Abstract
X-linked sideroblastic anemia (XLSA) is caused by mutations in the erythroid-specific 5-aminolevulinate synthase gene (ALAS2). XLSA was diagnosed in a 32-year-old woman with a mild phenotype and moderately late onset. Pyridoxine therapy had no effect in the proband, but in her affected son engendered a modest increase in hemoglobin concentration and a 4-fold reduction in ferritin iron. Molecular analysis identified a C to G transversion at nucleotide -206 from the transcription start site, as defined by primer extension, in the proximal promoter region of ALAS2. No other mutations were found in the promoter region, the flanking intronic sequences, the exons, or the 3′ genomic region. The same mutation was found in her affected son but not in any other of her unaffected relatives. The mutation resulted in a 94% loss of activity relative to the wild-type sequence for a luciferase reporter construct containing the proximal 293 nucleotides (nt's) of the ALAS2 promoter when transfected into human erythroid K562 cells. Confirming the mutation's deleterious effect, the ALAS2 mRNA level in the proband's erythroid precursors was reduced 87%. The mutation occurred in or near 3 different putative transcription factor binding sites of unknown erythroid importance. The dramatic decreases in reporter activity and mRNA level suggest that the region of the mutation may bind a novel and important erythroid regulatory element.
Introduction
X-linked sideroblastic anemia (XLSA; OMIM 301300)1 is caused by the deficient activity of the erythroid-specific form of the mitochondrial enzyme, 5-aminolevulinate synthase (E.C. 2.3.1.37; ALAS2).2-4 More than 25 different mutations in the erythroid ALAS2 gene have been identified in XLSA patients.5,6 All of these mutations have been single base substitutions within the region of the enzyme conserved in prokaryotes (encoded by exons 5-11), leading to either an altered amino acid and reduced ALAS2 activity and/or stability or in one case, to an early stop codon resulting in a truncated protein with little or no enzymatic activity.5,7
The clinical phenotype for XLSA is microcytic, hypochromic anemia with iron accumulation. Severity varies widely depending on the amount of residual enzymatic activity, with onset of anemia typically during youth but ranging from birth3,8 to the ninth decade of life.9,10 While probands are frequently males because of hemizygosity of the X-linked defect, females are occasionally affected, most likely because of skewed X-chromosome inactivation in favor of sparing the mutant allele.9
The enzymatic reaction catalyzed by ALAS2 requires pyridoxal 5′-phosphate as a cofactor, and most published ALAS2 mutations have resulted in pyridoxine-responsive phenotypes as measured by consistent, even if slight, increases in hemoglobin (Hb) concentration following vitamin supplementation.5,7 This is particularly apparent if iron overload has been ameliorated by phlebotomy or chelation therapy.6
In this report, we describe a female proband with XLSA who had a mutation in the proximal promoter region of the ALAS2 gene. Her affected son also had this mutation and was mildly pyridoxine-responsive. This mutation is the first promoter mutation identified in XLSA patients and occurred in a potential transcription factor binding site, dramatically reducing the activity of a luciferase reporter construct in erythroid K562 cells and the level of ALAS2 mRNA in the proband's erythroid precursors.
Patients and methods
Case report: family 10
Families 1 to 9 were identified in this laboratory as previously described.6 The proband of family 10 (II-4; Figure 1) was a female of Welsh descent (date of birth, May 16, 1944) who presented at the University Hospital of Wales at age 32 with a long-standing history of tiredness that had worsened over the previous year, breathlessness with physical exertion such as climbing stairs, and excessive perspiration.
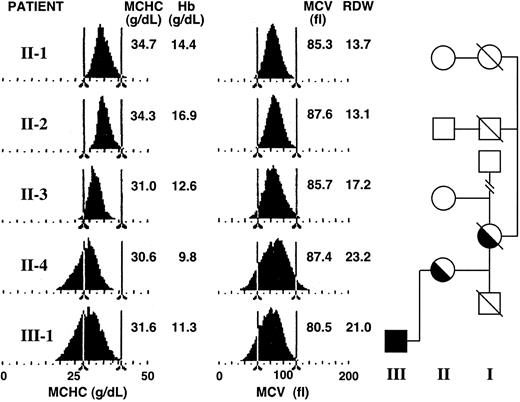
Hematologic profiles of family 10. Red blood cell hemoglobin concentration and red cell volume histograms for members of the proband's (II-4) family.
Hematologic profiles of family 10. Red blood cell hemoglobin concentration and red cell volume histograms for members of the proband's (II-4) family.
The blood film showed anisocytosis, hypochromia, and microcytosis. Laboratory analyses revealed hypochromic anemia with an Hb of 106 g/L (normal range [nl] = 115 to 155) and mean corpuscular hemoglobin (MCH) of 26.0 pg (nl = 27 to 34). There was slight microcytosis (mean corpuscular volume [MCV], 80 fl; nl = 80 to 99). The size distribution of the red cells (Coulter Z-B1 counter and the C2000 Channelyzer; Beckman Coulter, Fullerton, CA) was broad, with a shift toward microcytic cells. The Hb distribution was also broad and strongly shifted toward hypochromic cells. Hemoglobin electrophoresis profile and globin chain synthesis ratio were normal. Her platelet count was low and her white cell count was at the lower limit of normal. Her hematologic picture was essentially unchanged from records at age 26 (Hb, 101 g/L; MCH, 25.9 pg; MCV, 82 fl) after which time she had received at least 20 administrations of oral iron. Analysis of iron at presentation (age, 32 years) showed increased stores (serum ferritin, 982 μg/L, nl = 15 to 200; serum iron, 46 μM, nl = 5 to 25; total iron binding capacity [TIBC], 60 μM, nl = 49 to 78; transferrin saturation, 78%, nl mean = 30%), possibly related to the prior iron supplementation.
The bone marrow was hypererythroid with dyserythropoiesis and 70% ringed sideroblasts. Myelopoiesis and thrombopoiesis were normal. Ferrokinetics showed increased marrow iron turnover (369 μM blood/d; nl = 70 to 140) with 84% ineffective erythropoiesis (nl = 20% to 30%) and a red cell lifespan of 95 days (nl = 65 to 145).11 A putative diagnosis of hereditary sideroblastic anemia was made (based on her relatively young age, her persistently low MCH, and her borderline normal/low MCV) and was confirmed by the presence of ringed sideroblasts in her son at age 8. She was placed on a regimen of folic acid and 150 to 300 mg/d pyridoxine with no change in Hb. Perhaps the response was attenuated because of inhibition by her excess iron. In previous studies, it has been noted that pyridoxine responsiveness can be recovered after removing excess iron stores.6
When seen again around age 49 years, her anemia was unchanged. Representative distribution profiles for her (II-4) and selected relatives are shown in Figure 1. The broadened and shifted values are clear in comparison with her unaffected cousins (II-1 and II-2). Her iron stores were somewhat reduced from their previously elevated levels (serum iron, 14.2 μM; TIBC, 46.2 μM; serum ferritin, 370 μg/L; and transferrin saturation, 31%). The total erythrocyte protoporphyrin was raised (4.7 μM; nl = 0.4 to 1.7). Zinc erythrocyte protoporphyrin was approximately 1.9 μM. Thyroid function, liver function, urea, and electrolytes were normal.
The proband's son (III-1) was born with cleft lip and palate but was hematologically normal (date of birth: December 16, 1973). At 11 years his Hb (122 g/L), MCH (26.0 pg), and MCV (75 fl) were borderline normal/low for his age, while his iron stores were normal (serum iron, 20 μM; serum ferritin, 42 μg/L [nl = 7 to 150], and transferrin saturation, 33%). However, 50% of his erythroblasts were ringed sideroblasts, indicating some excess iron in the marrow. By age 19 his transferrin saturation had increased slightly to about 48% and his serum ferritin was about 193 μg/L (nl = 15 to 300 μg/L). During a subsequent one-year period of pyridoxine supplementation, his serum ferritin decreased (53 μg/L), while his Hb increased slightly from 114 to 129 g/L (Figure 2). This is within the range of response seen in numerous XLSA patients (15-90 μg/L).7 The presence of a fraction of apo-ALAS2 in his reduced level of normal enzyme would be responsive to B6 supplementation. His red cells now show broad size and hemoglobin distributions (Figure 1). Total erythrocyte protoporphyrin was found increased at ages 7 (3 μM) and 19 (9.0 μM). Zinc erythrocyte protoporphyrin at age 19 was approximately 2.3 μM.
Effect of pyridoxine supplementation on hemoglobin and iron stores of the proband's son. In the upper panel, blood hemoglobin concentrations are indicated by closed circles and serum ferritin concentrations by closed squares. In the lower panel, the bar indicates a one-year period of supplementation with 100 mg per day oral pyridoxine.
Effect of pyridoxine supplementation on hemoglobin and iron stores of the proband's son. In the upper panel, blood hemoglobin concentrations are indicated by closed circles and serum ferritin concentrations by closed squares. In the lower panel, the bar indicates a one-year period of supplementation with 100 mg per day oral pyridoxine.
The proband's mother had been anemic (possibly aggravated by a gastric ulcer) and took iron supplements. Her red cell size distribution was broad also—somewhat similar to, but not as marked as, her daughter's. She died at 83 years of age. No other relative had any clinical evidence of XLSA.
Characterization of red cells
MCV and MCHC histograms were determined using the H3 blood cell analyzer (Bayer-Technicon, Tarrytown, NY). Total erythrocyte protoporphyrin was analyzed by the method of Piomelli.12 Zinc protoporphyrin–heme ratios were measured using the ProtoFlour-Z (Helena Laboratories, Gates-head, United Kingdom), and the mean intracellular zinc protoporphyrin concentration was calculated from the MCHC.
Determination of the ALAS2 transcription initiation site by primer extension
A genomic clone (λ5X-E0) was isolated from an unamplified human lymphoid genomic library (designated λ5X)13 by hybridization to the human ALAS2 cDNA,14 and the 12-kilobase (kb) insert was subcloned into pGEM9Z. Portions of the clone were sequenced in both orientations by the dideoxynucleotide chain termination method.15
The human erythroleukemic cell line, K562, was obtained from the American Type Culture Collection (ATCC CCL 243; Manassas, VA) and subcloned on semisoft agar. Individual clones were isolated and assayed for butyrate induction of hemoglobin. A subclone (JC2) was selected that gave 40% to 50% induced cells after 4 days of treatment (data not shown). Cells were maintained at a density of 2 to 5 × 105 cells/mL in Glasgow modified Eagle medium (MEM; Life Technologies, Rockville, MD) with 10% fetal calf serum, 2 mM glutamine, 100 U/mL sodium penicillin G, and 100 μg/mL streptomycin sulfate (all media components were from Life Technologies). Induction was carried out for 3 days in roller bottles with 1.5 mM sodium butyrate. For purification of poly (A)+ RNA, the harvested cells were washed with saline/10 mM EDTA (ethylenediaminetetraacetic acid), and 2.5 × 108 cells were homogenized with 7 mL of 4 M guanidine thiocyanate, 0.5% N-lauroyl sarcosine, 25 mM sodium citrate (pH 7.0), 0.7% 2-mercaptoethanol, and 0.1% antifoam A (Dow-Corning, Midland, MI) in a Potter-Elveham homogenizer.16 The solution was overlaid on 4 mL of 5.7 M cesium chloride in 25 mM sodium citrate, pH 5.0, and centrifuged at 35 000 rpm in an SW41 rotor (Beckman, Palo Alto, CA) for 20 hours at 20°C. The RNA pellets were washed once with 400 μL EtOH and air-dried. Following solution in 3 mL H2O, 10 mL TE (10 mM Tris [tris(hydroxymethyl)aminomethane]–HCl [pH 8.5] and 1 mM EDTA) was added and the RNA extracted twice with phenol-chloroform and once with chloroform. The RNA was precipitated at -20°C overnight after addition of 1:10 volume of 3 N NaOAc and 2 volume EtOH. The precipitate was collected at 10 000g for 20 minutes, washed with 70% EtOH, air-dried, and redissolved in 2 mL TE. Poly (A)+ RNA was selected from 4 to 10 mg total RNA with oligo dT cellulose (Type 3; Collaborative Research, Bedford, MA) as described except for the omission of the 0.1 M NaCl wash.17
For each primer extension annealing reaction, 20 μg poly (A)+ RNA was precipitated with EtOH and then dissolved in 30 μL hybridization buffer containing 10 mM Tris-HCl (pH 7.4), 0.4 M NaCl, 1 mM EDTA, and 5 to 7 pmol end-labeled (γ-32P] adenosine triphosphate [ATP]) ALAS2 primer no. 2060 (Table 1). After incubation at 90°C for 3 minutes, the sample was annealed at 59°C for 2 hours and then quickly chilled in ice water. The primer-annealed RNA was precipitated for one hour at 4°C after the addition of 170 μLH2O and 400 μL EtOH. The pellet was washed with 70% EtOH and air-dried. For primer extension, the pellet was dissolved in 20 μL reverse transcriptase reaction buffer (50 mM Tris-HCl (pH 8.0), 50 mM KCl, 10 mM MgCl2, 5 mM dithiothreitol, 1 mM each deoxynucleoside triphosphate [dNTP], 1 U/μL RNasin, and 50 μg/mL actinomycin D). The reaction was initiated with the addition of 50 units of Super AMV reverse transcriptase (Molecular Genetic Resources, Tampa, FL) and incubated at 42°C for 2 hours. The reaction mixture was then precipitated for one hour with 2 vol EtOH at 4°C. The precipitated pellet was resuspended in 4 μL TE, pH 7.4, and 6 μL loading buffer (95% formamide containing 20 mM EDTA, 0.05% bromophenol blue, and 0.05% xylene cyanol) was added. The primer extension products were analyzed on a 0.4 mm × 40 cm 8% polyacrylamide sequencing gel. The size standard consisted of a sequencing reaction using the identical end-labeled primer as sequencing primer and an ALAS2 genomic clone template (3.3-kb XbaI fragment in pGEM4Z). Sequencing was conducted according to the manufacturer's protocol for Sequenase (USB, Cleveland, OH) with the following modifications: (a) Plasmid DNA (5 μg) was denatured in 20 μL of 0.2 N NaOH. (b) All 4 dNTPs were added and the radioisotope was supplied by 3 pmol of the 5′-end labeled primer. (c) Autoradiography was for 36 hours with one Cronex Lightening Plus (Dow Chemical, Midland, MI) intensifying screen, or for 5 days with no screen.
PCR primers and conditions for amplification of ALAS2 from genomic DNA
Amplified region . | Primer coordinates* . | Primer no. . | Temperature, °C . | Amplicon, bp . | Oligonucleotides† . |
|---|---|---|---|---|---|
| Proximal promoter | UP17888 | 768 | 55 | 367 | 5′-GCCGCCAAGCTT-TTGAGGAGATCTATAGTCAGAGAGG-3′ |
| -293 to +28 | LP18179 | 769 | 55 | 367 | 5′-GCCGCCCCATGGTGGCTTTACCAACAGTACCGGAATGCCGAAC-GAATGACAGGTGGGTACTTGG-3′ |
| Mismatch PCR | UP17956 | 432 | 60 | 327 | 5′-TCCAAAGCCCAAATGAGCTAATCTT-3′ |
| promoter mutation | LP18252 | 128 | 60 | 327 | 5′-GCCGCCGAATT-CAGCTGGCAGACCAGAGATA-3′ |
| Primer extension | LP18203 | 2060 | 71 | NA | 5′-CTGTTGCCCTGCACTGAGGACG-3′ |
Amplified region . | Primer coordinates* . | Primer no. . | Temperature, °C . | Amplicon, bp . | Oligonucleotides† . |
|---|---|---|---|---|---|
| Proximal promoter | UP17888 | 768 | 55 | 367 | 5′-GCCGCCAAGCTT-TTGAGGAGATCTATAGTCAGAGAGG-3′ |
| -293 to +28 | LP18179 | 769 | 55 | 367 | 5′-GCCGCCCCATGGTGGCTTTACCAACAGTACCGGAATGCCGAAC-GAATGACAGGTGGGTACTTGG-3′ |
| Mismatch PCR | UP17956 | 432 | 60 | 327 | 5′-TCCAAAGCCCAAATGAGCTAATCTT-3′ |
| promoter mutation | LP18252 | 128 | 60 | 327 | 5′-GCCGCCGAATT-CAGCTGGCAGACCAGAGATA-3′ |
| Primer extension | LP18203 | 2060 | 71 | NA | 5′-CTGTTGCCCTGCACTGAGGACG-3′ |
UP indicates upper-strand primer; LP, lower-strand primer; and NA, not applicable
Coordinates corresponding to the 5′ ends of the primers are taken from the GenBank genomic clone Z83821
The hyphen internal to the sequence indicates the beginning of the ALAS2 genomic sequence. The annealing temperatures are for the regions to the right of the vertical bar. HindIII, NcoI, and EcoRI sites introduced for cloning purposes are underlined in primers no. 768, 769, and 128, respectively. The mismatched cytosine in primer no. 432 is indicated in bold
Mutation analysis in the proband's ALAS2 gene
Genomic DNA was isolated by standard techniques18 from peripheral blood of the proband and other family members. Amplification and sequencing of each ALAS2 gene exon with 50 to 100 nt of flanking intronic sequences, 1 kb of 5′ flanking sequence, and 350 base pair (bp) of 3′ flanking sequence were accomplished as previously described.3,6 For confirmation of the promoter mutation by restriction analysis, mismatch polymerase chain reaction (PCR) was designed using primers listed in Table 1 and the Expand High Fidelity System (Roche Molecular Biochemicals, Indianapolis, IN). The mismatch in the sense primer (no. 432) in the presence of the mutation created a DdeI site. The restriction digests were electrophoresed in 3.5% MetaPhor agarose (FMC Bioproducts, Rockland, ME). For polymorphism analysis, 120 alleles from healthy unrelated white females were assessed by mismatch PCR and digestion with DdeI.
Molecular analysis of the HFE gene
The Cys282Tyr and His63Asp mutations of the HFE gene were studied as previously described6 using HotSTarTaq DNA polymerase (Qiagen, Valencia, CA). The Cys282Tyr mutation created an additional RsaI site, while the His63Asp mutation resulted in the deletion of a Sau3AI site. The restriction digests were electrophoresed in 2.5% Ultrapure agarose (Life Technologies).
Promoter-reporter gene expression construct with the ALAS2-206G mutation
In order to study the effect of the promoter point mutation, constructs were generated using the promoter-less pGL3 basic luciferase reporter vector (Promega, Madison, WI). DNA from a healthy individual and the proband's son were used as templates to obtain the normal and the mutant promoter sequences, respectively. A GC clamp and a HindIII site were added to the sense primer, while a GC clamp, an NcoI site, and the pGL3 basic sequence from the NcoI site to the HindIII site were added to the antisense primer no. 769 (Table 1). The most proximal 293 bp of the ALAS2 promoter sequence including 28 bp of exon 1 was PCR amplified, and the product was digested by HindIII and NcoI and ligated to the HindIII-NcoI–digested pGL3 basic vector to generate the normal erythroid promoter (Nl-EPr293) and mutant (Mu-EPr293) reporter constructs. The insert and junction sequences in the construct were confirmed in both orientations.
Cell lines, DNA transfections, and reporter assays
The human erythroleukemia cell line K562 (subclone JM; selected on semisoft agar for maximum inducibility by butyrate; J. Ganguly and D.F.B., unpublished data, June 1998) was maintained in suspension in Glasgow Eagle media with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 2 mM glutamine. The 293T human embryonic kidney epithelial cells (ATCC CRL 11 268) were grown attached in Dulbecco modified Eagle medium (DMEM; Life Technologies) with 10% FBS, 1% penicillin-streptomycin, and 2 mM glutamine.
The cell lines were cotransfected with the firefly (Photinus) luciferase promoter-reporter gene constructs described in the previous section and with a Renilla luciferase tyrosine kinase promoter gene construct (pRL-TK; Promega) as an internal transfection control. The pGL3 control vector (Promega) containing the firefly luciferase gene with a tyrosine kinase promoter was used as a positive control. Plasmid DNA was purified using the Qiagen maxi-prep procedure. For K562 cells, transient transfection was performed with the DMRIE-C (1,2-dimyristyloxypropyl-3-dimethyl-hydroxyethyl ammonium bromide-cholesterol) reagent (Life Technologies) according to the manufacturer's instructions. Cells (2 × 106) were resuspended in 100 μL serum-free media with 2 μg firefly luciferase reporter plasmid and 50 ng Renilla luciferase vector. Lipofectamine reagent (Life Technologies) was used for transient transfection of 293T cells according to the manufacturer's recommendations. At 24 hours before transfection, 6-well plates were seeded with 2 × 105 cells/well in antibiotic-free DMEM such that they were approximately 70% confluent at the time of transfection. Firefly and Renilla luciferase vector cDNAs were used at concentrations of 1 μg/well and 10 ng/well, respectively.
Reporter gene expression was quantitated according to the instructions for the Dual Luciferase Reporter Assay System (Promega). Following transfection, cells were grown for 4.5 hours. After addition of 1 mL complete media, the plates were further incubated for a period of 48 hours. Cells were harvested, washed, lysed in a volume of 200 μL, and frozen at -95°C prior to assay. Aliquots (20 μL) were quantitated in 96-well microtiter plates in a DYNEX Model MLX luminometer (Thermo Lab-systems, Chantilly, VA) with a 10-second integration time for each luciferase reaction. Firefly luciferase activity was normalized to the activity of the Renilla luciferase transfection control and expressed as the fold change from the normal promoter activity. Extracts from each transfection were assayed in triplicate for at least 3 independent transfection experiments. The results were expressed as means ± SD.
Quantitation of ALAS2 and ferrochelatase (FECH) mRNA in cultured peripheral erythroid precursors
Mononuclear cells were isolated from 20 mL platelet-depleted peripheral blood obtained from the proband and healthy individuals by Histopaque (Sigma-Aldrich, Dorset, United Kingdom) density centrifugation. After 2 hours of incubation in 200-mL plastic flasks, nonadherent mononuclear cells were cultured at a density of 5 × 105/mL in 0.5 mL volumes of serum-free medium19 containing 0.83% methylcellulose, 1.9 ng/mL interleukin-3, and 2 units/mL recombinant erythropoietin. After 12 days, erythroid bursts from 8 to 16 wells were harvested into ice-cold phosphate-buffered saline (PBS), washed twice with PBS, and counted. Cells were deposited onto slides by cytocentrifugation for morphologic examination, and the remainder (approximately 500 000 cells) were resuspended in 10 volumes of ice-cold RNAlater (Ambion, Austin, TX), left on ice for 1 to 4 hours, and stored at ambient temperature. RNA isolation was performed using Trizol reagent (Invitrogen, Groningen, the Netherlands) following the manufacturer's instructions. K562 RNA for optimization of real-time PCR was isolated using 107 cells.
The reverse transciptase (RT)–PCR primers were designed using mRNA sequences for the ALAS2, FECH, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) genes obtained from the National Center for Biological Information website (http://www.ncbi.nlm.nih.gov) and Primer Express software (Applied Biosystems, Weiterstadt, Germany). The primer sets crossed intron/exon boundaries of genomic sequences ensuring amplification of mRNA only. The respective forward and reverse primers for ALAS2 were CTGCCAGGGTGCGAGATT and TTGGCTGCTCCACTGTTACG; for FECH they were TTGTTCTCTAAGGCCCTGGC and GCGGACAGCTCAGGGTCA; and for GAPDH they were TCATGGGTGTGAACCATGAGA and GCTTAAGCAGTTGGTGGTGCA. cDNA was synthesized from 500 ng to 1 μg of total RNA in 100 μL using random hexamers and TaqMan Reverse Transcription Reagents (Applied Biosystems). Quantitative PCR was performed using 5 μL cDNA with 400 nM of both sense and antisense primers (Invitrogen) in a final volume of 25 μL using the Sybr Green PCR core reagents in an ABI PRISM 7000 Sequence Detection System instrument (Applied Biosystems). Fluorescence was generated by laser excitation of Sybr Green bound to double-stranded DNA. Single transition dissociation curves for all 3 amplicons indicated the absence of nonspecific side reactions and primer dimer artifacts. PCR conditions were as follows: 2 minutes (50°C), 10 minutes (95°C), and 40 cycles of 15 seconds (95°C), and 1 minute (60°C). Measurements were carried out in triplicate for each sample. GAPDH cDNA amplifications were used to control for variability in the initial quantities of cDNA.
To evaluate PCR efficiency for each set of primers, serial dilutions of reverse transcribed K562 RNA (1:10; 1:100; 1:1000) were amplified, and the slopes of plots of ΔCT (test sample cycle threshold (CT) minus control sample CT) versus log cDNA input were found to be 0.1 or less, indicating that amplification efficiency was similar and that relative quantitation of ALAS2, FECH, and GAPDH was valid. The ALAS2-to-GAPDH and the FECH/GAPDH ratios in each sample were calculated using the formula 2-ΔΔCT as defined in the Applied Biosystems User Bulletin.20
Results
Confirmation of the transcription initiation site for human ALAS2
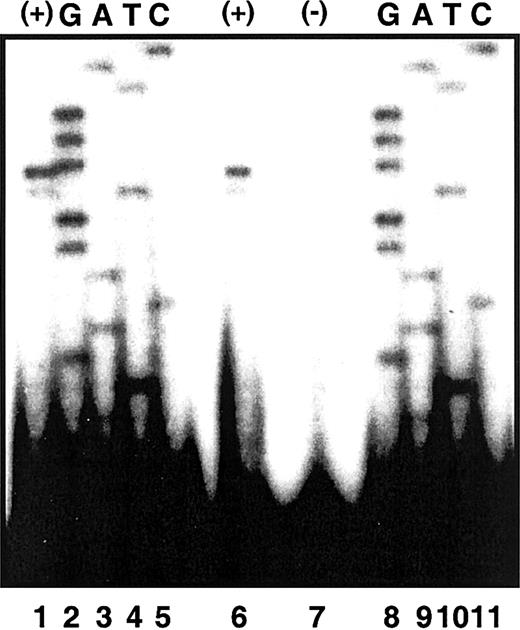
Previously, Cox et al21 had identified 2 ALAS2 initiation sites of equal use 52 and 53 nt upstream from the translation initiation site in mRNA from human fetal liver. To confirm this result, we performed primer extension analyses using poly(A)+ RNA isolated from butyrate-induced K562 cells (“Patients and methods”). A high degree of accuracy in the determination of the extension size was achieved by using the identical end-labeled extension primer (hybridizing to ALAS2 exon 1) to generate a sequencing ladder from an ALAS2 genomic clone. Hence, the identity of the terminal nucleotides of the extension products could be read directly from the antisense sequence of the genomic ladder. Major and minor extension products were observed corresponding to 5′ untranslated regions (UTRs) of 52 and 51 nt, respectively (Figure 3). These results establish the cytosine 52 nt upstream from the translation initiation site as the transcription start site in K562 mRNA. Although the previous study differed from these results in that signals of equal intensity were observed corresponding to 5′ UTRs of 53 and 52 in fetal liver mRNA,21 there too, the same nucleotide was designated as the transcription start site.
Primer extension of ALAS2 poly (A)+ RNA. Electrophoretic separation of primer extension products was carried out in denaturing polyacrylamide gel electrophoresis as described in “Patients and methods.” Lanes 1 and 6 are primer extension products of poly (A)+ RNA by an ALAS2 exon 1 primer. Lane 7 is an extension control with no template. Lanes 2 to 5 and 8 to 11 are sequencing extension products from a genomic ALAS2 clone.
Primer extension of ALAS2 poly (A)+ RNA. Electrophoretic separation of primer extension products was carried out in denaturing polyacrylamide gel electrophoresis as described in “Patients and methods.” Lanes 1 and 6 are primer extension products of poly (A)+ RNA by an ALAS2 exon 1 primer. Lane 7 is an extension control with no template. Lanes 2 to 5 and 8 to 11 are sequencing extension products from a genomic ALAS2 clone.
Characterization of genomic ALAS2 and identification of a point mutation in the ALAS2 promoter
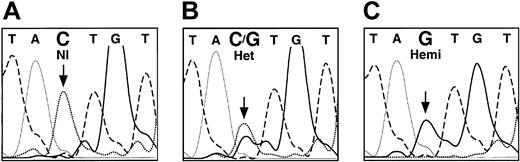
Sequence analysis of a genomic ALAS2 clone (λ5X-E0) demonstrated that 899 bases 5′ of the transcription start site and 109 bases 3′ were identical to nucleotides 17289 to 18297 of GenBank accession number Z83821. The analysis of the 11 exons, the intron-exon junctions, and the 5′ and 3′ flanking sequences of the ALAS2 genomic DNA from the proband's son led to the identification of a single point mutation, a C to G transversion in the ALAS2 promoter (Figure 4). This mutation was 206 bp upstream from the transcription start site (-206C>G, Figure 5). Mismatch PCR analysis of DNA from the proband and additional family members was used to confirm the presence of the mutation. One of the PCR primers (no. 432; Table 1) contained a mismatch near the 3′ end such that in combination with the -206C>G mutation, a DdeI site was created. This analysis showed that the proband was heterozygous and her affected son was hemizygous for the mutation, while the other unaffected family members were healthy (Figure 6A). This mutation was not found in any of 120 alleles in unrelated white females, indicating that this mutation was not a polymorphism (data not shown).
Fluorescent dideoxy sequence analysis of the ALAS2 gene promoter. The bases are distinguished by heavy solid lines (G), light solid lines (A), dashed lines (T), and dotted lines (C). Panel A shows healthy female control; panel B, heterozygous female (proband); and panel C, hemizygous son of the proband. The mutated base is indicated by an arrow.
Fluorescent dideoxy sequence analysis of the ALAS2 gene promoter. The bases are distinguished by heavy solid lines (G), light solid lines (A), dashed lines (T), and dotted lines (C). Panel A shows healthy female control; panel B, heterozygous female (proband); and panel C, hemizygous son of the proband. The mutated base is indicated by an arrow.
Transcription factor motif analysis of the ALAS2 promoter. (A) Sequence of the ALAS2 promoter, exon 1 and part of intron 1. Putative transcription factor binding sites are boxed with the factor identified above and an arrow indicating the orientation relative to the normal convention. The mutation -206C>G is boxed. HpaII sites are underlined. (B) Alignments of consensus sequences (MatInspector matrices) for (i) Cdx2, (ii) RORE, and (iii) MEF2. Dots indicate bases in the normal ALAS2 promoter aligning with the motif consensus. A vertical arrow marks the C that is mutated to a G in the proband.
Transcription factor motif analysis of the ALAS2 promoter. (A) Sequence of the ALAS2 promoter, exon 1 and part of intron 1. Putative transcription factor binding sites are boxed with the factor identified above and an arrow indicating the orientation relative to the normal convention. The mutation -206C>G is boxed. HpaII sites are underlined. (B) Alignments of consensus sequences (MatInspector matrices) for (i) Cdx2, (ii) RORE, and (iii) MEF2. Dots indicate bases in the normal ALAS2 promoter aligning with the motif consensus. A vertical arrow marks the C that is mutated to a G in the proband.
Restriction analysis of the proband and family members. (A) The presence of the -206C>G mutation was detected by mismatch PCR and restriction analysis with DdeI as described in “Patients and methods.” The PCR product in the presence of the mutation had an additional DdeI restriction site. Lanes 1, 2, and 3 showed the restriction fragments from healthy individuals (128, 75, 66, and 58 bp). Lane 4 shows the restricted PCR products of the heterozygous proband (128, 105, 75, 66, 58, and 23 bp), while lane 5 shows the restriction fragments from her hemizygous son (105, 75, 66, 58, and 23 bp). The 23-bp fragment is not resolved on this gel. (B) Lanes 1 and 5 show a normal pattern for Sau3AI digestion of the PCR-amplified region of the His63Asp mutation of the HFE gene (138 and 70 bp; the weak 208 bp band is due to incomplete digestion). Lanes 2 to 4 demonstrate that 3 family members are heterozygous for the His63Asp mutation (208, 138, and 70 bp).
Restriction analysis of the proband and family members. (A) The presence of the -206C>G mutation was detected by mismatch PCR and restriction analysis with DdeI as described in “Patients and methods.” The PCR product in the presence of the mutation had an additional DdeI restriction site. Lanes 1, 2, and 3 showed the restriction fragments from healthy individuals (128, 75, 66, and 58 bp). Lane 4 shows the restricted PCR products of the heterozygous proband (128, 105, 75, 66, 58, and 23 bp), while lane 5 shows the restriction fragments from her hemizygous son (105, 75, 66, 58, and 23 bp). The 23-bp fragment is not resolved on this gel. (B) Lanes 1 and 5 show a normal pattern for Sau3AI digestion of the PCR-amplified region of the His63Asp mutation of the HFE gene (138 and 70 bp; the weak 208 bp band is due to incomplete digestion). Lanes 2 to 4 demonstrate that 3 family members are heterozygous for the His63Asp mutation (208, 138, and 70 bp).
Hemochromatosis (HFE) gene mutation analysis
The proband, son, and 3 family members were screened for HFE mutations Cys282Tyr and His63Asp by PCR and restriction site analysis. No Cys282Tyr mutation was found (data not shown), while the DNA from the proband, her half-sister, and a male cousin all showed a heterozygous pattern for the His63Asp mutation (208, 138, and 70 bp; Figure 6B). Her son and female cousin had the normal pattern (138 and 70 bp). Thus, genetic hemochromatosis was absent in this family.
Functional analyses of the promoter point mutation
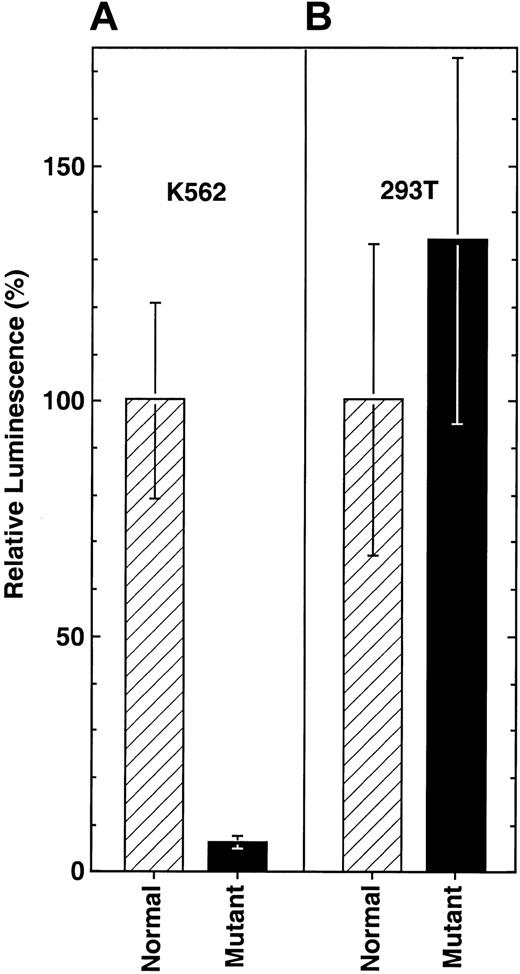
To investigate the effect of the -206C>G point mutation on the function of the ALAS2 promoter, reporter constructs Nl-EPr293 and Mu-EPr293 were used in transient transfections of butyrate-induced human erythroid (K562) cells and untreated human embryonic kidney (293T) cells. For human erythroid K562 cells in the presence of the -206C>G mutation, ALAS2 promoter activity was dramatically decreased to 6.2% of the activity of the normal promoter construct (Figure 7A). In contrast, in nonerythroid 293T cells the -206C>G mutation was associated with a slight increase (34%, but with overlapping standard deviations) in promoter activity relative to the normal promoter construct (Figure 7B).
Transient expression of luciferase reporter constructs for the ALAS2 promoter. Firefly luciferase activity was normalized to the activity of the Renilla luciferase as described in “Patients and methods” and expressed as the fold change from the normal promoter activity in the same assay. (A) Expression in K562 cells. (B) Expression in 293T cells. Results are the average of 2 independent experiments with each sample assayed in triplicate. Error bars indicate one standard deviation from the mean.
Transient expression of luciferase reporter constructs for the ALAS2 promoter. Firefly luciferase activity was normalized to the activity of the Renilla luciferase as described in “Patients and methods” and expressed as the fold change from the normal promoter activity in the same assay. (A) Expression in K562 cells. (B) Expression in 293T cells. Results are the average of 2 independent experiments with each sample assayed in triplicate. Error bars indicate one standard deviation from the mean.
The levels of ALAS2 and FECH mRNA relative to endogeneous control GAPDH mRNA were determined by real-time quantitative PCR as described in “Patients and methods.” ALAS2 mRNA from erythroblasts obtained from the proband's peripheral blood erythroid burst-forming units (BFU-Es) that had been differentiated in culture for 12 days was reduced, on average, 87 ± 8% (n = 3) from that in cells from 2 unaffected individuals. Of interest, FECH mRNA in erythroblasts derived from the proband was reduced 61 ± 28% (n = 3) from that in the cells from 2 unaffected individuals.
Motif analysis of the ALAS2 proximal promoter region
The sequence of the region including the promoter point mutation was analyzed for putative transcription factor binding motifs using the MatInspector program version 5 (Genomatix Software, Munich, Germany) based on perfect matches to the core sequences and optimized overall homology. Alignments of putative transcription factor binding sites with the sequence surrounding the -206 mutation (core similarity, 0.75) were identified for 3 factors including Cdx2, the mammalian caudal-related intestinal transcription factor; MEF2, the myocyte enhancer factor; and a half-site for RORE, a hormone response element (Figure 5B). Additional putative and known sites in the sequence are noted that may play a role in the regulation of ALAS2 transcription, possibly in relationship with the -206 site.
Discussion
It is not common for point mutations in the promoter region to cause human disease. As of March 2003, the statistical tables of the Human Gene Mutation Database indicated that they accounted for 266 (0.8%) of 33 252 known mutations.22 Presumably, because multiple factors support the optimal transcriptional complex, a defect in a single transcription factor's binding may not substantially affect the enzymatic activity of the gene product.23 On the other hand, increasing attention is being focused on regulatory mutations that cause disease. For example, point mutations in 5 different sites in or adjacent to the erythroid Krüppel-like factor (EKLF) binding motif, CACCC, and in all positions of the TATA motif have been found in patients with β-thalassemia.24 Mutations in the erythroid-specific GATA1 binding motif, WGATAR, in gene promoters have been shown to cause δ-thalassemia25 and congenital erythropoietic porphyria.26
In this report, the first regulatory mutation in the ALAS2 gene to cause XLSA was identified. Position -206 in the proximal promoter region of a sideroblastic anemia proband containedaCto G transversion that resulted in moderately late onset of a classical XLSA clinical phenotype in the proband. The proband and her son were the only individuals in this family to have the -206 mutation and to display clinical phenotypes consistent with a defect in heme biosynthesis.
To characterize the mutation's effect with respect to cellular regulation, pGL3 reporter constructs were made containing -293 to +28 of ALAS2 genomic sequence with and without the C to G transversion. Transfection of these constructs in butyrate-induced erythroid K562 cells resulted in a strong (94%) decrease of the activity of the mutated proximal promoter with respect to the wild-type construct. In contrast, this mutation did not lower the luciferase expression in the 293T embryonic kidney cells, indicating that the unmutated site bound (or failed to bind) an erythroid-specific factor. Confirming the functional relevance of the mutation, real-time quantitative PCR demonstrated that the proband's ALAS2 mRNA level in peripheral erythroblasts was reduced 87% relative to 2 healthy control patients.
Thus, the coincidence of the disease phenotype with the presence of the -206 mutation, the lack of detection of any other mutations in the ALAS2 gene, the absence of this mutation in 120 normal alleles, the functional deficit of promoter activity caused by this mutation in luciferase reporter assays, and the 87% reduction in erythroblast ALAS2 mRNA levels all support the -206C>G mutation as the cause of XLSA in this family.
Whereas red cells from female heterozygotes with pyridoxine-responsive XLSA can frequently be separated into 2 distinct populations of red cells of different size and/or cell density (ie, intracellular hemoglobin concentration), the red cell size and hemoglobin density histograms for both the proband and her son showed similar broad distributions (Figure 1). This is consistent with a nearly complete lyonization of the proband's marrow erythrocytes to the mutant genotype, the finding of an 87% reduction in ALAS2 mRNA, and the clinical expression of the XLSA phenotype.
The modest elevation of free erythrocyte protoporphyrin observed in both the proband and her affected son may be due to the observed 61% reduction in FECH mRNA in peripheral erythroblasts cultivated from the proband. The cause of this reduction is unexplained. It is interesting that total protoporphyrin is also somewhat elevated in the homozygous porphyrias27 where, as in this case of promoter down-regulated ALAS2, heme pathway intermediates would be expected to be in low concentration. Nonetheless, most studies have found that free erythrocyte protoporphyrin is low in pyridoxine-responsive anemias (eg, Horrigan and Harris28 ).
The promoter mutation described in this report altered a region of the human ALAS2 gene in which transcription factor binding sites had not previously been identified. Analysis of the mutation site and nearby sequences using motif search programs such as MatInspector Professional (Genomatix Software) and SIGSCAN29 identified 3 candidate transcription factor motifs including Cdx2, MEF2, and RORE (Figure 5B).
Cdx2 is intestine-specific and a member of the caudal-related homeobox gene family. It participates in the development and differentiation of the intestinal epithelium.30-32 With the exception of -206G, the surrounding bases were a perfect match to the matrix consensus (Figure 5B). Although there are no reports showing erythroid expression of Cdx2, one could speculate that some related protein plays a permissive role in ALAS2 expression.
The RORE binding protein (Figure 5B2) is a member of a subgroup of the nuclear hormone receptor superfamily for which no ligands have been identified. It can bind to RORE half-sites as a monomer because of the increased specificity of adjacent AT-rich sequences.33,34 Adjacent ROREs similar to those shown in Figure 5B have been shown to serve as binding sites for retinoic acid receptor and retinoid X receptor heterodimers.35 The expression of nuclear orphan receptor TR4 in hematopoietic cells36 suggests that some nuclear hormone receptor may be important for ALAS2 transcription.
Members of the MEF2 family were originally shown to regulate the transcriptional activity of most muscle-specific genes in myogenesis.37 Subsequently, MEF2 was found to play roles in proliferation and apoptosis in additional cell types.38 The -206G mutation created a nearly perfect match to the MEF2 consensus (Figure 5B), suggesting that an erythroid analog of MEF2 might be a negative regulatory factor.
It had previously been demonstrated that the region between -124 and -293 bp of the human ALAS2 promoter increased reporter gene expression 3-fold in MEL cells and 1.5-fold in K562 cells.39 Thus, some as-yet-unidentified transcription factor could be binding to a response element at the -206C site and activating ALAS2. Further studies of the promoter mutation region will require specific binding assays, such as electrophoretic shift and footprinting in erythroid cells, to uncover the mechanism by which this mutation disrupts the transcription of the ALAS2 gene, perhaps revealing important new functions of the ALAS2 promoter and potentially those of other erythroid-specific genes.
Prepublished online as Blood First Edition Paper, March 27, 2003; DOI 10.1182/blood-2002-06-1623.
Supported in part by grants (R01 DK40895 and DK26824) from the National Institutes of Health (D.F.B.), a grant (584) from the March of Dimes Birth Defects Foundation (D.F.B.), a grant (5897) from the Association Française Contre les Myopathies (S.B.), and a grant (99/57/RG) from the Leukaemia Research Appeal for Wales (A.M.); P.D.C. was the recipient of a March of Dimes Birth Defects Foundation Predoctoral Graduate Research Training Fellowship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Dr Yannick Lemarchand-Brustel for providing resources for real-time quantitative PCR; the Department of Medical Biochemistry, University Hospital of Wales, for erythrocyte protoporphyrin measurements; Roseanne Greenburg, Jayati Ganguly, and Ying Mao for technical assistance; and the proband and family members for their participation in these studies.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal