Abstract
The liver, skin, and gastrointestinal tract are major target organs of acute graft-versus-host disease (GVHD), the major complication of allogeneic bone marrow transplantation (BMT). In order to gain a better understanding of acute GVHD in the liver, we compared the gene expression profiles of livers after experimental allogeneic and syngeneic BMT using oligonucleotide microarray. At 35 days after allogeneic BMT when hepatic GVHD was histologically evident, genes related to cellular effectors and acute-phase proteins were up-regulated, whereas genes largely related to metabolism and endocrine function were down-regulated. At day 7 after BMT before the development of histologic changes in the liver, interferon γ (IFN-γ)–inducible genes, major histocompatibility (MHC) class II molecules, and genes related to leukocyte trafficking had been up-regulated. Immunohistochemistry demonstrated that expression of IFN-γ protein itself was increased in the spleen but not in hepatic tissue. These results suggest that the increased expression of genes associated with the attraction and activation of donor T cells induced by IFN-γ early after BMT is important in the initiation of hepatic GVHD in this model and provide new potential molecular targets for early detection and intervention of acute GVHD.
Introduction
Allogeneic bone marrow transplantation (BMT) is the treatment of choice for a number of malignant conditions as well as hematologic and metabolic disorders. Acute graft-versus-host disease (GVHD), a major complication of allogeneic BMT, is a rapidly progressing systemic illness characterized by tissue injury in various organs, including the liver, skin, gut, and lymphoid tissues.1,2
Acute GVHD is initiated by donor T cells in response to host antigen-presenting cells (APCs).3 Damage to the intestinal mucosa by the conditioning regimen of BMT, such as irradiation or chemotherapy, allows the translocation of lipopolysaccharide (LPS) into the systemic circulation, which activates host APCs and stimulates monocytes and macrophages to secrete inflammatory cytokines.4 This cytokine cascade synergies with cellular effectors such as cytotoxic T-lymphocytes (CTLs) and natural killer (NK) cells, and results in the amplification of tissue injury.5 In the liver, the histologic signature of acute GVHD is inflammatory cell infiltration into portal areas, bile duct epithelial damage, cholestasis, and endothelialitis.6
We hypothesized that the global quantitative analysis of gene expression would help to illuminate the mechanisms of the complex and multifactorial process of acute GVHD in the liver, particularly early during the disease process when histology is rarely evaluable because of the morbidity of hepatic biopsies early after transplantation. We therefore analyzed hepatic tissue at 2 different time points after BMT in a well-characterized murine model to evaluate the kinetics of changes in gene expression.
Materials and methods
Mice
Female C57BL/6 (B6, H-2b) and B6D2F1 (H-2b/d) mice were purchased from the Jackson Laboratories (Bar Harbor, ME). The age of mice used as BM transplant recipients ranged between 9 and 15 weeks, while the age of donors was 9 to 11 weeks.
BMT and isolation of the liver
Mice underwent transplantation according to a standard protocol described previously.7 On day 0, B6D2F1 mice received 13 Gy total body irradiation (TBI; 137Cs source), split into 2 doses separated by 3 hours to minimize gastrointestinal toxicity. After TBI, 5 × 106 bone marrow cells (BMCs) and 2 × 106 nylon wool–purified splenic donor T cells from allogeneic B6 donors or syngeneic B6D2F1 donors were injected intravenously into recipients.
Mice were housed in sterilized microisolator cages. They received normal chow and autoclaved hyperchlorinated drinking water for the first 3 weeks after BMT, and filtered water thereafter. The liver was harvested at both day 7 and day 35 after syngeneic BMT or allogeneic BMT. Naive nonirradiated mice were also used as controls. Since each group contained 3 mice, a total of 15 mice were evaluated independently.
Histology
Formalin-preserved liver or spleen was embedded in paraffin, and 5-μm thick sections were stained with hematoxylin and eosin for histologic examination. Slides were coded and examined in a blinded fashion by one individual (C.L.) using a semiquantitative scoring system for abnormalities known to be associated with hepatic GVHD.7 The scoring system denoted 0 as normal, 0.5 as focal and rare, 1.0 as focal and mild, 2.0 as diffuse and mild, 3.0 as diffuse and moderate, and 4.0 as diffuse and severe. Scores were added to provide a total score for each specimen.
Preparation of cRNA and microarray hybridizations
Total RNA was isolated from the frozen liver using Trizol reagent (Invitrogen, Carlsbad, CA), followed by cleanup on an RNeasy spin column (Qiagen, Valencia, CA), and then used to generate cRNA probes. cRNA synthesis and hybridizations to Mouse genome U74A Ver.1 Arrays (Affymetrix, Santa Clara, CA) were performed as previously described.8
Microarray analysis
Arrays were scanned and probe intensities extracted from each image (GeneArray scanner and Microarray Suite 4.0; Affymetrix). Each probe-set on the arrays typically consisted of sixteen 25–base pair oligonucleotides complementary to a specific cRNA (perfect match = PM features), and 16 identical probes whose sequence was altered at the central base (mismatch = MM features). Publicly available software was used to process the probe intensities (code and documentation at http://dot.ped.med.umich.edu:2000/pub/BMT/index.html) as previously described.9
An array from a naive mouse sample was chosen as the standard. Probe-pairs for which PM-MM was less than 1000 or for which PM or MM were saturated on the standard were excluded from the analysis. One-sided Wilcoxon signed-rank tests of PM-MM differences for each probe-set on each array were used to help judge whether transcripts were detectable. Gene expression levels were computed to be the mean of the PM-MM differences for each probe-set on each microarray, after discarding the 25% highest and lowest such differences. The standard was scaled to give average expression levels of 1500 units for the 12 654 probe-sets represented on the arrays. The expression levels for each array were then normalized using a piecewise linear function that made 99 evenly spaced quantiles agree with the corresponding quantiles on the standard. Normalized expression levels were log-transformed by mapping x to log(max(x+1,00,0)+100). Fold changes (FCs) were computed as the ratio of group means, after first replacing means less than 100 by 100. One-way analysis of variance models were fit to log-transformed data and pairs of groups compared by the resulting contrast tests. There were 9996 probe-sets analyzed in the correct orientation; the remainder were assay controls or sequences with reversed orientations.
Ribonuclease protection assay (RPA)
RPA was performed using RiboQuant RPA kit (PharMingen, San Diego, CA) according to the manufacturer's instructions. The template sets we used included 3 categories of genes, including caspase families (mAPO-1), chemokines (mCK-5b), and murine customized probe-sets containing intercellular adhesion molecule 1 (ICAM-1) and vascular cell adhesion molecule 1 (VCAM-1). The sample loading was normalized by the housekeeping gene, L32, included in each template set. RPA was performed on 10 μg total RNA hybridized with probes labeled with α-32P uridine triphosphate (UTP). After digestion of ssRNA, the RNA pellet was solubilized and resolved on 6% polyacrylamide gels. Gels were exposed by phosphorimaging and analyzed on a PhosphoImager system, Storm 860 (Amersham Biosciences, Piscataway, NJ). Radioactivity in individual bands (after background subtraction) in comparison with L32 was assessed with ImageQuant software (Amersham Biosciences).
Immunohistochemistry
Livers and spleens fixed in 10% formalin before being embedded in paraffin were subsequently deparaffinized and rehydrated. The tissue sections were heated at 95°C for 25 minutes in Antigen Retrieval Citra solution (BioGenex, San Ramon, CA) to rescue antigen for ICAM-1 staining. Next, they were treated with either a goat antimouse ICAM-1 primary antibody (Ab; Santa Cruz Biotechnology, Santa Cruz, CA) at 1:500 dilution or a rabbit antimouse interferon γ (IFN-γ) primary Ab (Biosource International, Camarillo, CA) at 1:250 dilution. Antibody binding was visualized by either secondary antigoat or antirabbit Vectastain ABC Elite kits (Vector Laboratories, Burlingame, CA) with diaminobenzidine as substrate followed by light counterstaining with methylgreen.
Results
Development of hepatic GVHD after BMT
We first studied the histologic changes of hepatic GVHD in a well-established mouse BMT system,7 in which GVHD is induced by both MHC and minor histocompatibility antigens' differences between donor and recipient. B6D2F1 mice (H-2b/d) received 13 Gy TBI and were injected with 5 × 106 BMCs with 2 × 106 splenic T cells from either allogeneic B6 donors or syngeneic B6D2F1 donors. Livers were harvested at both day 7 and day 35 after BMT and cut into 2 pieces, one for histologic examination and the other for RNA extraction. As shown in Table 1, histologic GVHD was quantitated for 16 individual parameters. Damage at day 35 was readily apparent in all 4 major categories (portal triads, bile ducts/ductules, vessels, and hepatocytes), whereas syngeneic controls without GVHD were identical to naive recipients that did not undergo transplantation. We also evaluated livers at day 7, a time when acute GVHD is histologically evident in other visceral organs, such as the gastrointestinal tract.7 As expected and in contrast to the findings on day 35, liver specimens at day 7 after BMT were identical to controls that did not undergo transplantation and did not show significant evidence of acute GVHD (Table 1).
Liver histopathology before and after BMT
. | Naive . | Syn day 35 . | Allo day 7 . | Allo day 35 . |
|---|---|---|---|---|
| Portal triads | ||||
| Portal tract expansion | 0.3±0.6 | 0.3±0.6 | 0±0 | 1.7±0.7† |
| Neutrophil infiltrate | 0±0 | 0±0 | 0±0 | 1.3±0.6*† |
| Mononuclear cell infiltrate | 0±0 | 0.3±0.6 | 0±0 | 1.3±0.6† |
| Bile ducts/ductules | ||||
| Mononuclear cell infiltrates | 0±0 | 0±0 | 0±0 | 1±0*† |
| Nuclear pleomorphism | 0.3±0.6 | 0±0 | 0±0 | 1±0*† |
| Cytoplasmic eosinophilia | 0±0 | 0±0 | 0±0 | 1±0*† |
| Nuclear multilayering | 0±0 | 0±0 | 0±0 | 0.7±0.6 |
| Pyknotic duct cells | 0±0 | 0±0 | 0±0 | 1±0*† |
| Intraluminal epithelial cells | 0±0 | 0±0 | 0±0 | 0.3±0.6 |
| Vessels | ||||
| Endotheliatis | 0±0 | 0±0 | 0±0 | 1.7±0.6*† |
| Mononuclear cells around CV | 0±0 | 0±0 | 0±0 | 0.7±0.6 |
| Hepatocytes | ||||
| Confluent necrosis | 0±0 | 0±0 | 0.3±0.6 | 0±0 |
| Acidophilic bodies | 0.3±0.6 | 0.7±0.6 | 1.3±0.6 | 1.7±1.2 |
| Mitotic figures | 0±0 | 0±0 | 0±0 | 1±0*† |
| Neutrophil accumulations | 0.7±0.6 | 0±0 | 0±0 | 1±0*† |
| Macrophage aggregates | 0.3±0.6 | 0±0 | 0.3±0.6 | 1±0* |
| Total score | 2.0±2.6 | 1.3±1.5 | 2.0±1.7 | 16.3±2.9*† |
. | Naive . | Syn day 35 . | Allo day 7 . | Allo day 35 . |
|---|---|---|---|---|
| Portal triads | ||||
| Portal tract expansion | 0.3±0.6 | 0.3±0.6 | 0±0 | 1.7±0.7† |
| Neutrophil infiltrate | 0±0 | 0±0 | 0±0 | 1.3±0.6*† |
| Mononuclear cell infiltrate | 0±0 | 0.3±0.6 | 0±0 | 1.3±0.6† |
| Bile ducts/ductules | ||||
| Mononuclear cell infiltrates | 0±0 | 0±0 | 0±0 | 1±0*† |
| Nuclear pleomorphism | 0.3±0.6 | 0±0 | 0±0 | 1±0*† |
| Cytoplasmic eosinophilia | 0±0 | 0±0 | 0±0 | 1±0*† |
| Nuclear multilayering | 0±0 | 0±0 | 0±0 | 0.7±0.6 |
| Pyknotic duct cells | 0±0 | 0±0 | 0±0 | 1±0*† |
| Intraluminal epithelial cells | 0±0 | 0±0 | 0±0 | 0.3±0.6 |
| Vessels | ||||
| Endotheliatis | 0±0 | 0±0 | 0±0 | 1.7±0.6*† |
| Mononuclear cells around CV | 0±0 | 0±0 | 0±0 | 0.7±0.6 |
| Hepatocytes | ||||
| Confluent necrosis | 0±0 | 0±0 | 0.3±0.6 | 0±0 |
| Acidophilic bodies | 0.3±0.6 | 0.7±0.6 | 1.3±0.6 | 1.7±1.2 |
| Mitotic figures | 0±0 | 0±0 | 0±0 | 1±0*† |
| Neutrophil accumulations | 0.7±0.6 | 0±0 | 0±0 | 1±0*† |
| Macrophage aggregates | 0.3±0.6 | 0±0 | 0.3±0.6 | 1±0* |
| Total score | 2.0±2.6 | 1.3±1.5 | 2.0±1.7 | 16.3±2.9*† |
Parameters of liver histopathology from naive mice or BMT recipients at either day 7 or day 35 after BMT were scored from 0 to 4 depending on extent and severity of injury as described in “Materials and methods.” Results represent means ± standard deviations. The Wilcoxon rank-sum test was used for the statistical analysis. Most parameters of injury are significantly worse in allo day 35 group versus syn day 35 group or versus allo day 7 group
All values were 0±0 for syn day 7 group
Syn indicates syngeneic; allo, allogeneic; and CV, central vein
P = .05
P = .05
Gene expression profile in the liver at day 7 and day 35
We next evaluated gene expression changes in the liver at both days 7 and 35 after BMT using oligonucleotide microarray. The Affymetrix MG_U74A(v1) chips contain 9996 probe-sets in correct orientation. On day 7, one syngeneic array with high background was excluded from analysis. Preliminary experiments using duplicate arrays for the same target (labeled cDNA) showed extremely high correlations averaging 0.995. The average correlation coefficient between arrays of the 3 biologic replicates was also very high at 0.975.
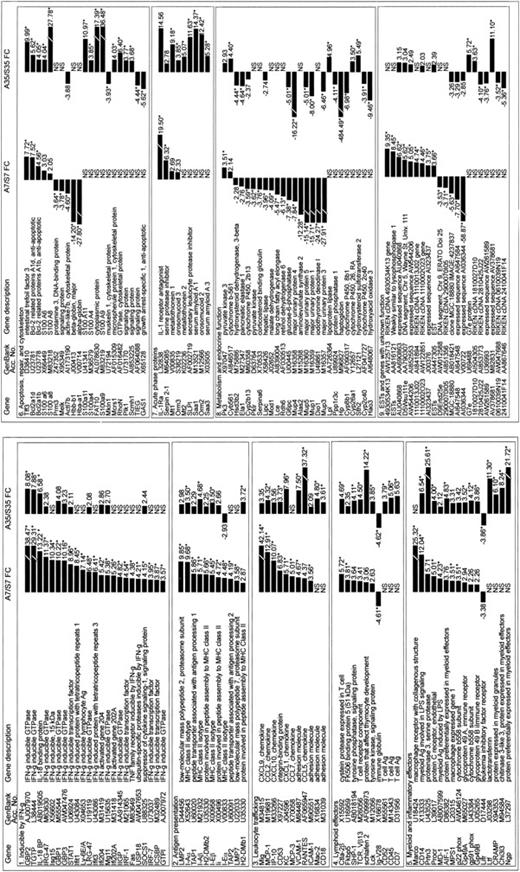
We analyzed only those probe-sets for which at least 2 of 14 arrays were statistically significant (P < .05) for a one-sided signed-rank test of PM-MM higher than 0, reducing the analysis to 7329 probe-sets. We then fit a one-way analysis of variance model to the 5 groups. Allogeneic and syngeneic BMT samples were compared for the expression level of each gene at either day 7 or day 35. We obtained significant (P < .01) differences in 456 probe-sets for day 7 and 554 probe-sets for day 35. We expected differences in only 73 (0.01 × 7329) probe-sets by chance alone, less than one sixth of the number actually obtained. In order to select only the largest transcriptional alterations, and to reduce further the possibility of false positives, we report here only those genes that gave 3.5 FCs or greater in addition to giving a P value less than .01.
A total of 108 nonredundant genes were up-regulated at either day 7 or day 35 in the liver after allogeneic BMT by these criteria, whereas 40 nonredundant genes were down-regulated. More than half of the genes that differed at one time point showed the evidence of similar changes at the other time points. Of 95 probe-sets altered (P < .01, FC > 3.5) at day 7, only 24 failed to yield a difference (P < .05) at day 35, whereas for the 87 probe-sets altered at day 35, 41 were not different (P < .05) at day 7.
The list of genes that differed by 2-fold or greater giving P value less than .01, which contains 356 probe-sets and thus may cover a broad range of GVHD-related genes, is available on the web (http://dot.ped.med.umich.edu:2000/pub/BMT/index.html).
Ribonuclease protection assay of genes represented in the oligonucleotide array
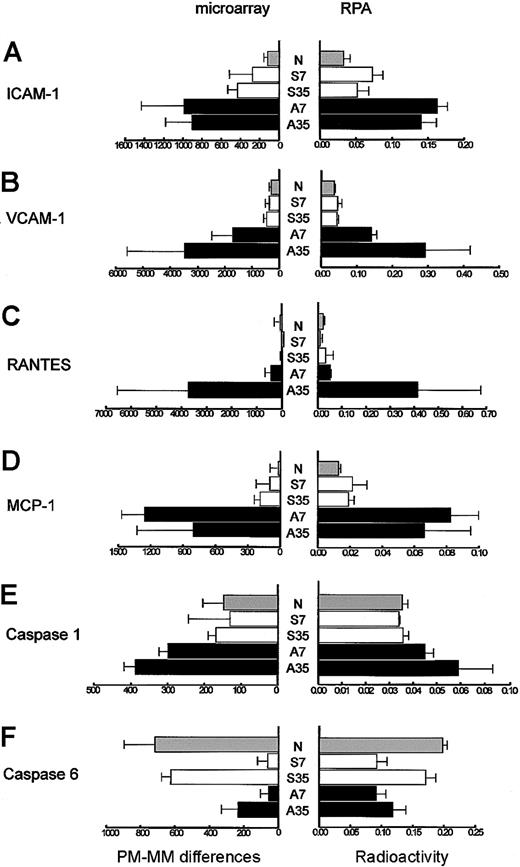
To validate the results of oligonucleotide array analysis, expression levels of 6 individual genes from 3 different categories (adhesion molecules, chemokines, and caspases) were analyzed by RPA (Figure 1). Regression analysis of these 6 separate genes demonstrated a close correlation between these 2 methods of measurement at both time points analyzed (P < .01). The magnitude of the decrease for caspase-6 gene expression was greater as measured by microarray than RPA, although there was a statistically significant correlation between the 2 methods.
A comparative analysis of microarray and RPA of a subset of genes increased in the liver after allogeneic BMT. (A) ICAM-1. (B) VCAM-1. (C) RANTES. (D) MCP-1. (E) Caspase-1. (F) Caspase-6. mRNA was prepared from the livers of naive mice (N), mice after syngeneic BMT at day 7 (S7) or day 35 (S35), and mice after allogeneic BMT at day 7 (A7) or day 35 (A35). The data represent the average of microarray and RPA, and error bars indicate standard deviation. The x-axis of microarray represents PM-MM differences of each gene, while the x-axis of RPA represents the radioactivity of each gene relative to housekeeping gene L32. Statistically significant correlation of each gene was verified from the comparison of expression level between microarray and RPA using the linear regression analysis (P < .01).
A comparative analysis of microarray and RPA of a subset of genes increased in the liver after allogeneic BMT. (A) ICAM-1. (B) VCAM-1. (C) RANTES. (D) MCP-1. (E) Caspase-1. (F) Caspase-6. mRNA was prepared from the livers of naive mice (N), mice after syngeneic BMT at day 7 (S7) or day 35 (S35), and mice after allogeneic BMT at day 7 (A7) or day 35 (A35). The data represent the average of microarray and RPA, and error bars indicate standard deviation. The x-axis of microarray represents PM-MM differences of each gene, while the x-axis of RPA represents the radioactivity of each gene relative to housekeeping gene L32. Statistically significant correlation of each gene was verified from the comparison of expression level between microarray and RPA using the linear regression analysis (P < .01).
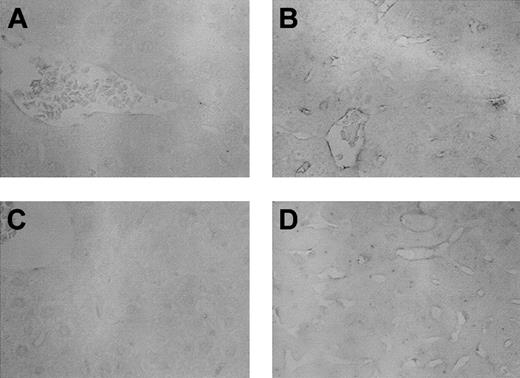
Immunohistochemistry of ICAM-1
The changes in gene expression were further evaluated by immunohistochemistry of ICAM-1 protein. Increased gene expression of ICAM-1 was detected by RPA and microarray as described in Figure 2 on both day 7 and day 35 after allogeneic BMT. Using the immunohistochemical techniques described in “Materials and methods,” increased expression of ICAM-1 protein was observed in portal vein endothelium, and sinusoidal endothelial cells in the liver after allogeneic BMT of the animal (Figure 2).
Immunohistochemistry of ICAM-1. Liver sections were obtained after BMT at either day 7 or day 35 and tested for the protein for ICAM-1 as described in “Materials and methods.” (A) Day 7 after syngeneic BMT; (B) day 7 after allogeneic BMT; (C) day 35 after syngeneic BMT; and (D) day 35 after allogeneic BMT. ICAM-1 protein was clearly detected in the endothelium of portal vein and hepatic sinusoids after allogeneic BMT (B,D). No unspecific binding of the isotype control was detected (not shown). Methylgreen counterstain; original magnifications, × 400.
Immunohistochemistry of ICAM-1. Liver sections were obtained after BMT at either day 7 or day 35 and tested for the protein for ICAM-1 as described in “Materials and methods.” (A) Day 7 after syngeneic BMT; (B) day 7 after allogeneic BMT; (C) day 35 after syngeneic BMT; and (D) day 35 after allogeneic BMT. ICAM-1 protein was clearly detected in the endothelium of portal vein and hepatic sinusoids after allogeneic BMT (B,D). No unspecific binding of the isotype control was detected (not shown). Methylgreen counterstain; original magnifications, × 400.
Patterns of gene expression change following BMT
We divided genes that were up- or down-regulated after allogeneic BMT into 9 different categories: (1) inducible by IFN-γ; (2) antigen presentation; (3) leukocyte trafficking; (4) lymphoid effectors; (5) myeloid and inflammatory effectors; (6) apoptosis, repair, and cytoskeleton; (7) acute phase proteins; (8) metabolism and endocrine function; (9) expressed sequence tags (ESTs) and genes with unknown function.
Inducible by IFN-γ
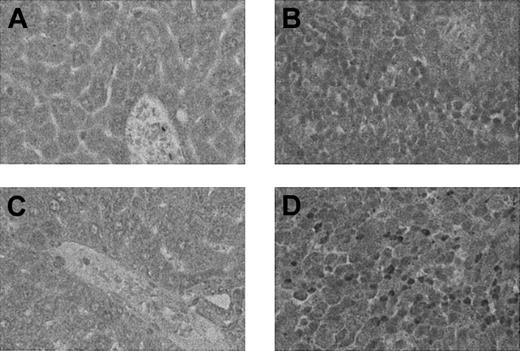
Of the 9 different categories evaluated, the category in which the largest number of genes was up-regulated at day 7 was that of IFN-γ–inducible genes, such as transcription factors and signaling proteins (Figure 3). For example, the expression of signal transducer and activator of transcription-1 (STAT1), which translocates to nucleus upon phosphorylation by IFN-γ receptor, was increased almost 9-fold on day 7 after allogeneic BMT. Activated STAT1 sequentially promotes the expression of a number of transcription factors such as interferon regulatory factor-1 (IRF-1), IRF-7, and IFN consensus sequence binding protein (ICSBP),10 all of which were also increased at this time. Interestingly, we could not detect increased expression of the IFN-γ gene itself, although many IFN-γ–inducible genes were up-regulated in the liver at day 7. We therefore performed immunostaining for IFN-γ at day 7 after BMT in both the liver and spleen. As shown in Figure 4, IFN-γ protein was not detected in the liver, although it was clearly evident in the spleen after allogeneic BMT, consistent with earlier observations demonstrating expansion of IFN-γ–secreting donor T cells by day 7 after BMT in this model.11,12 Thus, IFN-γ induces the expression of genes in an endocrine-type fashion, acting at a distance.
Differentially expressed genes after BMT. Recipient mice underwent transplantation and their livers were harvested on days 7 and 35 after BMT as described above. Total RNA was isolated from the harvested liver samples, and microarray analysis was performed using the mouse genome U74A Ver.1 Arrays oligonucleotide microarrays as described in “ Materials and methods. ” Allogeneic and syngeneic BMT samples were compared for the expression level of each gene at either day 7 or day 35. Those genes demonstrating the largest transcriptional alterations, a 3.5-fold change (FC) or greater, are shown (*P < .01).
Differentially expressed genes after BMT. Recipient mice underwent transplantation and their livers were harvested on days 7 and 35 after BMT as described above. Total RNA was isolated from the harvested liver samples, and microarray analysis was performed using the mouse genome U74A Ver.1 Arrays oligonucleotide microarrays as described in “ Materials and methods. ” Allogeneic and syngeneic BMT samples were compared for the expression level of each gene at either day 7 or day 35. Those genes demonstrating the largest transcriptional alterations, a 3.5-fold change (FC) or greater, are shown (*P < .01).
Immunohistochemistry of IFN-γ. Liver (A,C) and spleen (B,D) sections were obtained at day 7 after syngeneic BMT (A-B) and allogeneic BMT (C-D) and tested for IFN-γ protein by immunohistochemistry as described in “Materials and methods.” Abundant IFN-γ protein was detected with peroxidase staining as brown granules in the spleen (D) but not in the liver (C) after allogeneic BMT. IFN-γ staining was detected in neither liver nor spleen from syngeneic BMT (A-B). Methylgreen counterstain; original magnifications, × 400.
Immunohistochemistry of IFN-γ. Liver (A,C) and spleen (B,D) sections were obtained at day 7 after syngeneic BMT (A-B) and allogeneic BMT (C-D) and tested for IFN-γ protein by immunohistochemistry as described in “Materials and methods.” Abundant IFN-γ protein was detected with peroxidase staining as brown granules in the spleen (D) but not in the liver (C) after allogeneic BMT. IFN-γ staining was detected in neither liver nor spleen from syngeneic BMT (A-B). Methylgreen counterstain; original magnifications, × 400.
IFN-γ–inducible GTPases, such as guanylate-binding protein 2 (GBP2), T-cell–specific guanine nucleotide triphosphate-binding protein (TGTP), IRG-47, GBP1, GBP3, LRG-47, Mg11, IIGP, and GTPI,10,13 comprise the largest group in this category of IFN-γ–inducible genes. Of interest is the 13-fold increase of interleukin-18 binding protein (IL-18 BP), which specifically binds to and neutralizes IL-18, an inducer of IFN-γ.14 Administration of IL-18 after allogeneic BMT increases IFN-γ–dependent activation-induced cell death of donor T cells, suggesting that IL-18 may act as a regulatory mechanism for acute GVHD.12 Ifi204 and Ifi202A, both of which inhibit cell proliferation, were also increased and may partially mediate the antiproliferative effect of IFN-γ.15 Similarly, suppressor of cytokine signaling-1 (SOCS1) inhibits Janus kinase (JAK) activities and negatively regulates signaling of cytokines such as IFNs, IL-2, IL-4, IL-6, IL-13, leukemia-inhibitory factor, stem cell factor, and erythropoietin.16 The Fas/Fas ligand (FasL) pathway is one of the principle components of T-cell cytotoxicity.17 The vast majority of genes in this category showed increased expression at day 7, but did not remain elevated in the liver when histologic damage was evident (day 35).
Antigen presentation
Mice possess 2 classical MHC class II isotypes, the I-A and I-E molecules, each composed of α and β chains.18 In the livers of allogeneic BMT recipients, I-Aα, I-Aβ, I-Eα, and I-Eβ were all up-regulated at day 7 and several genes remained elevated at day 35, whereas no increase of MHC class I gene expression was observed. Gene expression was also increased for low-molecular-mass polypeptide 2 (LMP2) and LMP7, proteasome subunits that enhance the production of peptides capable of association with MHC class I molecules.19 The transporter associated with antigen processing 1 and 2 (TAP1 and 2) genes, which translocates peptides generated by proteasomes into endoplasmic reticulum, was also increased. In addition, there was increased expression of MHC-DMs, whose presence in lysosomes facilitates the assembly of MHC class II molecules with peptides from exogenous proteins.18,20 Thus, genes related to peptide processing for both MHC class I and class II were up-regulated.
Leukocyte trafficking
Most leukocyte trafficking–related genes involving adhesion molecules and chemokines were up-regulated after allogeneic BMT on both day 7 and day 35. There was increased expression of several chemokines that activate leukocyte cell-surface integrins, which specifically bind to adhesion molecules.21 VCAM-1 and ICAM-1, adhesion molecules expressed on endothelial cells, are critical for the migration of leukocytes into inflamed tissue.21 CD18, a common component of lymphocyte function-associated antigen-1 (LFA-1) and Mac-1, is the ligand for ICAM-1.21
Both CC chemokines such as monocyte chemoattractant protein-1 (MCP-1) (CCL2), MCP-3 (CCL7), and regulated by activation, normal T-cell expressed and secreted (RANTES; CCL5), and CXC chemokines such as Mig (CXCL9), IP-10 (CXCL10), and KC (CXCL1), were up-regulated in hepatic GVHD. These chemokines are classified as inflammatory chemokines because they are induced by inflammatory cytokines such as IFN-γ and tumor necrosis factor α (TNF-α), and are necessary for the recruitment of effector cells into tissues.22,23 RANTES was profoundly up-regulated (37-fold) at day 35, consistent with the infiltration of effector cells into liver parenchyma observed at that time (Table 1).
Lymphoid effectors
As expected, many T-cell antigens such as TCR, CD7, CD45, and CD52 were up-regulated in livers after allogeneic BMT. FK506 binding protein 5 (Fkbp5), a T-cell–specific immunophilin family member that inhibits calcineurin formation, causes decreased production of IL-2 after T-cell activation.24 SHP-1 is a cytosolic tyrosine phosphatase that negatively regulates T- and B-cell receptor activation.25 Down-regulation of immunoglobulin gene V28 (Ig-V28) is consistent with the immunosuppression and B-cell lymphopenia associated with GVHD.26 Schlafen 2 is preferentially expressed by lymphoid cells during positive selection in the thymus.27 Most genes in this category were predominantly expressed at day 35, again consistent with the infiltration of effector cells observed at this time (Table 1).
Myeloid and inflammatory effectors
Antigens or cell-surface receptors expressed in myeloid cells and genes related to their functions were up-regulated in livers after allogeneic BMT. Marco, a member of the scavenger receptor family, is induced by LPS,28 an inflammatory molecule that is found in the systemic circulation in this GVHD model.7 CD14 binds to LPS and stimulates TNF-α secretion from macrophages.29 Expression of MD-1, another gene implicated in LPS signaling,30 was also increased at day 7. Gp49A and 49B, which have close sequence similarity to the human killer inhibitory receptor of NK cells, are expressed in myeloid cells,31,32 mast cells,33,34 and activated NK cells31 in mice and were increased on day 35 when tissue damage was evident.
Prominent increases were observed for the genes encoding proteinase 3,35 lactotransferrin (Ltf),36 cathelin-related antimicrobial peptide (CRAMP),37 and neutrophilic granule protein (Ngp),38 which are abundantly present in the granules of myeloid cells and mediate antimicrobial activity of granulocytes. Phagocyte cytochrome b558, a heterodimer composed of p22phox39 and gp91phox,40 generates the superoxide radical (
Most genes in this category were predominantly up-regulated at day 35, again consistent with the inflammatory infiltrates that were observed at this time. Genes induced by LPS or implicated in LPS signaling, such as Marco, CD14, and MD-1, were preferentially expressed at day 7, when LPS translocation across the injured gastrointestinal tract during acute GVHD.7 Activation of the innate immune response by molecules such as LPS may be important for the subsequent activation of adaptive immunity, and both play roles in acute GVHD.41,42
Apoptosis, repair, and cytoskeleton
Apoptosis is the primary histologic feature of GVHD target tissue destruction. Bcl-2–related proteins A1d (Bcl2a1d) and A1b (Bcl2a1b) are antiapoptotic Bcl-2 family proteins induced by inflammatory cytokines43 and were up-regulated in the liver after allogeneic BMT. Intestinal trefoil factor 3 (Tff3) is induced during the biliary disease in the liver44 and facilitates tissue repair.45,46 S100 family proteins regulate cytoskeleton constituents and modulate cell growth and differentiation at the site of chronic inflammation.47 Among S100 family proteins, S100a8 and S100a9 form heterodimers that regulate several aspects of inflammatory response, and their expression is restricted to neutrophils, activated macrophages, and endothelial cells.48 RhoN,49 a member of Rho family GTPases, controls cytoskelton organization through stimulating the polymerization of actin, and may thus control cell growth and differentiation.
Acute-phase proteins
As a consequence of the inflammation that occurs during GVHD, hepatocytes produce a number of acute-phase proteins such as protease inhibitors, amyloid, orosomucoids, and metallothioneins.50 IL-1 receptor antagonist (IL-1Ra), serine protease inhibitor (Serpin), metallothioneins,51 orosomucoids,52 secretary leukocyte protease inhibitor (SLPI),53 and serum amyloid A proteins54 inhibit inflammation. The up-regulation of these proteins may therefore suggest a potential regulatory response to the ongoing GVHD.
Metabolism and endocrine function
In contrast to all the other categories, most genes associated with metabolism and endocrine functions were down-regulated after allogeneic BMT, probably reflecting the stress response of hepatic tissue. Down-regulation of these genes is known to be caused by hepatotoxins such as LPS and thioacetamide.55 Interestingly, these genes were primarily down-regulated at day 7, demonstrating that stress in hepatic tissue was evident long before there was histologic evidence of hepatic damage.
ESTs and genes with unknown function
Some genes in this category have sequence similarities to genes with known function in the other species. Down-regulation of clone MGC:18880 IMAGE:4237837 (MGC:18880), which is similar to cytochrome P450, 4a10, may indicate the stress of the liver. Expressed sequence AV378681 (AV378681), up-regulated at day 35, is very similar to human matrix metalloproteinase 12, a macrophage elastase that plays an important role in inflammation associated with tissue remodeling and destruction.56
Discussion
GVHD is an alloimmune response that begins when donor T cells recognize host alloantigens. Since host APCs are critical to this process,3,5 the alloimmune response may require secondary lymphoid organs, which provide the proper environment for APCs, to interact with naive T cells.57 Donor T cells activated by host APCs produce Th1 cytokines and trigger the cytokine cascade of GVHD. Although there was no histologic evidence of donor T-cell infiltration in the liver 7 days after allogeneic BMT in our model, many IFN-γ–inducible genes were up-regulated at this time despite the lack of up-regulation of the IFN-γ gene itself in the liver. IFN-γ production is an early event in the cytokine cascade of GVHD with peak serum levels by day 7 after allogeneic BMT.58-60 We detected significant expression of IFN-γ in the spleen, but not the liver, consistent with a previous kinetic study of organ-specific cytokine expression in GVHD demonstrating that the spleen is the principal source of inflammatory cytokines including IFN-γ and TNF-α.61 Other secondary lymphoid organs are also sites where donor T cells encounter host alloantigens because removal of recipients' spleens does not prevent GVHD in this model.62 Thus IFN-γ produced by allogeneic donor T cells in the secondary lymphoid organs transactivates genes on target organs, stimulating the recruitment of effector cells to target organs and eventually rendering them vulnerable to attack by cellular and inflammatory mediators.
IFN-γ plays an important role in several aspects of pathophysiology during GVHD. First, IFN-γ increases the expression of MHC molecules and genes related to antigen processing. A consistent observation in hepatic GVHD is the aberrant expression of MHC class II molecules on bile duct epithelium, a site of lymphocytic infiltration.63-66 We found increased expression of MHC class II molecules before the infiltration of effector cells, consistent with the hypothesis that increased MHC class II expression on bile duct epithelium might render it vulnerable to immunologic attack by allogeneic CD4 T cells. However, expression of MHC class II alloantigen on host epithelium is not required for the induction of CD4-mediated GVHD,5 and thus a causal role for increased expression of MHC class II is controversial.
Second, IFN-γ increases the expression of adhesion molecules. An increased number of LFA-1–positive cells and an enhanced expression of ICAM-1 and VCAM-1 on bile duct epithelial cells are observed during the development of hepatic GVHD.67-69 This increased expression of ICAM-1 and VCAM-1 occurred prior to the infiltration of effector cells, suggesting that blockade of interaction between adhesion molecules and their ligands may be a possible prophylactic strategy. Indeed, administration of monclonal antibodies (mAbs) against ICAM-1, VCAM-1, and LFA-1 can reduce hepatic GVHD, although the magnitude of the effect varies depending on the experimental system and antibody used.68-70 It should be noted that disruption of the ICAM-1 gene in the recipient does not improve the histologic findings of the liver, suggesting that host ICAM-1 expression is not necessary for the development of hepatic GVHD.71
Third, IFN-γ stimulates the production of inflammatory chemokines that control the migration of effector cells toward sites of inflammation.22,23 Early after allogeneic BMT, we found increased expression of inflammatory chemokines, including Mig and IP-10, and CXCR3 ligands. Up-regulation of Mig and IP-10 was also part of the early pathogenesis of allograft cardiac rejection.72,73 The exact role of those inflammatory chemokines in the initiation of hepatic GVHD is currently unknown.
In contrast to the early up-regulation of most inflammatory chemokines, RANTES, a CCR5 ligand, was significantly up-regulated at day 35 with the infiltration of effector cells in the liver. In a nonirradiated GVHD model, macrophage inflammatory protein 1 α (MIP-1α), another ligand for CCR5, was up-regulated in the liver 2 weeks after BMT even though the expression of RANTES was unchanged.74 In that study, the administration of mAbs to CCR5 prevented the accumulation of T cells to the liver.74 Similar late induction of RANTES has been observed in experimental models of the heart75 and lung76 allograft rejection, suggesting RANTES-CCR5 interaction might also be a suitable therapeutic target for these transplanted organs.
Fourth, IFN-γ increases Fas expression in target tissues.77 Accumulating evidence suggests that Fas-mediated T-cell cytotoxicity plays an essential role in the development of hepatic GVHD. Use of FasL-deficient donor mice78 and administration of anti-FasL mAb79 can significantly reduce hepatic GVHD, whereas Fas-deficient recipients are resistant to the induction of GVHD specifically in the liver.80 Our study demonstrated that Fas was up-regulated at day 7 in the liver, suggesting that the liver becomes vulnerable to Fas-L–mediated CTL attack prior to hepatic damage.
Lastly, IFN-γ renders monocytes and macrophages sensitive to endogenous LPS, resulting in the increased production of TNF-α and IL-1,7,81 which share overlapping inflammatory properties in GVHD pathophysiology.82 In addition to the proinflammatory activity it shares with IFN-γ, TNF-α can directly induce apoptosis or necrosis83 in target tissue. TNF-α–mediated cytotoxicity plays a crucial role in the development of intestinal GVHD.79,84 Recent studies have demonstrated that intensive blockade of TNF-α and IL-1 can prevent hepatic GVHD, probably limiting effector cell migration to the liver by the suppression of inflammatory chemokine production.5
Although our results suggest that IFN-γ recruits effector cells to the liver during acute GVHD, lack of IFN-γ secretion by donors does not prevent the hepatic infiltration of lymphocytes.85,86 Similarly, IFN-γ knock-out mice (GKO) can reject cardiac allografts.87 Microarray analysis of GKO recipients in a cardiac allograft model reveals that some chemokines and their receptors such as MCP-1, MIP-1, stromal cell–derived factor-1 (SDF-1), C10, and CXCR4 were induced as efficiently as in normal recipients despite the absence of IFN-γ function even though the expression of Mig, IP-10, and RANTES was decreased in the heart.88 It should be noted, however, that disruption of the IFN-γ receptor gene in a transplanted heart can prevent cardiac allograft rejection, suggesting a critical role of IFN-γ–inducible genes in the alloresponse.89
Systemic IFN-γ is clearly induced in many models of acute GVHD,61,90 but it is usually not observed in the plasma in this model later than 2 weeks after transplantation (data not shown). Nevertheless, the lack of IFN-γ gene expression in the liver on day 35 at the time of tissue damage was curious, although several explanations are possible. The mononuclear cell infiltration of the liver in GVHD is usually highly localized and confined to very specific locations such as the periportal space. The sampling method used here did not target such areas specifically, and the amount of infiltrate present in any sample is thus likely to be small. Infiltrates are also likely to be composed of several cell types that could further dilute the concentration of single cytokine mRNA in any given sample. Such a mixture of cell types is supported by the increased expression of genes related to myeloid effector functions (proteinase 3, p22 phox, and gp91 phox) and is consistent with the prominence of myeloid lineage cells recently reported during the later phases of experimental GVHD.91 It is also possible that the cells producing IFN-γ are present in target organs only transiently; they may initiate damage that is then amplified by other soluble or cellular effectors.5,91 During GVHD IFN-γ can regulate T-cell expansion through activation-induced cell death.12 Thus even though they may have initiated the tissue destruction, the cells producing IFN-γ may have largely eliminated themselves in a kind of “clonal contraction” by the time histopathologic damage is evident. A more detailed kinetic analysis of the samples using techniques such as laser capture microdissection will be required to address this issue.
Based on the studies of murine GVHD directed selectively to class I versus class II differences, CD4+ and CD8+ donor T cells differently induce GVHD. CD4-mediated GVHD is rapid, whereas CD8-mediated GVHD is slower.92 CD4+ T cells are primarily responsible for the induction of acute GVHD in this donor/recipient strain combination since depletion of CD4+ T cells, but not CD8+ T cells, from donor inoculum markedly inhibits GVHD mortality.93 Similarly, murine GVHD directed to other strain combinations with full MHC differences involves predominantly CD4+ T cells,92,94,95 whereas CD8+ T cells are mostly responsible for the induction of GVHD across minor histocompatibity barriers.92 Future studies will focus on gene profiles of CD8-mediated GVHD in order to help clarify the molecular mechanisms of GVHD induced by different T-cell subsets.
In summary, we have identified gene expression changes during the development of acute GVHD in the liver after allogeneic BMT. The results suggested that the expression of many genes, particularly those encoding IFN-γ–inducible proteins, was increased prior to histologic evidence of GVHD. The increased expression of genes in target tissue required for antigen presentation and recruitment of effector cells preceded their actual infiltration, suggesting that those genetic changes are important prerequisites for the migration of effector cells to their targets. Our study underscores the role of IFN-γ in the initiation of GVHD in this model, and provides the many new potential molecular targets for early diagnosis and therapeutic intervention of acute GVHD.
Prepublished online as Blood First Edition Paper, March 27, 2003; DOI 10.1182/blood-2002-09-2748.
Supported by National Institutes of Health grant CA4592 (J.L.M.F.)
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Toshihide Iwashita for technical suggestions.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal