Abstract
Increased risk of graft-versus-host disease (GVHD) has been described in recipients of hematopoietic stem cell transplantations when the donor is a parous woman. Cells from prior pregnancies are now known to persist in women and could contribute to GVHD. We asked whether male DNA (presumed fetal microchimerism) is present in apheresis products of female donors. A total of 50 samples were studied by using real-time quantitative polymerase chain reaction (PCR) for the Y chromosome-specific sequence DYS14. Among 29 growth factor-mobilized peripheral blood mononuclear cell (G-PBMC) products, 34% were positive for male DNA. Quantitative results, expressed as DNA genome equivalent of male cells per million host cells (gEq/mil), ranged from 0 to 35 gEq/mil. Among 21 CD34-enriched cell fractions, 48% were positive with a range of 0 to 357 gEq/mil. In summary, male DNA was frequently detected in G-PBMC and CD34-enriched products from female donors. Whether fetal microchimerism contributes to GVHD merits further investigation. (Blood. 2003;102: 3845-3847)
Introduction
Graft-versus-host disease (GVHD) is a major complication of allogeneic hematopoietic stem cell transplantation (HSCT) and a leading cause of mortality and morbidity. HLA incompatibility is the strongest risk factor for GVHD, but other factors, including donor parity, are thought to play a role.1-4 The reason(s) for increased risk of GVHD when the donor is a parous woman is unknown. Flowers et al1 suggested that allosensitization of maternal T cells to fetal minor histocompatibility antigens during pregnancy could prime cells to similar antigens in a transplant recipient. However, the discovery that fetal cells persist in the circulation of parous women decades after childbirth raises the question whether non-HLA-matched fetal cells are present in apheresis products and could contribute to GVHD. Fetal microchimerism (FMc) refers to low levels of fetal cells harbored by the mother, which persist at least 38 years postpartum.5,6 FMc has also been described in cellular subsets, including CD34+CD38+, CD3 (including CD4 and CD8), CD19, CD14, and CD56/16 subsets.6,7 Our objective was to determine whether male DNA (probable FMc) is present in apheresis products from female donors. Growth factor-mobilized peripheral blood mononuclear cells (G-PBMCs) and CD34-enriched apheresis products of female donors were tested using real-time quantitative polymerase chain reaction (QPCR) for DYS14, a Y chromosome-specific sequence.
Study design
Study design and subjects
A total of 50 G-PBMC and CD34-enriched apheresis products from 46 women collected between March 2001 and April 2003 at the Fred Hutchinson Cancer Research Center (FHCRC, Seattle, WA) were studied. The FHCRC Institutional Review Board approved the study. Samples from female donors were unidentified and obtained randomly from the Cellular Therapy Laboratory on the day of apheresis (G-PBMC, n = 25) or from a cryopreserved research repository (CD34-enriched, n = 12). Pregnancy and transfusion history in these women was unknown. Additional G-PBMC apheresis products were obtained from women with scleroderma (systemic sclerosis; SSc) undergoing autologous transplantation (G-PBMC, n = 4; CD34-enriched, n = 9). For these subjects, pregnancy and transfusion history was known.
Peripheral blood mononuclear cell (PBMC) mobilization and CD34-enrichment
PBMC mobilization was performed by using either recombinant granulocyte colony-stimulating factor (G-CSF) alone or intermediate-dose chemotherapy followed by G-CSF. Donors underwent a 16- to 20-L leukapheresis by using a continuous-flow blood separator (Cobe Spectra; Cobe Laboratories, Lakewood, CO). CD34-enrichment was performed by using the Baxter 300 Isolex System (Baxter, Irvine, CA) or Cellpro Ceprate System (Cellpro, Seattle, WA).
Quantitation of male DNA by real-time QPCR
Genomic DNA was extracted using Promega Wizard Purification Kits (Promega, Madison, WI) according to manufacturer's instructions. QPCR methods and β-globin primers have previously been described.8,9 We previously reported QPCR using DYS14 as a Y chromosome-specific target.10 Twelve aliquots of DNA were tested from each donor. Two additional DNA aliquots were amplified with β-globin primers for quantitation of host DNA. Standard curves for β-globin and DYS14 were run simultaneously, and the quantitation threshold was set whereby amplifications were consistently in a linear range (Figure 1). For ease of expression, DNA quantities were reported as the DNA genome equivalent number of male cells per million host cells (gEq/mil) by using a conversion factor of 6.6 pg DNA per cell. The male genome equivalents (gEq's) for each sample was calculated by summing quantities of male DNA over aliquots and dividing by the total host gEq assayed. Assuming that the presence of male DNA in female donor apheresis products follows a Poisson process, the result represents the maximum likelihood estimate of the Poisson mean rate for each subject. Testing 10 000 gEq per aliquot (approximately 120 000 total gEq) was experimentally determined to be optimal for the QPCR assay (data not shown). Excessive host DNA is inhibitory for QPCR amplifications, and testing too little host DNA decreases quantitation sensitivity. An acceptable range of total host DNA tested per subject was considered to be 50 000 to 240 000 gEq.
Representative QPCR amplification plot of male DNA in G-PBMCs of a female donor. The x-axis represents the number of cycles of a PCR reaction, and the y-axis is fluorescence intensity over background. As the number of cycles increases, smaller DNA quantities amplify. Each sample is tested for β-globin (measure of total DNA) and DYS14 (measure of male DNA), with quantitative results assessed by the intersection of the amplification plots with the threshold.
Representative QPCR amplification plot of male DNA in G-PBMCs of a female donor. The x-axis represents the number of cycles of a PCR reaction, and the y-axis is fluorescence intensity over background. As the number of cycles increases, smaller DNA quantities amplify. Each sample is tested for β-globin (measure of total DNA) and DYS14 (measure of male DNA), with quantitative results assessed by the intersection of the amplification plots with the threshold.
Each PCR reaction contained 5 μL DNA, 5 μL 10 × platinum buffer, 300 nM of each amplification primer, 100 nM dual-labeled probe, 200 nM of each deoxynucleoside triphosphate (dNTP), 3.5 mM MgCl2, 1.5 U platinum Taq (Gibco BRL, Burlington, ON), and 60 nM Rox reference dye (Synthegen, Houston, TX) for a total reaction volume of 50 μL. Initial incubation was at 50°C for 2 minutes, followed by 95°C for 10 minutes, 45 cycles of 95°C for 15 seconds, and 60°C for 1 minute. A Perkin-Elmer Applied Biosystems 7000 sequence detector collected the amplification data; data were analyzed using Sequence Detection System software (PE Applied Biosystems, Foster City, CA).
Results and discussion
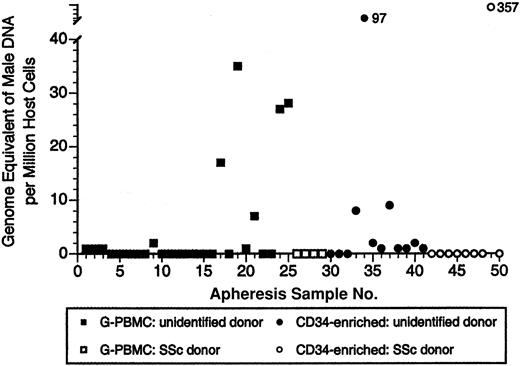
Overall, we found male DNA in 34% of G-PBMC samples and 48% of CD34-selected samples. The mean number of gEq's assayed per patient was 121 717 (range, 59 640-222 600). The quantitative range of male DNA detected in G-PBMCs was 0 to 35 gEq/mil, and in CD34-enriched samples it was 0 to 357 gEq/mil (Figure 2). The highest level of male DNA detected was in the CD34-enriched apheresis product of a woman with SSc (357 male gEq/mil). She gave birth to 3 sons and had no history of blood transfusion prior to autologous transplantation. No male DNA was detected in any of the other 8 women with SSc, including 3 who had previously given birth to sons. Given the low frequency of events that we are detecting and the finite amount of DNA tested, our results may be an underestimate of the true prevalence of male DNA in female donor apheresis and CD34-enriched products.
Male DNA in G-PBMC and CD34-enriched apheresis products of female donors. Each symbol represents results for 1 of a total of 50 apheresis products tested from 46 women. Four women with SSc had both G-PBMC and CD34-enriched products tested (all samples negative).
Male DNA in G-PBMC and CD34-enriched apheresis products of female donors. Each symbol represents results for 1 of a total of 50 apheresis products tested from 46 women. Four women with SSc had both G-PBMC and CD34-enriched products tested (all samples negative).
We present data that male DNA (probable FMc) is not uncommon in G-PBMC and CD34-enriched apheresis products from female donors. The male DNA most likely resulted from a prior pregnancy, although in some subjects it could derive from a blood transfusion, as we lacked this history in unidentified subjects. However, the highest level of male DNA was presumably of fetal origin, as the woman had no blood transfusions and had given birth to 3 sons. Thus, our results suggest donor FMc is present both in G-PBMC and CD34-enriched apheresis products. FMc in these products could derive from a variety of cell lineages. Estimating from the lowest quantity of male DNA detected (1 gEq/mil), this would represent approximately 10 000 to 40 000 male cells infused during a typical HSCT using G-PBMCs (S.H., personal communication, May 2003).
Although immunoregulatory mechanisms must exist to allow tolerance of microchimeric cells in the donor, transplantation into immunocompromised recipients could change their propensity to cause disease. Maternal cells, for example, may cause GVHD when they engraft in children with immunodeficiency.11 Either HLA-disparate fetal T cells transferred in apheresis products or the T-cell progeny of engrafted fetal stem cells could initiate GVHD. Alternatively, fetal stem cells could give rise to antigen-presenting cells, which could activate donor and/or fetal T cells.
In addition to FMc, other sources of chimerism may be transferred during HSCT. Nonirradiated blood transfusions can result in persistent microchimerism or lead to transfusion-associated GVHD.12,13 Cells from the mother traffic into the fetus during pregnancy and persist into adult life.14 Chimerism may also develop with cell transfer between twins in utero.5 These sources of chimerism could contribute to GVHD with male or nulligravid female donors. To our knowledge, no prior studies have addressed the hypothesis that donor FMc is transferred during HSCT or may be implicated in GVHD. The effect of transplanting a small population of non-HLA-matched cells on the development of GVHD is unknown and merits further study.
Prepublished online as Blood First Edition Paper, July 17, 2003; DOI 10.1182/blood-2003-05-1570.
Supported by National Institutes of Health grants HD01264, AI41721, AR48084, CA15704, and CA18029.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal