Abstract
Differentiation of pluripotent embryonic stem (ES) cells is associated with expression of fate-specifying gene products. Coordinated development, however, must involve modifying factors that enable differentiation and growth to adjust in response to local microenvironmental determinants. We report here that the ephrin receptor, EphB4, known to be spatially restricted in expression and critical for organized vessel formation, modifies the rate and magnitude of ES cells acquiring genotypic and phenotypic characteristics of mesodermal tissues. Hemangioblast, blood cell, cardiomyocyte, and vascular differentiation was impaired in EphB4–/– ES cells in conjunction with decreased expression of mesoderm-associated, but not neuroectoderm-associated, genes. Therefore, EphB4 modulates the response to mesoderm induction signals. These data add differentiation kinetics to the known effects of ephrin receptors on mammalian cell migration and adhesion. We propose that modifying sensitivity to differentiation cues is a further means for ephrin receptors to contribute to tissue patterning and organization.
Introduction
Embryonic stem (ES) cells are pluripotent and their in vitro differentiation provides an experimental window to define the molecular mechanisms controlling germ layer fate determination and tissue formation. Mesoderm induction of ES cells is particularly amenable to in vitro study, and differentiation into blood and vasculature is generally accepted as recapitulating aspects of normal mammalian development. The hematopoietic and vascular systems of the mouse develop from aggregates of extraembryonic mesoderm cells, which colonize the yolk sac at approximately 7.5 days of gestation1,2 and establish blood islands. Blood islands are thought to derive from hemangioblasts,3-5 a common precursor of hematopoietic and endothelial cells. They are organized into an outer layer of endothelial precursors or angioblasts, surrounding an inner cluster of primitive erythroblasts producing embryonic hemoglobin.6-8 The existence of hemangioblasts has been supported by the observation that the hematopoietic and endothelial lineages express a number of genes in common including receptor tyrosine kinases (RTKs), Flk-1 (VEGF-R2) and Tie-2; transcription factors, GATA-2, Runx1, and Scl; and cell surface markers, Sca-1 and CD34.9-14 Recent studies have demonstrated that Flk-1 can serve as a molecular marker for hemangioblasts during early development since Flk-1+ populations can give rise to both hematopoietic and endothelial cells.15-17 Further, Flk-1 is required for hematopoietic and vascular endothelial development within blood islands of the yolk sac.18,19 Molecular regulators of the formation of hemangioblasts are unknown and of considerable interest given the developmental position of the hemangioblast between earliest germ layer formation and tissue-specific stem cell types.
Over the past decade, essential roles for Eph receptor tyrosine kinases and their ligands, the ephrins, have emerged from studies of embryonic development.20 Fourteen different Eph receptors have been catalogued into EphA or EphB subclasses based on their affinity for ligands.21 All 8 identified ephrins are membrane proteins of either glycerophosphatidylinositol (GPI)–linked (ephrinA) or transmembrane (ephrinB) proteins.21 Rather than long-range communication, signaling between Eph receptors and their ligands is restricted to sites of direct cell-cell contact and is capable of inducing reciprocal bidirectional events between interacting cells.22-24 Localized expression of ephrins and their receptors are thought to play critical roles in defining tissue patterning and the organizing of highly spatially restricted cell locations. In particular, they participate in modifying cell migration, adhesion, and somite formation.25 EphB4 (or HTK) and its cognate ligand ephrinB2 (or HTKL) play critical roles in determining vascular networks. EphB4 is specifically expressed at the venous endothelium, while ephrinB2 is specifically and reciprocally expressed on arterial endothelial cells at the early stages of vascular development.26 Mice lacking either EphB4 or ephrinB2 are embryonic lethal and display identical defects in forming capillary connections between arterial and venous networks of the head and yolk sac.26-28 EphB4 and ephrinB2 are coexpressed in the yolk sac,27 the first site of hematopoiesis and vascular development during embryogenesis. Our previous studies indicated that EphB4 participates in adult hematopoiesis with primitive human CD34+ hematopoietic cells undergoing accelerated differentiation in the context of activated EphB4.29 We therefore examined EphB4 effects on the hemangioblast, where the processes of hematopoiesis and vasculogenesis intersect.
Identification and characterization of hemangioblasts have been hampered by difficulties in accessing the embryo prior to the establishment of the blood islands. In vitro ES differentiation is an alternative experimental system that has provided compelling, direct evidence for the presence of the hemangioblast.30 ES cells differentiate efficiently in vitro and give rise to a differentiated cell mass called embryoid bodies (EBs) upon removal of leukemia inhibitory factor (LIF).31,32 Although the embryoid body is far less organized than the actual embryo, it can partially mimic the spatial organization in the embryo. ES cells can be used to study the differentiation of the mammalian embryo into its earliest recognizable tissue lineages. Many different lineages have been reported within developing EBs including epidermis,33 neuronal and glial cells,34,35 muscle cells,36,37 endothelial cells,38,39 and hematopoietic cells.40,41 It has been reported that the developmental mechanisms of vascular and hematopoietic systems in EBs are similar to that in the yolk sac.16,30,42,43 Recent studies have shown that blast colony–forming cells (BL-CFCs), generated from EBs, represent the long sought common progenitors of hematopoietic and endothelial cells, the hemangioblast. BL-CFCs give rise to primitive, definitive hematopoietic and endothelial cells when plated in medium with hematopoietic and endothelial cell growth factors.30,44 In this study, we report that BL-CFCs and Flk-1+ cells in early-stage EBs were significantly reduced in EphB4-deficient ES cells. Moreover, EphB4 appears to be required for ES cell differentiation into spontaneously beating myocytes and for the induction of alphacardiac mycosin heavy chain (MHC). Analysis of EphB4-deficient ES cells indicated that EphB4 is essential for proper hematopoietic, endothelial, hemangioblast, and primitive mesoderm development. EphB4 affects the rate and magnitude of differentiation suggesting it participates in modifying developmental signals and may be important in coordinating integrated tissue formation.
Materials and methods
ES cell culture
EphB4-deficient ES cells were kindly provided by Dr David Anderson (California Institute of Technology, Pasadena, CA). EphB4–/– ES cells were generated by a high concentration of G418 selection from EphB4+/– cells that gave germ line transmission for EphB4+/– mice.28 Two clones of undifferentiated EphB4–/– ES cell lines (no. 49 and no. 71 as EphB4–/–1 and EphB4–/–2, respectively) were maintained on a mouse feeder cell line, SNL, in ES medium containing Dulbecco modified Eagle medium (DMEM), 10 ng/mL murine leukemia inhibitory factor (mLIF; Chemicon International, Temecula, CA), 15% fetal calf serum (FCS; HyClone, Logan, UT), 1 mM sodium pyruvate, 2 mM glutamine, 0.1 mM nonessential amino acid, 100 μM monothioglycerol (MTG; Sigma, St Louis, MO), 50 U/mL penicillin, and 50 μg/mL streptomycin. ES cells were cultured on gelatincoated plates for one week prior to EB induction.
ES cell differentiation and BL-CFC assay
ES cell differentiation into EBs were carried out as described.30,40,44 Briefly, EBs were generated either in liquid or 1% methylcellulose cultures (1 × 104 ES cells per 35-mm Petri dish) in ES differentiation medium containing Iscove modified Dulbecco medium (IMDM), 15% FCS (StemCell Technologies, Vancouver, BC, Canada), 2 mM glutamine, 450 μM MTG, 50 μg/mL ascorbic acid, and 20% BIT (1% bovine serum albumin [BSA], 10 μg/mL insulin, and 200 μg/mL transferrin; StemCell Technologies). After 6 days of differentiation, 50 ng/mL murine stem cell factor (mSCF), 1 ng/mL interleukin 3 (IL-3), 5 ng/mL IL-11, and 2 U/mL human erythropoietin (hEPO; Amgen, Thousand Oaks, CA) were added to cultures to promote hematopoietic differentiation. To assess BL-CFCs, day-3.5 to -4.0 EBs were dissociated by trypsin treatment and by passing through 20-gauge needle to generate single cells. To generate blast colonies from hemangioblasts, 1 × 104/mL EB cells were replated in 1% methylcellulose in the presence of IMDM, 10% PDS (bovine platelet-poor plasma-derived serum; Biomedical Technologies, Stoughton, MA), 2 mM glutamine, 450 μM MTG, 25 μg/mL ascorbic acid, 20% BIT, 5 ng/mL human vascular endothelial growth factor (hVEGF), 50 ng/mL SCF, 10 ng/mL human fibroblast growth factor 2 (hFGF-2), and 2 U/mL hEPO. BL-CFCs can be recognized as loose clusters of cells after 4 days of culture. To analyze the hematopoietic and endothelial developmental potential of the BL-CFC, blast colonies from either EphB4+/– or EphB4–/– were transferred into matrigel-coated plates and cultured in the presence of growth factors known to support both hematopoietic and endothelial cells.30 Following 3 to 4 days in culture, nonadherent hematopoietic and adherent endothelial cells were harvested for reverse transcriptase–polymerase chain reaction (RT-PCR) analysis. Primitive erythroid progenitors (EryP) were analyzed from day-6 EBs in 1% methylcellulose cultures containing IMDM, 10% PDS, 2 mM glutamine, 450 μM MTG, 5% PFHM-II (protein-free hybridoma media II; Gibco-BRL, Carlsbad, CA), and 4 U/mL hEPO. EryP colonies can be identified as small brilliant red colonies and are scored after 5 to 7 days replating.44 Other definitive myeloid progenitors were analyzed from day-10 to -12 EBs in 1% methylcellulose cultures containing IMDM, 10% PDS, 2 mM glutamine, 450 μM MTG, 5% PFHM-II, 4 U/mL hEPO, 50 ng/mL SCF, 1 ng/mL IL-3, 5 ng/mL macrophage colony-stimulating factor (M-CSF), 5 ng/mL thrombopoietin (TPO), 5 ng/mL IL-11, 30 ng/mL granulocyte-CSF (G-CSF), 3 ng/mL GM-CSF, and 5 ng/mL IL-6. Hematopoietic colonies were counted 7 to 10 days after replating. SCF, IL-3, M-CSF, TPO, IL-11, G-CSF, GM-CSF, IL-6, hVEGF, and hFGF-2 were purchased from R&D Systems (Minneapolis, MN).
Sprouting EB induction
ES cell differentiation into endothelial cells was performed as described.43,45 EBs were initially generated in 1% methylcellulose cultures (1 × 103 ES cells/mL in 2 mL in 35-mm Petri dish) in IMDM containing 15% FCS, 2 mM glutamine, 450 μM MTG, 20% BIT, 50 ng/mL hVEGF, 100 ng/mL hFGF-2, 2 U/mL hEPO, and 10 ng/mL mIL-6. After 11 days of EB differentiation, EBs were harvested and resuspended at 50 EBs/mL in collage medium containing rat-tail collagen type I (Becton Dickinson, San Jose, CA) or Vitrogen (Cohesion Technologies, Vancouver, BC, Canada), 15% FCS, 450 μM MTG, 10 μg/mL insulin, 50 ng/mL hVEGF, 100 ng/mL hFGF-2, and 10 ng/mL mIL-6. After gently mixing EBs into collagen medium, 1 mL was dispensed into 35-mm Petri dishes or double-chamber slides and incubated at 37°C without CO2 for 30 minutes to form a stable collagen gel matrix. The cultures were then incubated for 3 to 6 days at 37°C and 5% CO2. The vascular spindlelike EBs were scored at day 3 on collagen matrix.
Flow cytometric analysis and immunostaining
EBs were trypsinized for 2 minutes and passed through a 20-gauge needle to produce a single cell suspension. Cells (1 × 106) were stained for β-galactosidase (β-gal) activity with a fluorescein di-β-d-galactopyranoside (FDG) kit (Molecular Probes, Eugene, OR) according to the manufacturer's instructions and then stained in 5% rat serum for 30 minutes at 4°C either with phycoerythrin (PE)–conjugated anti-Flk1 and allophycocyanin (APC)–conjugated anti–c-kit or with PE-conjugated anti-CD45 and APC-conjugated anti-Ter119. Dead cells were gated out by a DNA dye, DAPI (4,6-diamidino-2-phenylindole), staining. All antibodies were purchased from BD Sciences (Pharmingen, San Diego, CA). The β-gal activity in EBs was analyzed by staining with 5-bromo-4-chloro-3-indolyl β-galactoside (X-gal). EBs were washed with phosphate-buffered saline (PBS) and fixed with 0.5% glutaraldehyde and 0.02% nonidet P-40 (NP-40) at room temperature for 20 minutes. EBs were then stained with 1 mg/mL X-gal, 250 μM K3Fe(CN)6, 250 μM K4Fe(CN)6, 2 mM MgCl2, and 0.01% NP-40 for 2 hours in a CO2-free incubator at 37°C in the dark.
EB cultures in collagen in chamber slides were dehydrated and fixed in methanol-acetone (3:1) solution for 20 minutes. After air drying and rehydrating with PBS, capillary-sprouting EBs were incubated in 5% rat serum for 20 minutes to block nonspecific binding and stained with 1 μg/mL PE-conjugated antimouse platelet endothelial cell adhesion molecule 1 (PECAM1, CD31; Pharmingen) for 1 hour.
Gene expression analysis
Total RNA was isolated from EBs using Trizol (Invitrogen, Carlsbad, CA) according to manufacturer's instructions and then cleaned by RNeasy kit (Qiagen, Valencia, CA). One microgram RNA was used for reverse transcription using 200 units SuperScript II RNase H– reverse transcriptase (Invitrogen) in the presence of 50 mM Tris (tris(hydroxymethyl)aminomethane; pH 8.3), 75 mM KCl, 3 mM MgCl2, 5 mM dithiothreitol (DTT), 40 units ribonuclease inhibitor, 0.5 mM deoxynucleoside triphosphates (dNTPs), and 100 ng random primers in 20-μL volume. Specific primers used for PCR are listed in Table 1. All the genes were analyzed on more than one occasion using RNA from independently EB samples.
Oligonucleotide primers used for RT-PCR
Gene . | Primer sequence . | Tm °C . | Size, bp . |
|---|---|---|---|
| EphB4 | Sense 5′-TGGCTGATCACGAACTTGACCTAC-3′ | 61 | 323 |
| Antisense 5′-AGGCAGGACTCGTCTCCTATTT-3′ | 58 | ||
| EphB2 | Sense 5′-TAGTCTTCCTCATCGCTGTG-3′ | 53 | 366 |
| Antisense 5′-ATGATGGATGCCTCACTCAG-3′ | 54 | ||
| EphB3 | Sense 5′-AAGACTATCGGTTACCACCC-3′ | 53 | 354 |
| Antisense 5′-GTGACCCCAATCCTTAGCAG-3′ | 56 | ||
| ephrinB2 | Sense 5′-TCAAGAATTCAGCCCTAACCTC-3′ | 57 | 440 |
| Antisense 5′-TGACGATGAAGATGATGCATCC-3′ | 59 | ||
| CXCR4 | Sense 5′-GGCTGTAGAGCGAGTGTTGC-3′ | 57 | 390 |
| Antisense 5′-GTAGAGGTTGACAGTGTAGAT-3′ | 44 | ||
| Flk-1 | Sense 5′-CACCTGGCACTCTCCACCTTC-3′ | 60 | 239 |
| Antisense 5′-GATTTCATCCCACTACCGAA-3′ | 54 | ||
| CD31 | Sense 5′-GTCATGGCCATGGTCGAGTA-3′ | 58 | 261 |
| Antisense 5′-CTCCTCGGCGATCTTGCTGAA-3′ | 64 | ||
| β-H1 | Sense 5′-CTCAAGGAGACCTTTGCTCA-3′ | 54 | 309 |
| Antisense 5′-AGTCCCCATGGAGTCAAAGA-3′ | 56 | ||
| β-major | Sense 5′-TCCGATGAAGTTGGTGGTGA-3′ | 58 | 307 |
| Antisense 5′-AAATCCTTGCCAAGGTGGTG-3′ | 58 | ||
| Scl | Sense 5′-TATGAGATGGAGATTTCTGATG-3′ | 50 | 396 |
| Antisense 5′-GCTCCTCTGTGTAACTGTCC-3′ | 51 | ||
| GATA-2 | Sense 5′-GACGACAACCACCACCTTATG-3′ | 56 | 283 |
| Antisense 5′-GACTTAAAGGTGGGAGGTGTC-3′ | 54 | ||
| Oct-4 | Sense 5′-AAAGGTGTTCAGCCAGACCA-3′ | 56 | 369 |
| Antisense 5′-CTCATACTCTTCTCGTTGGG-3′ | 52 | ||
| Rex-1 | Sense 5′-CCAGTCCAGAATACCAGAGT-3′ | 50 | 338 |
| Antisense 5′-TGCAAGTAATGAGCTCGCCC-3′ | 60 | ||
| β-actin | Sense 5′-GATGACGATATCGCTGCGCTG-3′ | 63 | 439 |
| Antisense 5′-GTACGACCAGAGGCATACAGG-3′ | 56 |
Gene . | Primer sequence . | Tm °C . | Size, bp . |
|---|---|---|---|
| EphB4 | Sense 5′-TGGCTGATCACGAACTTGACCTAC-3′ | 61 | 323 |
| Antisense 5′-AGGCAGGACTCGTCTCCTATTT-3′ | 58 | ||
| EphB2 | Sense 5′-TAGTCTTCCTCATCGCTGTG-3′ | 53 | 366 |
| Antisense 5′-ATGATGGATGCCTCACTCAG-3′ | 54 | ||
| EphB3 | Sense 5′-AAGACTATCGGTTACCACCC-3′ | 53 | 354 |
| Antisense 5′-GTGACCCCAATCCTTAGCAG-3′ | 56 | ||
| ephrinB2 | Sense 5′-TCAAGAATTCAGCCCTAACCTC-3′ | 57 | 440 |
| Antisense 5′-TGACGATGAAGATGATGCATCC-3′ | 59 | ||
| CXCR4 | Sense 5′-GGCTGTAGAGCGAGTGTTGC-3′ | 57 | 390 |
| Antisense 5′-GTAGAGGTTGACAGTGTAGAT-3′ | 44 | ||
| Flk-1 | Sense 5′-CACCTGGCACTCTCCACCTTC-3′ | 60 | 239 |
| Antisense 5′-GATTTCATCCCACTACCGAA-3′ | 54 | ||
| CD31 | Sense 5′-GTCATGGCCATGGTCGAGTA-3′ | 58 | 261 |
| Antisense 5′-CTCCTCGGCGATCTTGCTGAA-3′ | 64 | ||
| β-H1 | Sense 5′-CTCAAGGAGACCTTTGCTCA-3′ | 54 | 309 |
| Antisense 5′-AGTCCCCATGGAGTCAAAGA-3′ | 56 | ||
| β-major | Sense 5′-TCCGATGAAGTTGGTGGTGA-3′ | 58 | 307 |
| Antisense 5′-AAATCCTTGCCAAGGTGGTG-3′ | 58 | ||
| Scl | Sense 5′-TATGAGATGGAGATTTCTGATG-3′ | 50 | 396 |
| Antisense 5′-GCTCCTCTGTGTAACTGTCC-3′ | 51 | ||
| GATA-2 | Sense 5′-GACGACAACCACCACCTTATG-3′ | 56 | 283 |
| Antisense 5′-GACTTAAAGGTGGGAGGTGTC-3′ | 54 | ||
| Oct-4 | Sense 5′-AAAGGTGTTCAGCCAGACCA-3′ | 56 | 369 |
| Antisense 5′-CTCATACTCTTCTCGTTGGG-3′ | 52 | ||
| Rex-1 | Sense 5′-CCAGTCCAGAATACCAGAGT-3′ | 50 | 338 |
| Antisense 5′-TGCAAGTAATGAGCTCGCCC-3′ | 60 | ||
| β-actin | Sense 5′-GATGACGATATCGCTGCGCTG-3′ | 63 | 439 |
| Antisense 5′-GTACGACCAGAGGCATACAGG-3′ | 56 |
Tm °C indicates annealing temperature.
DNA microarrays
cDNA synthesis, cRNA synthesis, and labeling were performed as described in the Affymetrix user's manual (Santa Clara, CA). Affymetrix mouse U74Av2 chips (about 12 000 genes) were hybridized on a GeneChip system (Affymetrix) at the Center for Genomics Research of Harvard University and the Massachusetts General Hospital (MGH) Cancer Center according to the manufacturer's instruction. All arrays were globally scaled to a target value of 150 using the average signal from all gene features. Scanned chip images were analyzed using the Microarray Suite Software version 5.0 (Affymetrix). Microsoft Excel was used for further analyses. Four experiments from independent EB samples were used at the Center for Genomics Research of Harvard University and the MGH Cancer Center. A transcript was considered “changed” when the fold change was at least 2.
Results
EphB4–/– ES cells organize into embryoid bodies with altered composition
EphB4-null ES cells (EphB4–/–) were generated from EphB4+/– cells by culturing in high concentrations of G418. Two individual clones of both EphB4+/– (EphB4+/–1 and EphB4+/–2) and EphB4–/– (EphB4–/–1 and EphB4–/–2) were originally derived and used in our experiments. However, because identical results were obtained from the 2 clones, subsequent experiments used only one of the clones (EphB4+/–1 and EphB4–/–1), and one clone for each genotype is reported here. The doubling time of undifferentiated ES cells from wild-type, EphB4+/–, and EphB4–/– remained the same (about 18 hours). Gene expression profiles and flow cytometric analyses of wild-type ES cells were very similar to EphB4+/– ES cells (data not shown). Because the wild-type cells had not gone through the transduction and selection processes of the genetically modified cells and we were concerned about alterations induced by these processes, we focused on comparing the EphB4+/– and EphB4–/– cells.
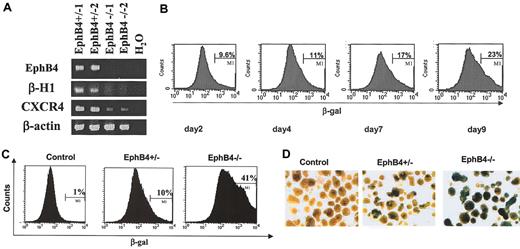
To induce spontaneous differentiation of ES cells into cystic EBs, ES cells were cultured in suspension in the absence of LIF for 4 days. RT-PCR was used to confirm that EphB4–/– EBs did not express EphB4 transcripts, excluding possible contamination by EphB4+/– or EphB4+/+ ES cells (Figure 1A). We initially examined the expression of a hematopoietic gene, β-H, as well as a gene expressed by both hematopoietic and endothelial cells, CXCR4, in the 2 EphB4+/– and 2 EphB4–/– cell lines. Expressions of both of these genes were down-regulated in EphB4-deficient ES cells, and data were consistent between both ES clones (Figure 1A). To determine the pattern of EphB4 expression during in vitro EB differentiation, we used β-gal expression as a histochemical marker since β-gal was transcribed from the EphB4 locus and reflected the expression of the endogenous EphB4 gene. The β-gal expression was assessed by staining with FDG and quantified with flow cytometry or by X-gal staining. Flow cytometric analysis demonstrated that the percentage of EphB4-expressing cells (β-gal+) was increased during ES cell differentiation (Figure 1B). The number of β-gal+ cells within EphB4–/– EBs (mean 40%) was significantly higher than those within EphB4+/– EBs (mean 10%; Figure 1C). By histochemical staining of β-gal, most of day 4 EphB4–/– EBs (more than 80%) were all strongly stained with β-gal, while fewer EBs from EphB4+/– (< 30%) were positive for β-gal staining (Figure 1D). No significant level of β-gal activity was detected in control, wild-type ES cells. There are several possible mechanisms accounting for the increase in β-gal–expressing cells. Either a negative feedback mechanism restricting transcriptional activity of the EphB4 promoter is absent in EphB4-deficient cells or diminished differentiation results in an expansion of primitive cells.
EphB4 gene expression indicated by β-galactosidase activity within EBs. (A) Two clones of each EphB4+/– (EphB4+/–1 and EphB4+/–2) and EphB4–/– ES cells (EphB4–/–1 and EphB4–/–2) were induced for EB differentiation for 4 days and analyzed for gene expression by RT-PCR. (B) Kinetics of β-gal expression in EphB4+/– EBs. EphB4+/– ES cells were allowed to differentiate into EBs in suspension cultures in Petri dishes. EB cells at day 2, 4, 7, and 9 were harvested, stained with FDG for β-galactosidase activity, and analyzed by flow cytometry. Dead cells were gated out by a DNA dye, DAPI, staining (data not shown). (C) Flow cytometric analysis of β-gal expression (β-gal+ cells) in day-4 EBs from control (wild-type), EphB4+/– and EphB4–/– ES cells. (D) X-gal staining β-galactosidase activity in day EBs was analyzed by staining with 5-bromo-4-chloro-3-indolyl b-galactoside (X-gal). Original magnification, × 4.
EphB4 gene expression indicated by β-galactosidase activity within EBs. (A) Two clones of each EphB4+/– (EphB4+/–1 and EphB4+/–2) and EphB4–/– ES cells (EphB4–/–1 and EphB4–/–2) were induced for EB differentiation for 4 days and analyzed for gene expression by RT-PCR. (B) Kinetics of β-gal expression in EphB4+/– EBs. EphB4+/– ES cells were allowed to differentiate into EBs in suspension cultures in Petri dishes. EB cells at day 2, 4, 7, and 9 were harvested, stained with FDG for β-galactosidase activity, and analyzed by flow cytometry. Dead cells were gated out by a DNA dye, DAPI, staining (data not shown). (C) Flow cytometric analysis of β-gal expression (β-gal+ cells) in day-4 EBs from control (wild-type), EphB4+/– and EphB4–/– ES cells. (D) X-gal staining β-galactosidase activity in day EBs was analyzed by staining with 5-bromo-4-chloro-3-indolyl b-galactoside (X-gal). Original magnification, × 4.
EphB4-deficient ES cells display a defect in hemangioblast development
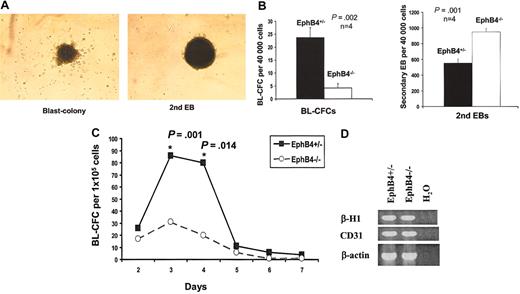
To evaluate the effects of EphB4 on the formation of the early mesoderm-associated hemangioblast, day-3 EBs of EphB4+/– and EphB4–/– ES cells were assayed for their potential to generate hemangioblast-containing BL-CFCs. Previous studies demonstrated that BL-CFCs could be detected as early as 2.5 days of EB differentiation. The maximum numbers of BL-CFCs could be reached from 3.25 to 4.25 days of EB differentiation.44 When cells from EBs are dissociated and replated on methylcellulose, BL-CFCs can be readily recognized as loose clusters compared with the compact secondary embryoid bodies (Figure 2A). BL-CFCs represent entry into a differentiation program where secondary EBs reflect cells retaining a primitive status. Relative to EphB4+/– ES cells, generation of BL-CFCs from EphB4–/– cells was significantly impaired (Figure 2B). EphB4-deficient cells gave rise to more secondary EBs, indicating that more EphB4-deficient cells remain undifferentiated (Figure 2B). To determine whether this decreased production was due to a delay in hemangioblast formation, a more detailed kinetic analysis of the formation of BL-CFCs from EphB4+/– and EphB4–/– ES cells was performed (Figure 2C). EphB4+/– and EphB4–/– ES cells demonstrated similar kinetics as previously described.44 However, the magnitude of BL-CFC formation was significantly lower in EphB4–/– ES cells. Therefore, the absence of EphB4 impairs hemangioblast differentiation.
Impaired BL-CFC formation of EphB4-deficient ES cells. (A) Examples of blast colony (BL-CFC) and secondary EB from EphB4+/– ES cells. BL-CFCs can be readily recognized as loose clusters compared with the compact secondary embryoid bodies. Original magnification, × 40. (B) Day-3 EBs of EphB4+/– and EphB4–/– ES cells were collected, and the cells were dissociated and assayed for their potential to generate BL-CFCs and secondary EB in methylcellulose cultures. Error bars represent standard deviation. P values were calculated with student t test. Data shown represent the average of 4 experiments. (C) Kinetic analysis of BL-CFC formation. The cells from EBs at indicated times were analyzed for BL-CFC generation. Data shown represent the average of 4 experiments. Error bars represent standard deviation. P values were calculated with Student t test. (D) RT-PCR analysis of developmental potential of the BL-CFCs. Day-3 EBs of EphB4+/– and EphB4–/– ES cells were used to generate BL-CFCs. Blast colonies from either EphB4+/– or EphB4–/– were transferred into matrigel-coated plates and cultured in the presence of growth factors known to support both hematopoietic and endothelial cells. Following 3 to 4 days in culture, nonadherent hematopoietic and adherent endothelial cells were harvested for RT-PCR analysis of lineage specific genes.
Impaired BL-CFC formation of EphB4-deficient ES cells. (A) Examples of blast colony (BL-CFC) and secondary EB from EphB4+/– ES cells. BL-CFCs can be readily recognized as loose clusters compared with the compact secondary embryoid bodies. Original magnification, × 40. (B) Day-3 EBs of EphB4+/– and EphB4–/– ES cells were collected, and the cells were dissociated and assayed for their potential to generate BL-CFCs and secondary EB in methylcellulose cultures. Error bars represent standard deviation. P values were calculated with student t test. Data shown represent the average of 4 experiments. (C) Kinetic analysis of BL-CFC formation. The cells from EBs at indicated times were analyzed for BL-CFC generation. Data shown represent the average of 4 experiments. Error bars represent standard deviation. P values were calculated with Student t test. (D) RT-PCR analysis of developmental potential of the BL-CFCs. Day-3 EBs of EphB4+/– and EphB4–/– ES cells were used to generate BL-CFCs. Blast colonies from either EphB4+/– or EphB4–/– were transferred into matrigel-coated plates and cultured in the presence of growth factors known to support both hematopoietic and endothelial cells. Following 3 to 4 days in culture, nonadherent hematopoietic and adherent endothelial cells were harvested for RT-PCR analysis of lineage specific genes.
To determine if the hematopoietic and endothelial developmental potential of EphB4-deficient BL-CFCs was also impaired, blast colonies of day-3 EB cells from either EphB4+/– or EphB4–/– were transferred into matrigel-coated plates and cultured in the presence of growth factors known to support both hematopoietic and endothelial cells. Following 3 to 4 days in culture, nonadherent hematopoietic and adherent endothelial cells were harvested for RT-PCR analysis (Figure 2D). The hematopoietic gene, β-H1, and endothelial gene, CD31 (PECAM1) were detected in EphB4-deficient BL-CFCs, indicating that despite the decreased number of BL-CFCs, EphB4-deficient BL-CFCs were able to further differentiate into hematopoietic and endothelial lineages.
Several studies have demonstrated that Flk-1 is a molecular marker for hemangioblasts during early development since Flk-1+ populations can give rise to both hematopoietic and endothelial cells.15-17 To determine whether the EphB4+ and Flk-1+ cells represent overlapping populations in early ES cell differentiation, cells from day-4 EBs were analyzed by flow cytometry. As shown in Figure 3A, half of the cells strongly positive for β-gal driven off the EphB4 promoter were Flk-1+, while no significant Flk-1+ cells (< 0.5%) were detected in the β-gal–negative population. Therefore, Flk-1+ cells are subsumed in the population of cells with an active EphB4 promoter. At an early stage of EB differentiation (day 4), Flk-1+ cells were significantly decreased in EphB4–/– ES cells (13%) compared with EphB4+/– ES cells (52%) (Figure 3B). By day 9, however, the frequency of Flk-1+ cells in EBs generated from both EphB4+/– and EphB4–/– ES cells became similar. These data demonstrate that the absence of EphB4 significantly decreases but does not completely block differentiation into Flk-1–expressing cells or BL-CFCs, both reflective of hemangioblasts.
Flow cytometric analysis of Flk-1+ cells and Sca-1+/c-kit+ cells in EBs. (A) The β-gal+ cells express Flk-1 in early EB differentiation. ES cells of EphB4+/– were induced into EB differentiation for 4 days and analyzed by flow cytometry. The β-gal–positive and –negative cells were gated respectively and analyzed for expression of Flk-1 and c-kit. (B-C) Both Flk-1+ and Sca-l+/c-kit+ cells decreased in EphB4–/– EBs. Day-4 and day-9 EB cells from either EphB4+/– or EphB4–/– ES cells were analyzed for Flk-1 (B) and Sca-1/c-kit (C) expression by flow cytometry. Dead cells were gated out by a DNA dye, DAPI, staining. Flow cytometric data are representative from 3 independent experiments.
Flow cytometric analysis of Flk-1+ cells and Sca-1+/c-kit+ cells in EBs. (A) The β-gal+ cells express Flk-1 in early EB differentiation. ES cells of EphB4+/– were induced into EB differentiation for 4 days and analyzed by flow cytometry. The β-gal–positive and –negative cells were gated respectively and analyzed for expression of Flk-1 and c-kit. (B-C) Both Flk-1+ and Sca-l+/c-kit+ cells decreased in EphB4–/– EBs. Day-4 and day-9 EB cells from either EphB4+/– or EphB4–/– ES cells were analyzed for Flk-1 (B) and Sca-1/c-kit (C) expression by flow cytometry. Dead cells were gated out by a DNA dye, DAPI, staining. Flow cytometric data are representative from 3 independent experiments.
EphB4 deficiency affects the developmental potential of both primitive and definitive hematopoiesis
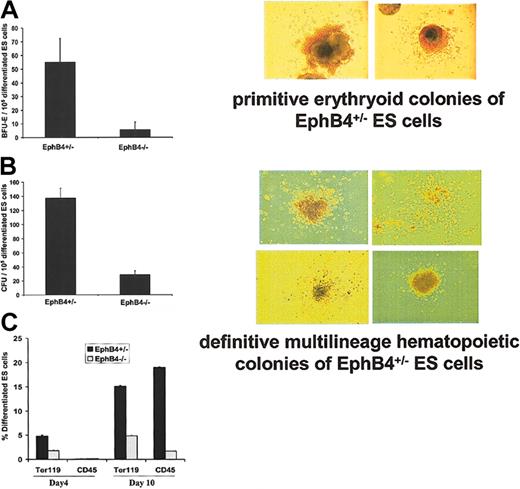
Previous studies demonstrated that BL-CFCs give rise to primitive and definitive hematopoietic cells as well as endothelial cells when plated in medium with hematopoietic and endothelial cell growth factors.30,44 To assess whether EphB4 signals also contribute to the differentiation of hematopoietic and endothelial cells, we used the in vitro ES cell system, which provides easy access to study both primitive and definitive hematopoiesis. At day 4 of EB induction, the number of cells staining for a hematopoietic stem cell immunophenotype (Scal-1+/c-kit+) was decreased in EphB4–/– ES cells and remained low at day 9 (Figure 3C). Decreases in Scal-1+/c-kit+ cells and down-regulation of βH1 globin in EphB4–/– EBs suggested a role for EphB4 in hematopoietic ES cell differentiation. Differentiated ES cells from day-5 to -7 or day-10 to -12 EBs were cultured in methylcellulose to generate primitive and definitive hematopoietic colonies, respectively.46 In the presence of EPO, the majority of primitive colonies at day 6 (Figure 4A) were erythroid precursors with brilliant red colonies. The definitive colonies from EB day 11 consisted of multilineage precursors (Figure 4B). As shown in Figure 4A-B, the EphB4–/– EB cells displayed a profound defect in forming both primitive and definitive hematopoietic colonies. To confirm an effect of EphB4 on hematopoietic development, we used flow cytometry to analyze cell surface markers (Figure 4C). Ter119 can be used as a marker for early erythroid differentiation, while CD45 is a marker for definitive multilineage hematopoietic cells. We observed that in early differentiation, EphB4–/– EBs produced substantially fewer Ter119+ cells (< 2%) compared with EphB4+/– EBs (approximately 5%), while CD45+ cells were low in both EphB4+/– and EphB4–/– EBs. By day 10 when definitive hematopoiesis occurs, EphB4–/– gave rise to 10-fold fewer CD45+ cells compared with EphB4+/– EBs (Figure 4C). Similar expression patterns of CD45+ were obtained when ES cells were cultured on fetal liver stromal cells for 10 days (data not shown). At day 10, Ter119+ cells from EphB4–/– EBs increased to 5%, while Ter119 cells of EphB4+/– EBs were 15%. These findings suggest that EphB4 participates in regulating primitive and definitive hematopoiesis during embryogenesis. In the absence of EphB4, differentiation of both is impaired.
EphB4 deficiency affects the developmental potential of both primitive and definitive hematopoiesis. (A) Primitive erythroid colonies (BFU-Es). Day-5 to -7 EB cells of EphB4+/– and EphB4–/– were assayed for their potential to generate primitive erythroid colonies in methylcellulose cultures in the presence of EPO. The right panel is morphology of the representative primitive erythroid colonies from EphB4+/– ES cells. Data shown represent the average of 4 experiments. Error bars represent standard deviation (P = .003). Original magnification, ×10. (B) Definitive hematopoietic colonies. Day-10 to -12 EB cells of EphB4+/– and EphB4–/– were assayed for their potential to generate multilineage hematopoietic colonies in methylcellulose cultures. The right panel is the representative of definitive multilineage hematopoietic colonies from EphB4+/– ES cells. Error bars represent standard deviation (n = 4, P = .001). (C) Decrease expression of Ter119 and CD45 in EphB4–/– ES cells. Day-4 and day-10 EB cells of EphB4+/– or EphB4–/– were analyzed by flow cytometry. Ter119 is a cell surface marker for erythroid cells and CD45 is a cell surface marker for definitive multilineage hematopoietic cells. Error bars represent standard deviation (n = 2).
EphB4 deficiency affects the developmental potential of both primitive and definitive hematopoiesis. (A) Primitive erythroid colonies (BFU-Es). Day-5 to -7 EB cells of EphB4+/– and EphB4–/– were assayed for their potential to generate primitive erythroid colonies in methylcellulose cultures in the presence of EPO. The right panel is morphology of the representative primitive erythroid colonies from EphB4+/– ES cells. Data shown represent the average of 4 experiments. Error bars represent standard deviation (P = .003). Original magnification, ×10. (B) Definitive hematopoietic colonies. Day-10 to -12 EB cells of EphB4+/– and EphB4–/– were assayed for their potential to generate multilineage hematopoietic colonies in methylcellulose cultures. The right panel is the representative of definitive multilineage hematopoietic colonies from EphB4+/– ES cells. Error bars represent standard deviation (n = 4, P = .001). (C) Decrease expression of Ter119 and CD45 in EphB4–/– ES cells. Day-4 and day-10 EB cells of EphB4+/– or EphB4–/– were analyzed by flow cytometry. Ter119 is a cell surface marker for erythroid cells and CD45 is a cell surface marker for definitive multilineage hematopoietic cells. Error bars represent standard deviation (n = 2).
EphB4 affects EB endothelial differentiation
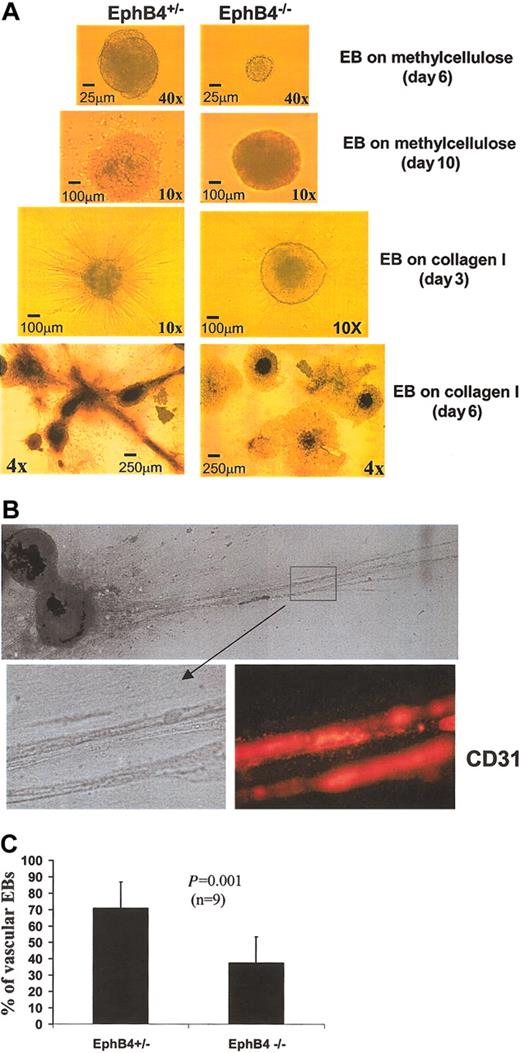
In vitro differentiation of ES cells has been used to investigate molecular mechanisms of vasculogenesis.38,43,47,48 To assess the role of EphB4 in vasculogenesis and angiogenesis, we used a model of in vitro differentiation using ES cells on a collagen I matrix to form a primitive vascular network. In this model, EBs develop endothelial sprouting in the presence of angiogenic growth factors.45 EphB4+/– and EphB4–/– ES cells were cultured on methylcellulose for 11 days in the presence of VEGF and basic FGF (bFGF) to induce a primitive vascular plexus.43 At day 6, smaller EBs were observed in the EphB4–/– ES cell culture (Figure 5A). After 10 to 11 days of culture, EphB4–/– EBs were of similar size as EphB4+/– EBs, but EphB4–/– EBs displayed a less well-defined morphology (Figure 5A). Subculture of these EBs on a collagen I matrix was used to assess sprouting angiogenesis as described by Feraud et al.45 After the standard 3-day culture on the collagen I matrix, the ability of EphB4–/– EBs to form microtubule outgrowths was significantly reduced (Figure 5). With additional culture, sprouts from individual EBs were able to connect and generate vascular network–like structures that were CD31+ by immunohistochemistry (Figure 5B). These data indicate a regulatory role for EphB4 in primitive vascular development.
Endothelial development in EphB4-deficient EBs is impaired. (A) EphB4–/– EBs display a less well defined vasculogenic morphology. ES cells of EphB4+/– (left) and EphB4–/– (right) were induced for vasculogenesis in the presence of VEGF, bFGF, EPO, and IL-6 in methylcellulose for 10 to 11 days. EBs were then transferred in collagen cultures for 3 to 6 days to develop sprouting EBs. (B) Capillary-like structures of EphB4+/– EBs were CD31+ by immunohistochemistry. EB cultures in collagen matrix were dehydrated and fixed in a methanol-acetone solution and stained with PE-conjugated antimouse PECAM1 (CD31; Pharmingen). Original magnification, × 4. (C) Total numbers of vascular-like EBs were significantly decreased in EphB4–/– EBs. EB in collagen matrix were analyzed for sprouting EB induction. Total EBs and capillary-like EBs (vascular EBs) were counted in EphB4+/– and EphB4–/– cultures. Error bars represent standard deviation (n = 9).
Endothelial development in EphB4-deficient EBs is impaired. (A) EphB4–/– EBs display a less well defined vasculogenic morphology. ES cells of EphB4+/– (left) and EphB4–/– (right) were induced for vasculogenesis in the presence of VEGF, bFGF, EPO, and IL-6 in methylcellulose for 10 to 11 days. EBs were then transferred in collagen cultures for 3 to 6 days to develop sprouting EBs. (B) Capillary-like structures of EphB4+/– EBs were CD31+ by immunohistochemistry. EB cultures in collagen matrix were dehydrated and fixed in a methanol-acetone solution and stained with PE-conjugated antimouse PECAM1 (CD31; Pharmingen). Original magnification, × 4. (C) Total numbers of vascular-like EBs were significantly decreased in EphB4–/– EBs. EB in collagen matrix were analyzed for sprouting EB induction. Total EBs and capillary-like EBs (vascular EBs) were counted in EphB4+/– and EphB4–/– cultures. Error bars represent standard deviation (n = 9).
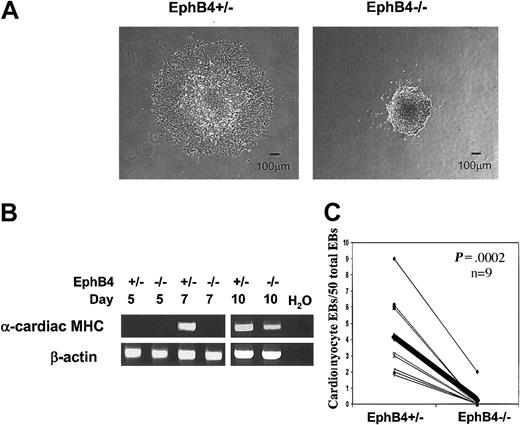
Differentiation of ES cells into beating cardiac myocytes is impaired
Hemangioblasts, blood cells, and vascular endothelial cells are of mesodermal origin. Since cardiomyocytes are also developed from mesoderm, we investigated whether EphB4 had any effect on cardiomyocyte formation by an in vitro differentiation assay. After 5 days in methylcellulose culture, individual EBs were transferred into separate wells of a 48-well plate coated with gelatin. After 24 hours, EphB4+/– EBs were attached to the surface of the wells and had a spread-out morphology. In contrast, EphB4–/– EBs had a smaller and more compact morphology (Figure 6A). Further, the expression of the muscle-specific gene, α-cardiac myosin heavy chain, was detected by RT-PCR on day 7 and day 10 in EphB4+/– EBs by RT-PCR, but only detectable in EphB4–/– day-10 EBs (Figure 6B), indicating muscle differentiation is also delayed in the absence of EphB4. After 9 days of culture (4 days after plating), spontaneously contracting EBs were observed in cells derived from EphB4+/– ES cells. The beating activity was significantly decreased in EphB4–/– EBs (Figure 6C), even with culture for an additional 10 days (data not shown). These results indicate that the formation of cardiac myocytes was impaired from EphB4-deficient ES cells.
Influence of EphB4 on cardiac myocyte differentiation. (A) Morphology of EB after attaching to gelatin-coated culture plates. (B) RT-PCR analysis of cardiac muscle–specific gene, α-cardiac myosin heavy chain (α-cardiac MHC). (C) EBs with beating cardiomyocytes. Paired data are linked by a line from the same experiment (n = 9). The thick line indicates the mean of 9 experiments.
Influence of EphB4 on cardiac myocyte differentiation. (A) Morphology of EB after attaching to gelatin-coated culture plates. (B) RT-PCR analysis of cardiac muscle–specific gene, α-cardiac myosin heavy chain (α-cardiac MHC). (C) EBs with beating cardiomyocytes. Paired data are linked by a line from the same experiment (n = 9). The thick line indicates the mean of 9 experiments.
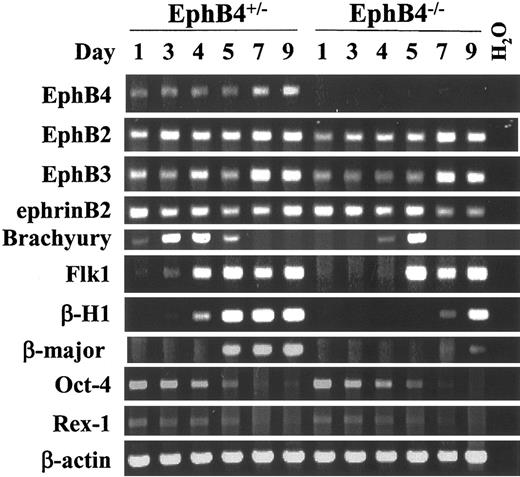
EphB4 deficiency alters expression time course of mesoderm-specific gene products
To examine the possible molecular mechanisms of EphB4 on ES cell differentiation, we analyzed gene expression of EBs by concentrating on several ES, hematopoietic, endothelial, and mesodermal-associated genes. EBs obtained from different time points were subjected to gene expression analysis using RT-PCR. Consistent with the results previously described by others,49 Rex1 and Oct-4, which are expressed in the inner cell mass and in undifferentiated ES cells, were down-regulated in EphB4+/– and EphB4–/– ES cells during EB differentiation over the course of 5 days (Figure 7). Both EphB4 and ephrinB2 gene expression was detectable in undifferentiated ES cells and in early EB development. The expression of EphB4 and ephrinB2 appeared earlier than the expression of Flk-1, raising the possibility that EphB4 and ephrinB2 may play a role in early EB differentiation. The absence of EphB4 did not effect gene expression of either EphB2 or EphB3, nor did it affect expression of the ligand, ephrinB2. However, expression of both the embryonic hemoglobin gene (β-H1) and adult hemoglobin gene (β-major) were down-regulated or delayed in EphB4–/– EBs. These data suggest that both primitive and definitive hematopoiesis were impaired in the absence of EphB4. Assessing genes associated with earlier developmental events, we also noted that Flk-1, the transcript regarded as the marker of hemangioblasts, was delayed in expression in the EphB4–/– EBs at the hemangioblast stage (day-3 to -4 EB). Further, expression of the T-box gene Brachyury, a molecular marker of primitive mesoderm, demonstrated altered kinetics. Brachyury expression peaked at EB day 3 and declined to undetectable levels by day 6 in EphB4+/– ES cells. In contrast, EphB4–/– ES cell expression of Brachyury was delayed, with peak levels not seen until day-5 EB. Therefore, the absence of EphB4 resulted in a consistent change in the time to expression of genes associated with formation of mesoderm and both blood and vascular derivatives.
EphB4 deficiency affects mesoderm gene expression during EB differentiation. RT-PCR of EBs at different time points. ES cells from EphB4+/–1 and EphB4–/–1 were induced for differentiation. RNA samples were harvested at day 1, 3, 4, 5, 7, and 9 of differentiated EBs and analyzed by RT-PCR. Size of each RT-PCR product is shown in Table 1. None of the samples showed genomic DNA amplification (not shown).
EphB4 deficiency affects mesoderm gene expression during EB differentiation. RT-PCR of EBs at different time points. ES cells from EphB4+/–1 and EphB4–/–1 were induced for differentiation. RNA samples were harvested at day 1, 3, 4, 5, 7, and 9 of differentiated EBs and analyzed by RT-PCR. Size of each RT-PCR product is shown in Table 1. None of the samples showed genomic DNA amplification (not shown).
To broadly assess gene expression influenced by EphB4, we used DNA microarray (Affymetrix U74Av2 chips) to compare day-4 EBs from EphB4+/– ES cells with those from EphB4–/– ES cells. We found that 571 genes were down-regulated and 873 genes were up-regulated in EBs on day 4 of EphB4–/–. Among genes down-regulated in EphB4–/– ES cells, there were genes involved in mesoderm formation (such as BMP-4, T-box2, TGF-β, and FGF-3), hematopoiesis (such as c-kit, SCF, GATA-2, and Scl), angiogenesis (such as VEGF and Tie2), and myogenesis (such as Myf6, myosin light chain) (Tables 2-3). These data are indicative of a broad-based effect on mesoderm-associated gene products. Of note, neural-related genes such as α-internexin, glutamic acid decarboxylase (GAD), nestin, N-CAM, and Zinc-finger cerebellum 2 demonstrated a different effect, being up-regulated in EphB4–/– ES cells. Therefore, the role of EphB4 in mesoderm differentiation may be inversely reflected in the neuroectoderm. These data suggest a potential role for EphB4 in affecting germ line choice, but further validation of this concept is needed and requires methods beyond the scope of this study.
Examples of genes down-regulated in EphB4-/- EBs by DNA array analysis
Gene name . | Annotation . |
|---|---|
| Developmental genes | |
| BMP-4 | Bone morphogenetic protein 4, mesodem induction |
| BMP-7 | BMP signaling |
| BMP-5 | BMP signaling |
| BMP-2 | BMP signaling |
| TGF-β1 | Transforming growth factor-β 1 |
| TGF-β2 | TGF-β signaling |
| TGF-βRI | TGF-β-type I receptor |
| TGF-βRII | TGF-β-type II receptor |
| Smad6 | Signal mediators of BMP and TGF-β signaling |
| LMP-1 | Lim protein-1, mediator of BMP-6 signaling |
| T-box 2 | T-box transcription factor, mesoderm development |
| IGF-2 | Insulin-like growth factor 2, mesoderm induction |
| IGFBP4 | Insulin-like growth factor binding protein 4 |
| FGF-3 | Early embryonic development, mesodermal marker |
| FGFR2 | FGF receptor 2 |
| Wnt5a | Frizzled ligand, Wnt signaling |
| wnt11 | Frizzled ligand, Wnt signaling |
| Pax3 | Paired-box transcription factor, myogenesis induction |
| Pax9 | Paired-box transcription factor |
| Endothelial lineage | |
| VEGF | Vascular endothelial growth factor, vasculogenesis |
| Tie2/Tek | Tyrosine kinase receptor, marker of endothelial cells |
| Hematopoietic lineage | |
| c-kit | Tyrosine kinase receptor of SCF |
| SCF | c-kit ligand (stem cell factor) |
| GATA-3 | Transcription factor, T-cell development |
| GATA-2 | Transcription factor, hematopoiesis |
| Scl/TAL-1 | Transcription factor, hematopoiesis |
| Runx1/AML1 | Transcription factor, hematopoiesis |
| CXCR4 | Chemokine receptor of SDF-1 |
| STAT5B | Transcription factor, hematopoiesis |
| GM-CSF-R | GM-CSF receptor |
| Myogenic lineage | |
| Myf6 | Myogenic transcription factor |
| Mylc2a | Myosin light chain regulatory A |
| Myla | Myosin light chain for cardiac atria |
| Mylf | Myosin light chain for fast skeletal muscle |
Gene name . | Annotation . |
|---|---|
| Developmental genes | |
| BMP-4 | Bone morphogenetic protein 4, mesodem induction |
| BMP-7 | BMP signaling |
| BMP-5 | BMP signaling |
| BMP-2 | BMP signaling |
| TGF-β1 | Transforming growth factor-β 1 |
| TGF-β2 | TGF-β signaling |
| TGF-βRI | TGF-β-type I receptor |
| TGF-βRII | TGF-β-type II receptor |
| Smad6 | Signal mediators of BMP and TGF-β signaling |
| LMP-1 | Lim protein-1, mediator of BMP-6 signaling |
| T-box 2 | T-box transcription factor, mesoderm development |
| IGF-2 | Insulin-like growth factor 2, mesoderm induction |
| IGFBP4 | Insulin-like growth factor binding protein 4 |
| FGF-3 | Early embryonic development, mesodermal marker |
| FGFR2 | FGF receptor 2 |
| Wnt5a | Frizzled ligand, Wnt signaling |
| wnt11 | Frizzled ligand, Wnt signaling |
| Pax3 | Paired-box transcription factor, myogenesis induction |
| Pax9 | Paired-box transcription factor |
| Endothelial lineage | |
| VEGF | Vascular endothelial growth factor, vasculogenesis |
| Tie2/Tek | Tyrosine kinase receptor, marker of endothelial cells |
| Hematopoietic lineage | |
| c-kit | Tyrosine kinase receptor of SCF |
| SCF | c-kit ligand (stem cell factor) |
| GATA-3 | Transcription factor, T-cell development |
| GATA-2 | Transcription factor, hematopoiesis |
| Scl/TAL-1 | Transcription factor, hematopoiesis |
| Runx1/AML1 | Transcription factor, hematopoiesis |
| CXCR4 | Chemokine receptor of SDF-1 |
| STAT5B | Transcription factor, hematopoiesis |
| GM-CSF-R | GM-CSF receptor |
| Myogenic lineage | |
| Myf6 | Myogenic transcription factor |
| Mylc2a | Myosin light chain regulatory A |
| Myla | Myosin light chain for cardiac atria |
| Mylf | Myosin light chain for fast skeletal muscle |
SDF-1 indicates stromal-derived factor 1; TGF-β, transforming growth factor β.
Examples of genes up-regulated in EphB4-/- EBs by DNA array analysis
Gene name . | Annotation . |
|---|---|
| Developmental genes | |
| cerebellum2 | Zinc-finger transcription factor |
| cerebellum4 | Zinc-finger transcription factor |
| α-internexin | Neuronal intermediate filament protein |
| nestin | Neuroectodermal marker |
| N-CAM1 | Neural cell adhesion molecular, neuroectodermal marker |
| N-CAM2 | Neural cell adhesion molecular |
| plp | Myelin proteolipid protein |
| Nfl | Neurofilament-medium |
| GAD | Glutamic acid decarboxylase |
Gene name . | Annotation . |
|---|---|
| Developmental genes | |
| cerebellum2 | Zinc-finger transcription factor |
| cerebellum4 | Zinc-finger transcription factor |
| α-internexin | Neuronal intermediate filament protein |
| nestin | Neuroectodermal marker |
| N-CAM1 | Neural cell adhesion molecular, neuroectodermal marker |
| N-CAM2 | Neural cell adhesion molecular |
| plp | Myelin proteolipid protein |
| Nfl | Neurofilament-medium |
| GAD | Glutamic acid decarboxylase |
Day-4 EBs of EphB4+/- and EphB4-/- were used for DNA microarray analysis. Affymetrix mouse U74Av2 chips (about 12 000 genes) were hybridized on a GeneChip system (Affymetrix) at the Center for Genomics Research of Harvard University. Scanned chip images were analyzed using the Microarray Suite Software version 5.0 (Affymetrix). A transcript was considered “change” when the fold change was at least 2. RNA samples for microarray were generated from 4 independent experiments.
Discussion
The data presented here indicate that EphB4 deficiency results in an alteration in the mesodermal differentiation outcome of ES cells. The effects observed were in the timing of expression of specific gene products, and these were validated as reflecting a substantial functional change by in vitro differentiation systems. EphB4 was not essential for completion of any of the differentiation programs assessed; rather, it modulated the rate and magnitude by which they were accomplished. The systems most well defined were those of very early transition into a multipotent hemangioblast and its descendents, resulting in fewer differentiated cells. These data indicate that EphB4 is an early-acting molecular regulator of hemangioblast formation, perhaps functioning temporally similar to Indian hedgehog or Scl.50,51 Its combined effect on the timing of expression of certain lineage-defining genes and the magnitude of specific phenotypically defined cells suggest that there is a critical window for coordinated gene expression that EphB4 may help orchestrate to achieve maximal mesodermal cell output. Absent signaling through this receptor, mesoderm differentiation programs can proceed but with much diminished efficiency in cell production.
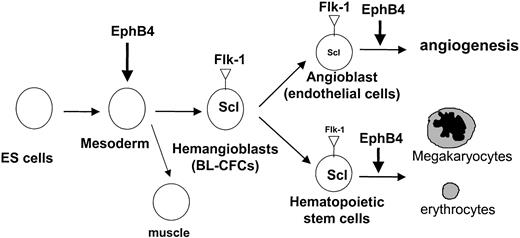
EphB4 appears to exert its effects prior to Flk-1 expression. Flk-1 is required for vascular and blood cell differentiation from hemangioblasts, and both EphB4 and Flk-1 appear to regulate this transition, but EphB4 appears to also act at an earlier step in differentiation. EphB4 affects the expression of the most primitive mesodermal gene products involved in hemangioblast and myocyte formation (Figure 8). These are the first data showing a role for EphB4 in myocyte development and place EphB4 uniquely early in mesoderm tissue development.
Model of EphB4 modulating hematopoiesis and angiogenesis. EphB4 plays a role in facilitating hemangioblast formation and enhancing subsequent hematopoietic and vasculogenic differentiation. EphB4 signaling may be involved in mesoderm induction.
Model of EphB4 modulating hematopoiesis and angiogenesis. EphB4 plays a role in facilitating hemangioblast formation and enhancing subsequent hematopoietic and vasculogenic differentiation. EphB4 signaling may be involved in mesoderm induction.
EphB4's ability to modify differentiation kinetics and outcome poses an interesting set of potential roles. In the simple differentiation systems tested, the time to achieve certain outcomes was delayed, accompanied by a change in the magnitude of cells bearing the more mature phenotype. Rate of differentiation was clearly associated with number of mature cells in settings such as that of embryoid body formation where a reciprocal relationship between mature and immature cells was apparent. The relative balance of mature to immature cells is an expected effect of a change in differentiation kinetics and may be of particular relevance for development. Ephrins and their receptors are often viewed from their association with guidance cues for cell migration, important for establishing organizational patterns of tissues. The data presented here of a differentiation-controlling effect of EphB4 suggests another possible means by which ephrins and Eph receptors may influence tissue pattern formation. While experiments with deficient cells cannot define the physiologic role of receptor activation, the data indicate that EphB4 receptor signaling can modulate the relative abundance of primitive and more differentiated cells. Since ephrins act locally, it may be a mechanism by which tissue boundaries achieve graduated differentiation subsets and perhaps geographically restrict stem cell populations. We previously noted that overexpression (activation) of EphB4 led to accelerated exit of primitive adult hematopoietic cells from a functional stem cell population.29 In the setting of the EphB4-deficient ES model used here, inhibition of differentiation and preservation of a primitive cell type was observed. Therefore the relative gradient of ligand/receptor expression and interaction may dictate the relative abundance of cells from specific differentiation stages.
Transition from a stem cell pool may be of particular interest in development at boundaries where ongoing stem cell function would have the potential to be particularly disruptive. Indeed, ectopic expression of EphB4 in mammary tissue resulted in disordered architecture, abnormal tissue function, and a predisposition to malignancy.52 Absence of EphB3 was associated with altered intestinal crypt architecture.53 These effects may be due to cell migratory effects, but containing primitive cells within borders of more differentiated populations may also reflect a graduated differentiation imposed by ephrin signaling. We propose a model by which ephrin expression is a means of creating a boundary of differentiated cells, effectively restricting primitive cells to locations where ephrin receptor activation is minimized. Data from Durbin et al25 in the zebrafish provide support for this possibility. They noted that altered expression of Ephrin A-L1, Ephrin B2, or EphA4 disrupted normal somite formation in conjunction with reduced expression of the muscle differentiation marker, MyoD.25 The data presented here provide further rationale for a link of differentiation to patterning, but other experimental systems are clearly required to definitively assess it in mammals.
Since the absence of EphB4 did result in an overabundance of more primitive cells, it raises the additional hypothesis that manipulation of the EphB4/ephrinB2 axis may provide a means of altering cell differentiation kinetics ex vivo. In particular, stem cell expansion has been an important but elusive goal for hematopoietic stem cells due to the propensity of the cells to differentiate when cultured. Whether inhibition of EphB4 signaling can favorably affect adult stem cell kinetics to preserve a stemlike pool is now reasonable to test.
In sum, the data presented here indicate that EphB4 is a new molecular modulator of mesodermal differentiation rate and for the first time define it as a regulator of hemangioblast and myocyte formation. The effect of EphB4 on differentiation kinetics, and thereby the relative abundance of mature to immature cells, may be hypothesized to participate in the known role of ephrins and their ligands in defining tissue organization during organogenesis.
Prepublished online as Blood First Edition Paper, September 4, 2003; DOI 10.1182/blood-2003-04-1063.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr David Anderson for the generous gift of EphB4-deficient ES cell lines.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal