Abstract
The tyrosine kinase inhibitor STI571 (imatinib) binds competitively to the adenosine triphosphate (ATP) binding site of the ABL kinase, thereby inhibiting auto- and substrate phosphorylation of the oncogenic protein BCR-ABL and preventing the activation of downstream signaling pathways. Comparative studies on leukemic cell samples obtained from chronic myelogenous leukemia (CML) and Philadelphia chromosome–positive (Ph+) acute lymphoblastic leukemia (ALL) patients before and after treatment with STI571 reported point mutations in resistant samples after a short time of therapy. The aim of this study was to determine whether patients with Ph+ ALL in whom resistance developed as a consequence of the Glu255Lys mutation already harbored this subclone prior to STI571 treatment. First, the migration pattern of cDNAs from 30 bone marrow samples from patients with Ph+ ALL was analyzed by polymerase chain reaction–single strand conformation polymorphism (PCR-SSCP). Thereafter, detailed mutational analysis using genomic DNA was performed on initial STI571-naive bone marrow samples of 4 individuals with Ph+ ALL, for whom the mutation Glu255Lys in association with STI571 treatment had been shown. A 166-bp PCR fragment spanning from nucleotide (nt) 862 to nt 1027 was cloned, and 108 clones per sample were analyzed by direct sequencing. This more sensitive technique revealed the presence of the Glu255Lys mutation in 2 initial samples, one clone each. We identified for the first time the mutation Glu255Lys in STI571-naive leukemic samples of Ph+ ALL patients. The findings suggest that the mutation exists in a very small subpopulation of leukemic cells at the beginning of STI571 therapy.
Introduction
The binding of adenosine triphosphate (ATP) to the catalytic domain of the ABL kinase is required for auto- and substrate phosphorylation of BCR-ABL. Increased phosphotyrosine residues on BCR-ABL itself allow it to interact with and phosphorylate effector molecules responsible for activating downstream signaling pathways. The ABL-selective tyrosine kinase inhibitor STI571 (imatinib) binds competitively to the ATP binding site of the ABL-tyrosine kinase,1 thereby preventing the signaling by this oncogenic protein and consequently inhibiting growth of the affected leukemic cells.2
Recently it was shown that in cases of chronic myelogenous leukemia (CML) and Philadelphia chromosome–positive (Ph+) acute lymphoblastic leukemia (ALL), resistance to STI571 is associated with 15 different single amino acid substitutions in 13 distinct positions, most frequently at positions Glu255 and Thr315, known to be important for STI571 binding within the ABL kinase domain.3-9 In particular, the mutation Glu255Lys was detected in one patient with CML and relapsed lymphoid blast crisis.9 In our previous work we performed mutational analysis by direct sequencing of a 714-bp region of ABL (GI6382056) encoding for the ATP binding site and the kinase activation loop.7 Using this technique we could not detect any of the known mutations in the samples naive for STI571. After treatment with STI571 we could detect mutations at the position 255 of ABL in 6 of the samples. Our findings are in very strong agreement with the results from 5 different groups reporting ABL mutations in STI571-resistant cells but not in initial samples obtained from patients with either CML or Ph+ ALL before any treatment with STI571.3-6,8 The short duration of STI571 treatment during which ABL mutations become detectable in resistant Ph+ ALL cells (median time to resistance, 8 weeks) raises the question about the mechanism leading to their development. Conversely it is possible that a small subpopulation of leukemic cells harboring the mutation, which is subsequently selected during treatment with STI571, pre-exists in Ph+ ALL patients. This has recently been shown in several patients with CML.10 To know this would strongly influence the direction of research in the field of resistance to STI571. Therefore, using techniques with increasing sensitivity to detect mutations, we stepwise analyzed STI571-naive bone marrow leukemic cells from patients with Ph+ ALL who had the mutation Glu255Lys in their STI571-resistant cells documented.
Study design
Patient samples
A total of 30 bone marrow samples from 21 patients (including 9 paired samples before and during treatment with STI571) were included in this work. This was a cohort of patients enrolled into consecutive “Phase II study to determine the safety and antileukemic effect of STI571 in adult patients with Ph+ acute leukemias” as described previously.7 Approval was obtained from the institutional review board of the University of Frankfurt/Main for these studies and informed consent was provided according to the Declaration of Helsinki. Since we were unable in our previous work7 to detect any mutation by direct sequencing of polymerase chain reaction (PCR) product in STI571-naive cells we now have reanalyzed the same sample set by more sensitive methods. For detailed mutational analysis by single-clone sequencing we investigated paired bone marrow samples from 4 patients before any treatment with STI571 and during the time of treatment when the patients showed resistance to STI571. The resistant cells of those patients were known to have the mutation Glu255Lys.
Low-density nonadherent bone marrow cells as well as selected CD34+ cells from 3 healthy individuals served as a negative control for the mutation analysis.
To avoid contamination of wild-type DNA samples with mutant DNA we performed the experiments in 2 different laboratories. The STI571-resistant cells were analyzed in the laboratory of H.P.K., Los Angeles, CA. The STI571-naive (initial) cells were analyzed in the laboratory of O.G.O., Frankfurt/Main, Germany.
Reverse transcription–polymerase chain reaction–single strand conformation polymorphism (PCR-SSCP)
Genomic DNA (gDNA) and RNA were extracted from nonadherent mononuclear cells using TRIzol reagent (Life Technologies, Grand Island, NY) according to the manufacturer's protocol. The content of gDNA was adjusted to 20 ng/μL for further analyses. Reverse transcription of RNA was performed as described previously.7 To avoid cross-contamination with gDNA the samples obtained prior to STI571 treatment and the samples obtained while the patients showed resistance to STI571 were processed separately. Negative controls were included in all reactions.
PCR-SSCP analysis using cDNA from all 21 patients including the 9 paired samples before and during treatment with STI571 was performed as previously reported.11 Primers specific for the ATP binding site of ABL were designed using gene bank information GI6382056: ATP–forward (F), 5′-GCG CAA CAA GCC CAC TGT CT-3′;ATP–reverse (R), 5′-GCA CTC CCT CAG GTA GTC CA-3′. Oligonucleotides were synthesized by Life Technologies.
Genomic polymerase chain reaction, cloning, and sequencing analysis
Primers for amplification of a 166-bp fragment of gDNA containing information of the ATP binding site (nucleotide [nt] 862 to nt 1027) of ABL were as follows: 255-F, 5′-CGC ACG GAC ATC ACC ATG AAG-3′; 255-R, 5′-AAT GCC AGC AGA CGC CTT GC-3′. PCR was performed as described previously7 using an annealing temperature of 55°C. PCR products were separated on a 2% agarose gel containing 0.3 mg/mL ethidium bromide and purified using the QIAquick purification system (Qiagen, Valencia, CA) according to the manufacturer's protocol. Purified amplification products were ligated into a pCR2.1-TA cloning vector (Invitrogen, Carlsbad, CA) as described by the manufacturer. DNA fragments were sequenced with an ABI Prism Dye Terminator Cycle Sequencing System (Perkin-Elmer, Foster City, CA) using the plasmid T7 primer binding site.
Results and discussion
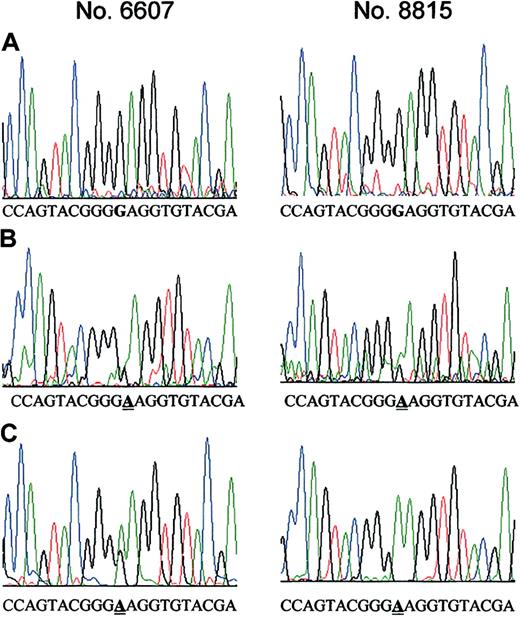
We analyzed 30 cDNAs (including 9 matched samples) obtained from the bone marrow of individuals with Ph+ ALL by PCR-SSCP. None of the samples prior to STI571 showed any change in the migration pattern of the amplified DNA, in agreement with our previous results by direct PCR-product sequencing. Furthermore, we identified identical shifted DNA bands in 4 of 6 STI571-resistant samples having the mutation Glu255Lys, confirming the results from our former analysis (data not shown). PCR-SSCP analysis can find conformation changes of DNA with a minimum detection level of about 2% to 5%. There were 2 STI571-resistant samples having the mutation Glu255Lys that did not show any shifted band. Therefore, we used a more sensitive methodology to clarify whether the Glu255Lys mutation was present in STI571-naive cells. The PCR products generated from gDNA from 4 patients were cloned. For each sample we initially analyzed 108 clones by direct sequencing. From each patient paired bone marrow samples collected prior to STI571 treatment and subsequently at the time of clinical resistance to STI571 were available. The samples from the time of resistance were known to have Glu255Lys from our previous analysis by direct sequencing of the PCR product.7 To ensure that this method was indeed suitable to identify clones from samples known to have the Glu255Lys mutation, we performed limited analysis of clones that were generated from STI571-resistant ALL cells. We identified Glu255Lys in 10% to 30% of these clones. Subsequent screening of a total of 432 clones from the samples obtained prior to STI571 treatment revealed the presence of one clone having the Glu255Lys mutation in 2 samples (6607, 8815; Figure 1). To more rigorously exclude that contamination with mutant DNA affected our experiments, we analyzed an additional 96 clones from each of the STI571-untreated clinical samples which harbored the Glu255Lys mutation in a minor subset. Moreover, low-density nonadherent bone marrow cells as well as selected CD34+ cells from 3 healthy individuals were analyzed. All of the additional clones were negative for the mutation described.
Point mutation Glu255Lys at the ATP binding site of the ABL kinase in bone marrow samples from patients with Ph+ ALL before treatment with STI571. A 166-bp region of ABL, which codes for the ATP binding site, was PCR-amplified using 2 primer pairs and cloned into TA-cloning vector. Sequencing of a total of 432 clones revealed the Glu255Lys mutation in 2 clones established from 2 different STI571-naive bone marrow cell samples. (A) DNA sequence of clones from the STI571-naive cells having the wild-type BCR-ABL sequence. The critical nucleotide (nt 910) is marked in bold. (B) DNA sequence of the 2 clones from the STI571-naive cells carrying the mutation Glu255Lys, which is caused by a G>A change at nt 910. The position of the mutation is underlined. (C) For comparison, clones that were derived from STI571-resistant cells from the same patients that were previously shown by direct DNA sequencing to have the Glu255Lys mutation were sequenced. The nucleotide change G>A at nt 910 was detectable in 10% to 30% of clones from STI571-resistant cells. The position of the mutation is underlined.
Point mutation Glu255Lys at the ATP binding site of the ABL kinase in bone marrow samples from patients with Ph+ ALL before treatment with STI571. A 166-bp region of ABL, which codes for the ATP binding site, was PCR-amplified using 2 primer pairs and cloned into TA-cloning vector. Sequencing of a total of 432 clones revealed the Glu255Lys mutation in 2 clones established from 2 different STI571-naive bone marrow cell samples. (A) DNA sequence of clones from the STI571-naive cells having the wild-type BCR-ABL sequence. The critical nucleotide (nt 910) is marked in bold. (B) DNA sequence of the 2 clones from the STI571-naive cells carrying the mutation Glu255Lys, which is caused by a G>A change at nt 910. The position of the mutation is underlined. (C) For comparison, clones that were derived from STI571-resistant cells from the same patients that were previously shown by direct DNA sequencing to have the Glu255Lys mutation were sequenced. The nucleotide change G>A at nt 910 was detectable in 10% to 30% of clones from STI571-resistant cells. The position of the mutation is underlined.
The development of mutations of ABL has been discussed extensively. So far the frequency and the distribution of such mutations in samples from patients with CML and Ph+ ALL remain uncertain because the number of patients investigated is small. Recently, a report describing BCR-ABL mutations in peripheral blood leukocytes from patients with CML prior to STI571 in 3 of 24 patients10 has been published. Our study, which demonstrates for the first time that the point mutation Glu255Lys in the ATP-binding site is already present prior to treatment with STI571 in Ph+ ALL, is in accordance with these results. The analysis of the clinical course of the patients having the Glu255Lys mutation in their initial bone marrow cells showed different kinetics of clonal selection. Patient no. 6607 initially responded to STI571 treatment, but recurrent leukemia was detectable on day 42 after initiation of STI571 therapy. Conversely, patient no. 8815 never responded to STI571 with 80% bone marrow blasts both prior to STI571 and on day 21 of treatment. One interpretation is that a transient response to STI571 of the initial leukemic population was counterbalanced by an expansion of the resistant mutant population. This shift of subpopulation could be missed clinically in the situation of rapid growth kinetics of the resistant cells. These data support the hypothesis that the inability of STI571 to eradicate ALL cells bearing the Glu255Lys mutation may result in selection of even a rare population of resistant leukemic cells. Furthermore, resistance against STI571 has been suggested to be multifactorial. Indeed, this assumption is in accordance with our previous data7 demonstrating resistance to STI571 in 2 of 9 patients with Ph+ ALL in whom no mutation could be detected. Nevertheless, our current data strongly suggest that the outgrowth of a subclone carrying the individual point mutation under the selective pressure of STI571 is one important mechanism of resistance to this agent. Because of the limited number of samples investigated, we cannot generalize our finding in advanced Ph+ ALL to de novo ALL and CML. A clinical consequence of this finding is the requirement of early combination therapy in which STI571 is given in conjunction with chemotherapeutic agents or other novel signal transduction modifiers.
Prepublished online as Blood First Edition Paper, March 27, 2003; DOI 10.1182/blood-2002-06-1756.
Supported by the Bundesministerium für Bildung und Forschung (BMBF) Competence Network Leukemias, the German Genom Research Network, and the Horst Jung Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank S. Wagner for technical assistance. We also thank A. Binckebanck for the help with collecting the bone marrow samples.
H.P.K. is a member of the Jonsson Comprehensive Cancer Center and holds the endowed Mark Goodson Chair of Oncology Research at Cedars Sinai Medical Center/UCLA School of Medicine.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal