Abstract
γ-Glutamylcysteine synthetase (γ-GCS) catalyzes the first and rate-limiting step in glutathione (GSH) biosynthesis: the adenosine triphosphate (ATP)–dependent ligation of glutamate and cysteine. γ-GCS consists of a catalytic (γ-GCSH) and modifier (γ-GCSL) subunit. Hereditary deficiency of γ-GCS has been reported in a small number of patients and is associated with low erythrocyte levels of γ-GCS and GSH leading to hemolytic anemia. Here we report a novel γ-GCSH mutation, isolated from the cDNA of 2 related patients diagnosed with γ-GCS deficiency. Each was found to be homozygous for a C>T missense mutation at nucleotide 379, encoding for a predicted Arg127Cys amino acid change. Computerized structure modeling identified that the mutated amino acid lies within a cleft on the protein surface of γ-GCSH, and the border of this cleft was shown to contain Cys249, an evolutionarily conserved residue that has been proven to lie near the binding site of γ-GCSH. Transfection studies showed that the mutation is associated with decreased GSH production, and binding studies using purified recombinant protein showed that the mutant protein has markedly decreased enzymatic activity compared to wild type.
Introduction
Glutathione (l-γ-glutamyl-l-cysteinyl-glycine; GSH) is an important intracellular antioxidant and represents the major cellular nonprotein thiol (> 90%). It is involved in a wide range of cellular reactions such as amino acid transport and protection of cells from damage by oxygen intermediates, free radicals, peroxides, and toxins of both endogenous and exogenous origin. As well, it can act as a source of cysteine for protein formation, reduces disulfides to sulfides, and is involved in the formation of deoxyribonucleotides from ribonucleotides.1-3
GSH is synthesized through the action of 2 enzymes, γ-glutamylcysteine synthetase, (γ-GCS; also known as glutamate-cysteine ligase, GCL) and glutathione synthetase. γ-GCS catalyzes the rate-limiting amide linkage between cysteine and the γ-carboxyl group of glutamate to form the dipeptide γ-glutamylcysteine. Next, glutathione synthetase catalyses the addition of glycine to the cysteine carboxyl group of γ-glutamylcysteine to form the tripeptide γ-glutamylcysteinyl-glycine (glutathione, GSH).
γ-GCS is a heterodimer composed of a heavy subunit (γ-GCSH) and a light subunit (γ-GCSL) that associate, through disulfide binding, to form the holoenzyme. This occurs in response to many cellular insults, particularly oxidative stress.4 The heavy subunit (73 kDa) contains all the catalytic activity for the enzyme and is also the site of feedback inhibition by GSH. It is encoded by a gene located on chromosome 6p12.5 The light subunit (31 kDa) serves a modifying function, increasing the affinity of γ-GCS for its substrates, glutamate and cysteine. It is encoded by a gene on chromosome 1p21.6
Hereditary γ-GCS deficiency is a rare disorder, having been reported in a total of 8 cases.7-11 Patients usually present with a history of recurring bouts of anemia and jaundice, and, in a few cases, neurologic abnormalities. Single-point mutations in the coding sequence of γ-GCSH have been identified in the 2 most recently reported cases of γ-GCS deficiency,8,11 and it is believed that these mutations are the underlying molecular cause of the condition. To further characterize this enzyme deficiency, we established cell lines from 2 sibling patients who were diagnosed with hereditary γ-GCS deficiency. We report here a novel mutation found in each of these patients, a single C>T transversion at cDNA nucleotide 379 in the γ-GCSH gene. We constructed a preliminary molecular model of the γ-GCSH protein, using its amino acid sequence and the chemical structure of the γ-GCS inhibitor, l-buthionine-(SR)-sulfoximine (BSO), and propose that the mutation occurs in the binding pocket of γ-GCSH. Enzymatic assays using purified recombinant catalytic subunit protein showed that the mutation is associated with a marked decrease in γ-GCSH activity, interfering with the enzymatic reaction it catalyses.
Materials and methods
Cell culture and treatments
Approval for the study was obtained from the institutional review board of McGill University. Dermal fibroblasts from each γ-GCS–deficient patient were obtained from Dr Schneider (University of California at San Diego). As control for our experiments, normal human fibroblasts were used. Cells were cultured in RPMI media (Wisent, St. Bruno, QC, Canada) containing 10% (vol/vol) fetal bovine serum and 1% (vol/vol) penicillin/streptomycin and incubated at 37°C and 5% CO2. Previous work with the γ-GCS– deficient cell lines showed slow growth characteristics and senescence after only a few passages. Therefore, to generate enough cellular material for experimentation, each cell line, as well as control, was immortalized using the E6/E7 region of the human papillomavirus.12,13
Sequence analysis
Total RNA was extracted from the fibroblast cells using the high pure RNA isolation kit (Boehringer Mannheim, Indianapolis, IN). cDNA was reverse transcribed from total RNA using a first-strand cDNA synthesis kit (MBI Fermentas, Vilnius, Lithuania). The human gene encoding for γ-GCSH was amplified via polymerase chain reaction (PCR) using primers and conditions according to Ristoff et al.11 Nucleotide sequences were determined on an Applied Biosystems (Foster City, CA) automatic sequencer using a dye terminator.
RNA isolation and analysis
Total RNA was isolated from near-confluent cultures of each cell line (see “Cell culture and treatments”). For Northern analysis, 20 μg total RNA was size fractionated on a 1% agarose-formaldehyde gel and then transferred overnight to a charged nylon membrane (Hybond N+; Pharmacia Biotech, Montreal, QC, Canada). RNA was membrane UV–cross-linked and the membranes were prehybridized for 2 hours at 42°C in hybridization buffer (50% vol/vol formamide, 5 × standard sodium citrate [SSC], 5 × Denhardt solution, 0.5 × sodium dodecyl sulfate [SDS], and 25 μg/mL single-stranded salmon sperm DNA). A PCR-generated probe for γ-GCSH was radiolabeled with 32P-deoxycytidine triphosphate (dCTP) using the oligolabeling kit (Pharmacia Biotech), and added to the blots at an activity of 106 cpm/mL hybridization buffer (dextran sulfate/prehybridization buffer 1:4 vol/vol) overnight at 42°C. Following hybridization, membranes were washed following an increasing stringency gradient in SSC and SDS. Membranes were briefly dried and exposed to radiographic film at -80°C. Autoradiograms were scanned and signals quantified using densitometry software (NIH Image, National Institutes of Health, Bethesda, MD). Stripped membranes were reprobed with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA to control for RNA loading.
Western blot analysis
Cells were grown in 10-cm dishes until 80% confluent. Media were removed and the cells were then lysed in RIPA solution containing 150 mM NaCl, 50 mM Tris (tris(hydroxymethyl)aminomethane)–HCL pH 7.5, 1% (vol/vol) Triton X-100, 0.1% SDS, 2 mM EDTA (ethylenediaminetetraacetic acid), 25 mM NAF, 1 mM Na orthovanadate, 1 mM phenylmethylsulfonyl fluoride (PMSF), 50 μg/mL aprotinin (Sigma, St Louis, MO), and 50 μg/mL leupeptin (Sigma). Protein concentration was determined using the Bradford assay with bovine serum albumin (BSA) as standard. Then, 50 μg protein was electrophoresed through a 10% SDS–polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto a nitrocellulose membrane. The membrane was incubated at 4°C overnight with a 1:20 000 rabbit polyclonal antibody raised against human γ-GCSH (courtesy of Dr Terrance Kavanagh, University of Washington, Seattle), followed by an incubation with a 1:20 000 secondary antirabbit horseradish peroxidase antibody (Biorad, Hercules, CA) for 1 hour at room temperature. Signals were detected following incubation with enhanced chemiluminescence (ECL) solutions (Amersham-Pharmacia Biotech, Piscataway, NJ) and exposure to radiographic film. Signals were scanned and quantified using densitometry software (NIH image). Membranes were reprobed with an anti–γ-GCSL polyclonal antibody (again courtesy of Dr Kavanagh) following the above protocol. Finally, membranes were stripped and similarly probed with an anti-GAPDH antibody to control for protein loading.
Protein modeling
Using the amino acid sequence of γ-GCSH, extensive database searches using BLAST and PSI-BLAST14 did not detect any significant homology matches with known protein structures. Therefore, the fold recognition method as implemented in PROSPECT15 (Oak Ridge National Laboratory, Oak Ridge, TN) was used to search for a structure template that would share similar folds with that of γ-GCSH. The predicted secondary structure from the PHD server16 was used to thread the sequence of γ-GCSH against all the templates in the FSSP database.17 Based on the top score template identified by PROSPECT (pdb entry: 1 gln), an initial 3-dimensional model of γ-GCSH catalytic subunit was built using MODELLER 4.0 (University of California at San Francisco).18 Here, 100 structure models were generated and the one with the lowest objective function was selected for further study. Residue Arg127 was found to lie within a deep cavity on the protein surface. To graphically indicate the cavity location, a γ-GCS inhibitor, BSO, was docked into the cavity. The complex was refined by molecular dynamics as implemented in DISCOVER module of Insight II package (Accelrys, San Diego, CA), using the slightly adapted molecular dynamic procedure demonstrated in our previous work.19 Twenty instant conformations were saved and energy minimized. The lowest-energy conformation was selected as the preliminary model.
Transfection studies
To avoid complications with endogenous γ-GCSH protein and GSH in our transfection experiments, murine γ-GCSH knockout cells were obtained from Dr Michael Lieberman (Baylor College of Medicine, Houston, TX), and were grown and maintained according to his recommendations.20 These cells can be grown only in the presence of GSH-enriched media. The complete cDNA sequence for γ-GCSH was kindly provided by Dr Tim Mulcahy (University of Wisconsin). The 379C>T mutation was introduced into the cDNA sequence using the site-directed mutagenesis kit (Stratagene, La Jolla, CA). The normal and mutated sequences were then cloned into the expression vector pcDNA 3.0 (Invitrogen, Burlington, ON, Canada), sequenced for accuracy, and transfected into the knockout cells using lipofectamine (Invitrogen). Stably expressing cells were selected with G418 and checked for protein expression through Western blotting. For GSH measurement experiments, 20 000 cells of each cell line (knockout, stable wild-type, and stable Arg127Cys) were seeded into 10-cm dishes and grown in complete media containing GSH. Once cells reached 50% confluency, GSH was removed from the media. Cell extracts were then harvested daily for 4 days using RIPA buffer. Protein measurements were performed using the Bradford assay. Equal protein amounts of each cell extract were pipetted into a 96-well plate (final volume 200 μL) and GSH concentrations were determined using monochlorobimane, a dye that fluoresces only after forming an adduct with GSH, and which has been shown to have high specificity for reduced GSH.21-23 Excitation was set at 375 nm and emission at 470 nm. Readings were measured on SpectraMax Gemini XS fluorimeter (Molecular Devices, Sunnyvale, CA). A standard curve was created using varying concentrations of GSH, and then the GSH concentration from each time point was determined. Values are expressed relative to total cell number.
Recombinant protein production
Wild-type and 379C>T cDNA sequences were cloned into the Nde1 and Xho1 sites of pet15b (Novagen, Madison, WI) using PCR-generated restriction sites. Pet-GCSH and pet-Arg127Cys were then used to transform Rosetta cells (Novagen), and 2 clones were selected, sequenced, and grown up for protein expression. Cells (1 L) were grown at 37°C in Luria Broth containing 100 μg/mL ampicillin, until an OD of 0.6 was reached. Isopropyl thiogalactose (IPTG; 0.5 mM) was then added to the culture media, and cells were grown for an additional 6 to 8 hours at 27°C. Cells were harvested following the protocol of Brekken and Phillips,24 except that a talon metal affinity resin (BD Biosciences Clontech, Palo Alto, CA) was used to recover the His-tagged protein, following the manufacturer's recommendations. Protein purity was assessed via Coomassie staining of samples run through a SDS-polyacrylamide gel. Protein purity was estimated to be more than 95%. Purified protein from empty pet15b-transformed Rosetta cells were used as a standard for comparison. Protein concentration was determined via Bradford assay.
Enzyme assays and kinetic measurements
Binding studies used a coupled biochemical reaction, as previously described,25 in which adenosine diphosphate (ADP) formation is coupled to reduced nicotinamide adenine dinucleotide (NADH) oxidation. Reaction buffer (100 mM Tris-HCl, pH 8.0, 150 mM KCl, 2 mM phosphoenolpyruvate, 20 mM MgCl2, 0.27 mM NADH) was mixed with 5 U type II rabbit muscle pyruvate kinase (500 U/mg; Sigma), 10 U type II rabbit muscle lactate dehydrogenase (1150 U/mg; Sigma), 10 mM l-glutamate (l-Glu), 10 mM aminobutyrate (a cysteine substitute), and 5 mM adenosine triphosphate (ATP) in a final volume of 1.0 mL. The reaction was initiated by the addition of purified γ-GCS protein. Reduction in absorbance of NADH at 340 nm and 37°C was followed over time. For kinetic measurements, the concentration of one substrate (l-Glu, 0.1-50 mM; l-aminobutyrate [l-Aba], 0.1-10 mM) was varied while all other reaction ingredients were held constant. For inhibition measurements by GSH, reaction rates were calculated over a range of l-glutamate concentrations (0.1-50 mM) in buffer containing a range of GSH concentrations (0-10 mM), using a competitive inhibition model analysis. Reaction blanks consisted of buffer plus enzyme, but without NADH. All analyses were performed using Sigma Plot 8.02 software (SPSS, Chicago, IL).
Results
γ-GCSH cDNA sequencing
The first step in characterizing γ-GCS deficiency in these 2 patients was to sequence the γ-GCSH gene. cDNA from each patient's cell line was sequenced from nucleotide -88 to 2041 and results compared to published and control (normal fibroblast) sequences. Each patient was found to be homozygous for a cDNA 379C>T mutation in the coding region of γ-GCSH, predicting an Arg127Cys amino acid change. To date, 2 other mutations and 2 polymorphisms have also been reported for γ-GCSH and the results of screening our samples for these are summarized in Table 1.
Summary of mutations and polymorphisms reported in the γ-GCSH gene and subsequent analysis in our γ-GCS-deficient patient-derived cell lines
Report . | Sequence . | Location . | Patient 1 . | Patient 2 . | References . |
|---|---|---|---|---|---|
| Polymorphic trinucleotide repeat | A1: 9 GAG | 5′ UTR cDNA - 10 to -30: 5 alleles | A1 | A2 | 5, 26 |
| A2: 8 GAG | |||||
| A3: 7 GAG | |||||
| A4: 10 GAG | |||||
| A5: 4 GAG | |||||
| Tetranucleotide insertion | CAGC | 3′ UTR cDNA 1972-1975 (f = 0.89) | + | + | 8 |
| Mutation | A > T: predicts an amino acid change of His370Leu | cDNA 1109 | - | - | 8 |
| Mutation | C > T: predicts an amino acid change of Pro158Leu | cDNA 473 | - | - | 11 |
Report . | Sequence . | Location . | Patient 1 . | Patient 2 . | References . |
|---|---|---|---|---|---|
| Polymorphic trinucleotide repeat | A1: 9 GAG | 5′ UTR cDNA - 10 to -30: 5 alleles | A1 | A2 | 5, 26 |
| A2: 8 GAG | |||||
| A3: 7 GAG | |||||
| A4: 10 GAG | |||||
| A5: 4 GAG | |||||
| Tetranucleotide insertion | CAGC | 3′ UTR cDNA 1972-1975 (f = 0.89) | + | + | 8 |
| Mutation | A > T: predicts an amino acid change of His370Leu | cDNA 1109 | - | - | 8 |
| Mutation | C > T: predicts an amino acid change of Pro158Leu | cDNA 473 | - | - | 11 |
RNA and protein analysis
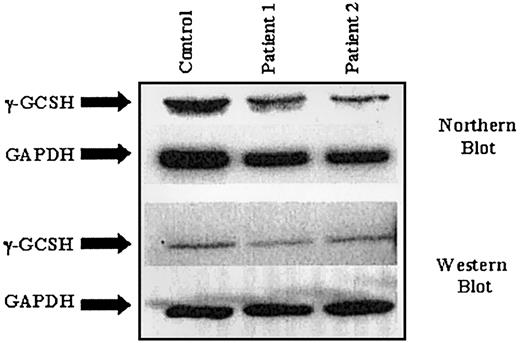
To determine whether γ-GCS deficiency was due to decreased levels of transcription, steady-state γ-GCSH RNA was examined by Northern blot analysis. RNA was isolated from each cell line and size fractionated through a polyacrylamide/formaldehyde gel. RNA membranes were probed using a 600–base pair (bp) 32P-labeled cDNA probe generated from a γ-GCSH–specific PCR. As shown in Figure 1, no significant difference in γ-GCSH RNA levels between either patient cell line and control was observed, when values were normalized to GAPDH. Similarly, no significant difference in γ-GCSL RNA levels was observed (not shown).
Northern and Western blot analysis on samples from 2 patients with γ-GCSH deficiency.
Northern and Western blot analysis on samples from 2 patients with γ-GCSH deficiency.
Similarly, protein expression in these 2 cell lines was studied to determine if the novel mutation causes a decrease in γ-GCSH protein levels or protein instability. Again, as shown in Figure 1, no significant difference was noted between patient and control samples when protein levels were normalized to GAPDH. Likewise, no difference was seen in protein levels of γ-GCSL (not shown).
Location of Arg127 in the γ-GCSH protein
To try and determine the location of the novel mutation and what, if any, effect the mutation might have on the protein structure of
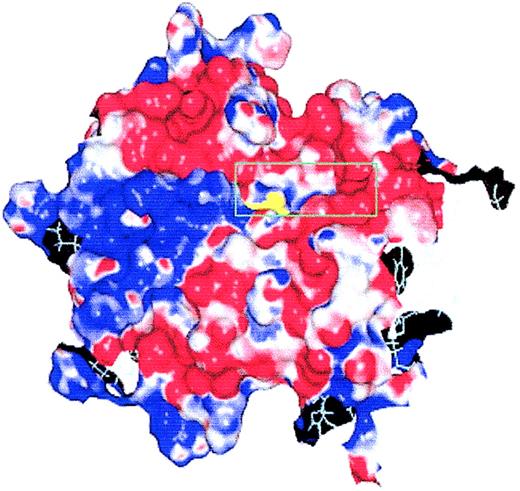
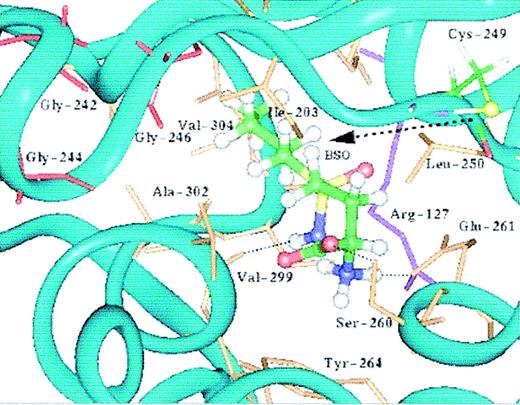
γ-GCSH, we constructed a preliminary computer model, based on the normal amino acid sequence of γ-GCSH, using the programs PROSPECT15 and MODELLER.18 We determined that our mutation lies within a deep cavity on the surface of the γ-GCSH protein, possibly a binding site in γ-GCS (Figure 2). To further delineate the cavity containing the mutation, we docked the γ-GCS inhibitor, BSO, into the cavity and refined the model using the program DISCOVER (Figure 3). Structurally, BSO resembles the dipeptide γ-glutamylcysteine, and it is thought that it inhibits γ-GCS through binding to the active site, thus blocking access to the substrates.26 As Figure 3 demonstrates, we were able to successfully dock BSO into the protein surface cavity and could predict which residues form hydrogen bonds with BSO. Our model identified that amino acid residue Cys249 borders the cleft of this cavity with its sulfur atom projecting toward the cavity interior.
The initial protein surface model of γ-GCSH. Except for Arg127, the molecular surface was colored according to the electrostatic potential in solution (positive in blue, negative in red, neutral in white). Arg127 is colored as yellow. For clarity, only of part of the molecular surface was shown. As shown, Arg127 was found to lie in a cavity on the protein surface.
The initial protein surface model of γ-GCSH. Except for Arg127, the molecular surface was colored according to the electrostatic potential in solution (positive in blue, negative in red, neutral in white). Arg127 is colored as yellow. For clarity, only of part of the molecular surface was shown. As shown, Arg127 was found to lie in a cavity on the protein surface.
The putative catalytic site of human γ-GCSH from the molecular dynamic-refined γ-GCSH-BSO complex. BSO is represented as ball-and-stick and colored according to the atomic coloring scheme (carbon in green, oxygen in red, nitrogen in blue, sulfur in yellow, and hydrogen in white). The side chains of Cys249 (ball-and-stick) and Arg127 (purple stick) as well as residues within 3 Å (0.3 nm) of the bound ligand, BSO, are also shown. The glycine-rich loop involves Gly242, Gly244, and Gly246 (red stick). The -SH group of Cys249 points toward BSO, which forms hydrogen bonds with Ser260, Glu261, and Val299. The -CH3(CH2)3 group of BSO is interacting with the hydrophobic pocket formed by residues Ala302, Val304, and Ile203. Except for BSO and Cys249, the hydrogen atoms are not shown.
The putative catalytic site of human γ-GCSH from the molecular dynamic-refined γ-GCSH-BSO complex. BSO is represented as ball-and-stick and colored according to the atomic coloring scheme (carbon in green, oxygen in red, nitrogen in blue, sulfur in yellow, and hydrogen in white). The side chains of Cys249 (ball-and-stick) and Arg127 (purple stick) as well as residues within 3 Å (0.3 nm) of the bound ligand, BSO, are also shown. The glycine-rich loop involves Gly242, Gly244, and Gly246 (red stick). The -SH group of Cys249 points toward BSO, which forms hydrogen bonds with Ser260, Glu261, and Val299. The -CH3(CH2)3 group of BSO is interacting with the hydrophobic pocket formed by residues Ala302, Val304, and Ile203. Except for BSO and Cys249, the hydrogen atoms are not shown.
Finally, our model identified that there is a glycine-rich loop (Gly242, 244, and 246) situated in close proximity to the cavity containing the mutation.
Glutathione production
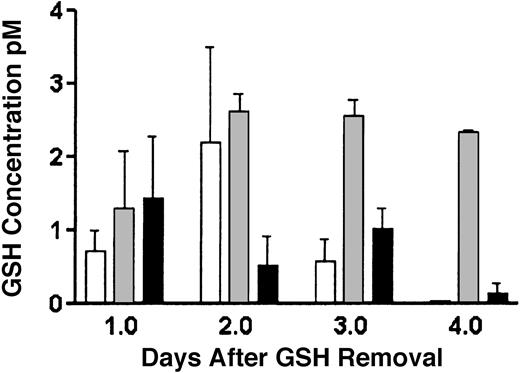
To determine if the novel mutation is associated with decreased glutathione production, stable transfectants of both wild-type and Arg127Cys transfected γ-GCSH knockout cells were made. Because these cells are endogenously deficient in both γ-GCSH protein and glutathione, any GSH production can be attributed to vector expression. As shown in Figure 4, transfection of Arg127Cys mutant γ-GCSH results in a marked decrease in GSH production, compared to wild-type, but maintains a GSH level above untransfected γ-GCSH knockout cells.
GSH determinations by monochlorobimane in γ-GCSH knockout (KO) and stably transfected wild-type (wt-GCSH) and mutant (Arg127Cys) cells. As shown, on GSH removal from culture media, γ-GCSH KO cells (□) quickly deplete in their GSH level. Wild-type γ-GCSH transfectants (▦) maintain an adequate level of GSH production over the 4-day period, whereas mutant Arg127Cys transfectants (▪) are intermediate in GSH production, approximately 10% to 30% of wild type. Values expressed are a ratio of GSH concentration to cell number and represent 3 measurements. Error bars represent SEM.
GSH determinations by monochlorobimane in γ-GCSH knockout (KO) and stably transfected wild-type (wt-GCSH) and mutant (Arg127Cys) cells. As shown, on GSH removal from culture media, γ-GCSH KO cells (□) quickly deplete in their GSH level. Wild-type γ-GCSH transfectants (▦) maintain an adequate level of GSH production over the 4-day period, whereas mutant Arg127Cys transfectants (▪) are intermediate in GSH production, approximately 10% to 30% of wild type. Values expressed are a ratio of GSH concentration to cell number and represent 3 measurements. Error bars represent SEM.
Kinetic measurements using recombinant γ-GCSH protein
To provide experimental evidence to support our hypothesis that the novel mutation may lie in the active binding site of γ-GCSH,we produced purified recombinant wild-type and Arg127Cys protein using a His-tag expression vector. Using this system, we were able to obtain very pure γ-GCSH protein at levels of 1 mg/mL elution buffer. Using a standard biochemical reaction that couples ADP production to NADH oxidation,25 it was found that the mutation is associated with about a 95% decrease in enzymatic activity (Vmax, wild-type [wt]: 531.75 ± 13.77 μmol/h/mg vs Arg127Cys: 26.48 ± 0.97 μmol/h/mg), using the substrates glutamate and aminobutyrate (Table 2). The mutant Arg127Cys enzyme is associated with a lowered Km for both glutamate and aminobutyrate, indicating an increased affinity for the 2 substrates. As well, the mutant enzyme is associated with a marked increased in the Ki for GSH (wt: 0.45 ± 0.04 mM vs Arg127Cys: > 1 M).
Kinetic constants for substrate binding for purified wild-type and Arg127Cys mutant γ-GCSH protein
. | Wild-type GCSH . | Arg127Cys . |
|---|---|---|
| Vmax, μmol/mg/h | 531.75 ± 13.77 | 26.48 ± 0.97 |
| Km glu, mM | 7.16 ± 0.62 | 0.63 ± 0.17 |
| Km aba, mM | 2.39 ± 0.63 | 0.25 ± 0.05 |
| Ki GSH, mM | 0.45 ± 0.04 | > 1000 |
. | Wild-type GCSH . | Arg127Cys . |
|---|---|---|
| Vmax, μmol/mg/h | 531.75 ± 13.77 | 26.48 ± 0.97 |
| Km glu, mM | 7.16 ± 0.62 | 0.63 ± 0.17 |
| Km aba, mM | 2.39 ± 0.63 | 0.25 ± 0.05 |
| Ki GSH, mM | 0.45 ± 0.04 | > 1000 |
Error represents the SEM calculated from 3 separate experiments
Discussion
Despite the importance of the glutathione system in normal and disease cellular biochemistry, little is known about the structure of the active binding site of γ-GCSH, the rate-limiting enzyme involved in GSH biosynthesis. Even though the number of reported cases of γ-GCS deficiency is small, the importance of the glutathione system makes this field of study very interesting and of paramount significance. The glutathione system has many components involved in drug detoxification and excretion including the glutathione-S-transferases (GSTs), the multidrug resistance protein (MRP)/glutathione S-conjugate export (GS-X) pump, and GSH itself. This system has been shown to play a role in a wide range of cellular reactions, particularly the protection of cells from damage by oxygen intermediates, free radicals, peroxides, and toxins of both endogenous and exogenous origin. As well, many components of this system have been implicated in the development of tumor chemoresistance, allowing neoplastic cells to metabolize and excrete chemotherapy drugs at a more efficient rate than their normal counterparts.
Patients with γ-GCS deficiency often present with a clinical history of repeated episodes of hemolytic anemia and jaundice and laboratory data usually reveal reticulocytosis, decreased erythrocyte survival time, erythrocyte GSH deficiency, and markedly diminished γ-GCS activity. The 2 patients studied here were clinically described by Konrad et al,10 and were siblings, brother and sister, of German descent. Each had markedly decreased erythrocyte GSH (∼3% of normal) and γ-GCS activity (∼2%-7% of normal) levels but normal levels of GSH synthetase. Leukocyte (∼40% of normal) and muscle (∼25% of normal) GSH levels were also decreased. Heterozygous family members were found to have intermediate levels of erythrocyte GSH (∼89%) and γ-GCS activity (∼63%). Each patient went on to develop neurologic disease including ataxia and myopathy, thought to be due to spinocerebellar degeneration.27 Because these were the first case reports of γ-GCS deficiency, it was believed that neurologic disease was part of the disorder, but this has not been seen in any further case reports.
In this study, we report a novel single nucleotide mutation, cDNA 379C>T, in the gene encoding the catalytic subunit of γ-GCS. To date, 2 other mutations and 2 polymorphisms have also been reported for γ-GCSH and are summarized in Table 1. The first reported mutation is an A>T transversion at cDNA nucleotide 1109, which predicts a His370Leu amino acid change.8 The second mutation is a C>T substitution at cDNA nucleotide 473, predicting a Pro158Leu amino acid change.11 Our patient cell lines contained neither of these mutations. As well, a series of GAG repeats has been identified in the 5′ untranslated region (UTR) located 10 to 30 bp upstream from the ATG translational start codon.5,28 Five alleles have been reported, each with a different frequency of occurrence depending on ethnic background: A1, 7 GAGs; A2, 8 GAGs; A3, 9 GAGs; A4, 10 GAGs; and A5, 4 GAGs. Of our 2 patient samples, one was found to have allele A1 and the other allele A2. Finally, a polymorphic tetranucleotide insertion (CAGC) has been reported in the 3′ UTR, located at cDNA nucleotide 1972, 61 bp 3′ to the TAG stop codon.8 This insertion appears to be very common because it was found in 85% of healthy people screened, and, in fact, it is likely that the tetranucleotide repeat represents the normal sequence and that the polymorphism is actually a tetranucleotide deletion. Each of our samples contained this CAGC insertion.
Our molecular model showed that the mutation lies in a deep cavity on the protein surface of γ-GCSH and is bordered by residue Cys249 and by a glycine-rich loop (Gly242, 244, and 246). Cys249 has been experimentally shown to lie in or near the glutamate-binding site of γ-GCSH and has been proposed as a putative active site residue of rat γ-GCSH.26 Interestingly, Cys319 in Trypanosoma brucei γ-GCS (which corresponds to mammalian Cys249) has been shown to be the site of enzyme inactivation by cystamine and it is believed that cystamine binds to the sulfur atom in Cys319 in T brucei, thus preventing substrate binding to the enzyme's active site.29 However, the same research has shown that Cys319 does not actually lie within the γ-GCS active site in T brucei because mutagenesis of this amino acid does not affect enzyme activity.24 Therefore, Cys319 is likely to lie near, but not within, the catalytic active site of γ-GCS in T brucei. These experimental data fit with our preliminary protein model of γ-GCSH, and if the same is true for mammalian γ-GCS, then the protein surface cavity outlined by Arg127 and Cys249 may be the γ-GCSH active binding site. As our model identified, the sulfur atom in Cys249 projects toward the cavity interior containing the novel Arg127Cys mutation, and therefore may act in the same way as in T brucei with regard to cystamine enzyme inactivation.
Another piece of our experimental work that supports our hypothesis that the novel mutation lies in and helps to define the catalytic active binding site of γ-GCSH, is the presence of the glycine-rich loop situated near the surface cavity. Because the reaction γ-GCS catalyses is dependent on ATP, one would expect an ATP-binding motif to be located near the active site. The enzymatic reaction is thought to proceed through the transfer of the ATP γ-phosphate group to glutamate to form an intermediate, to which cysteine binds to form the dipeptide.30 A common motif found in phosphate-binding sites is a glycine-rich loop (the P-loop).31 The amino acid sequence of γ-GCSH contains one glycine-rich area, and as stated, our model identified that this loop is located near the cavity in which the novel mutation is located.
Transfection studies and enzymatic kinetic measurements showed that the presence of the mutation is associated with decreased enzymatic activity and GSH production. Interestingly, although the Arg127Cys mutant enzyme had an increased affinity for the substrates glutamate and aminobutyrate (as is apparent from the lowered Km values), its maximum reaction velocity is decreased compared to wild type. This indicates that the enzyme is able to bind substrate but is less efficient at ligation of the substrates to form the dipeptide product, γ-glutamylcysteine. Arg127Cys enzyme had a much higher Ki value than wild-type for GSH, indicating that feedback inhibition by GSH is markedly reduced and would likely never occur at physiologic levels of GSH (1-10 mM). Because the γ-glutamyl moiety of GSH occupies the glutamate-binding site of γ-GCS to achieve its inhibition,26 the introduction of the novel mutation may interfere with this ability. It has been suggested that GSH acts as an inhibitor of the catalytic subunit at normal physiologic levels of GSH, but that this inhibition is lessened on formation of the holoenzyme.32 This is in agreement with our kinetic measurements for wild-type γ-GCSH.
γ-GCS catalyses the first and rate-limiting step in GSH biosynthesis and, as a result, has received much research attention regarding its activity, expression, and regulation. Although the number of reports of GSH synthetase deficiency are numerous,33-35 there are only 8 published reports of γ-GCS deficiency.7-11,27 Deficiency of either enzyme results in hemolytic anemia and jaundice, but patients with GSH synthetase deficiency present with a broader clinical picture, owing to differences in tissue distribution. Those with mild and moderate cases demonstrate GSH deficiency in the red cell compartment and suffer with hemolytic anemia and metabolic acidosis, whereas many of those with severe disease, in which the deficiency is more generalized, die in the neonatal period, suffering with neurologic symptoms, convulsions, and psychomotor development retardation.34 It is interesting that the same clinical spectrum of symptoms has not been reported for patients with γ-GCS deficiency, even though both conditions have the same result—decreased GSH. γ-GCS deficiency may have a more lethal phenotype, so that only mild cases will be diagnosed. Recently, 2 γ-GCS knockout mouse models have been created,20,36 and in each case, the knockout is lethal at embryonic day 8. The knockouts were created via a complete deletion of exon 120 or exons 4 to 637 in γ-GCSH. Therefore, it is not surprising that the knockout mouse models are embryonic lethal, because a large and important part of the γ-GCSH cDNA coding region is deleted in each case. These knockouts result in no, or severely truncated, γ-GCSH protein production. To date, reported cases of γ-GCS deficiency have identified single nucleotide mutations, and, as a result, the effect on γ-GCS is not complete as in the mouse knockouts, thereby allowing the mutated enzyme to demonstrate partial activity. This partial γ-GCS activity is compatible with life, but results in disease symptoms following cellular stress or oxidative damage. An interesting finding from clinical studies of GSH modulation has been the interindividual variation in basal GSH levels in surrogate normal tissues such as peripheral blood mononuclear cells.37 One wonders if some of this variation can be accounted for by heterozygous mutations or polymorphisms in γ-GCSH, such as in the case of family members of γ-GCS–deficient patients.
As is evident from this report and others, even single nucleotide changes in the γ-GCSH gene can affect enzymatic function. This fact in itself suggests the importance of this enzyme in GSH formation and its role in cellular biochemistry. Both of our sibling cases of γ-GCS deficiency were homozygous for a missense mutation in the γ-GCSH gene, and based on our preliminary protein model, we believe that the corresponding mutated amino acid lies in the enzyme catalytic active site. This hypothesis is supported both by our molecular model showing the neighboring location of the glycine-rich P-loop and the conserved Cys249 residue, and our experimental results showing that the mutation is associated with decreased enzymatic activity and glutathione production. We are currently refining our molecular model of γ-GCSH through site-directed mutagenesis studies and are investigating other methods of protein structure determination (eg, nuclear magnetic resonance, crystallography). As well, the crystal structure of Escherichia coli γ-GCS is expected to be published soon and this may give some further insight into the mammalian enzyme.38 Ultimately, detailed information about the catalytic active binding site of γ-GCSH would allow the development of more selective inhibitors of γ-GCS. We have used our molecular model to screen the National Cancer Institute's chemical database for compounds that can bind to the pocket outlined in our model, and thus can act as a potential inhibitor of γ-GCS. From roughly 90 000 compounds, we have identified 35 that are predicted to bind to this site with as much, or greater, affinity as BSO. We are currently testing these compounds with our recombinant protein and in vitro, hoping to develop a more potent, and ideally more selective inhibitor of γ-GCS. With ongoing research of γ-GCS–deficient patients, we can determine how this and other mutations in γ-GCSH lead to decreased enzymatic activity, and the knowledge gained from such research can aid in the development of novel inhibitors of this important enzyme to be used in the treatment of cancer or infectious diseases. As well, these patients offer a means to study the effects of GSH modulation on risk of disease development.
Prepublished online as Blood First Edition Paper, March 27, 2003; DOI 10.1182/blood-2002-11-3622.
Supported by grants from the Canadian Institutes for Health Research, the Cancer Research Society, and Valorization Recherche Québec.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal