafting human cells. Within the CD34+ population, short-term repopulation (3 weeks) is derived mainly from CD34+CD38+ cells, whereas 90% of long-term repopulation is from the more rare and primitive CD34+CD38- cells. Other investigators have demonstrated that all long-term engraftment in a similar immunodeficient mouse model, the NOD/SCID mouse, is generated by CD34+CD38- cells.20,21
Consistent with these studies, bioluminescence imaging showed that late-term engraftment predominated when purified CD34+CD38- cells were transplanted. These strikingly different kinetics of engraftment between CD34+ and CD34+CD38- cells were consistently seen in all animals studied and demonstrate the ability of bioluminescence imaging to detect populations with intrinsically different engraftment profiles in vivo.
Thus, by combining 2 novel tools, lentiviral vectors to efficiently and stably express luciferase in human HSCs and in vivo bioluminescence imaging, a dynamic profile of progenitor and HSC engraftment is now possible. All areas of the marrow can be simultaneously assessed and changes in donor engraftment and proliferation over time in individual animals can be appreciated. This approach will have great utility in studies of homing and migration of different stem and progenitor cells and will be particularly useful when studying cell populations for which the timing and distribution of engraftment are unknown.
Prepublished online as Blood First Edition Paper, August 28, 2003; DOI 10.1182/blood-2003-05-1432.
Supported by grants from the National Institutes of Health (P50HL54850, P01CA59318, P01HL07104) and the Martell Foundation (G.M.C. and D.B.K.). This work was made possible in part by a generous grant from the Henry L. Guenther Foundation. G.M.C. is a Scholar of the Leukemia and Lymphoma Society. D.B.K. is an Elizabeth Glaser Scientist from the Pediatric AIDS Foundation and is the recipient of a Distinguished Clinical Scientist Award from the Doris Duke Charitable Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank the labor and delivery staff of Kaiser Sunset Permanente Hospital and StemCyte Inc, Arcadia, CA, for their assistance in collection of cord blood samples and the Vector Core of the Childrens Hospital Los Angeles Research Institute for production of lentiviral vectors. Thanks also to Dr Chris Contag, Stanford University, for initial helpful discussions.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal