Abstract
The use of lentiviral vectors for the transduction of hematopoietic stem cells has evoked much interest owing to their ability to stably integrate into the genome of nondividing cells. However, published large animal studies have reported highly variable gene transfer rates of typically less than 1%. Here we report the use of lentiviral vectors for the transduction of canine CD34+ hematopoietic repopulating cells using a very short, 18-hour transduction protocol. We compared lentiviral transduction of hematopoietic repopulating cells from either stem cell factor (SCF)– and granulocyte-colony stimulating factor (G-CSF)–primed marrow or mobilized peripheral blood in a competitive repopulation assay in 3 dogs. All dogs engrafted rapidly within 9 days. Transgene expression was detected in all lineages (B cells, T cells, granulocytes, and red blood cells as well as platelets) indicating multilineage engraftment of transduced cells, with overall long-term marking levels of up to 12%. Gene transfer levels in mobilized peripheral blood cells were slightly higher than in primed marrow cells. In conclusion, we show efficient lentiviral transduction of canine repopulating cells using an overnight transduction protocol. These results have important implications for the design of stem cell gene therapy protocols, especially for those diseases in which the maintenance of stem cells in culture is a major limitation.
Introduction
Inefficient gene transfer to hematopoietic stem cells has been a significant obstacle to successful stem cell gene therapy. Although stem cell gene transfer efficiency has improved over the past few years, only diseases with a selective advantage of corrected cells, such as severe combined immunodeficiency (SCID)–X1, have been successfully treated by stem cell gene therapy.1 Most diseases, however, do not afford such a selective advantage of gene-corrected cells. Thus, higher gene transfer efficiencies and/or in vivo selection strategies will be required for successful stem cell gene therapy.
Oncoretroviral vectors have been the preferred vector systems for stem cell transductions, but these vectors require cell division for stable vector integration.2,3 Because most hematopoietic stem cells are quiescent,4 ex vivo prestimulation for at least 1 to 2 days has been necessary for efficient oncoretroviral transduction. Prestimulation is followed by an additional 1 or 2 days for the cell transduction, resulting in a total culture time of typically 3 to 4 days.5 Extended culture periods are associated with a loss of stem cells and potentially delayed engraftment, which in turn affect gene transfer levels in vivo.6 In addition, prolonged culture periods are not feasible for stem cell transductions in diseases with a marked fragility of stem cells, such as Fanconi anemia.7
Owing to their ability to transduce nondividing cells, human immunodeficiency virus type 1 (HIV-1)–based lentiviral vectors have great potential for the therapeutic delivery of genes to quiescent cells, such as hematopoietic stem cells.8,9 In addition, the preintegration complex of lentiviral vectors is more stable than that of oncoretroviral vectors. This could make possible the introduction of the lentiviral vector into stem cells during a very brief ex vivo culture period; vector integration would then occur in vivo during the early posttransplantation period.8 These properties of lentiviral vectors support the hypothesis that prolonged culture times are not needed for the stable transduction of target cells, although some activation of target cells is probably required.10
While lentiviral transduction of murine stem cells and human repopulating cells in nonobese diabetic (NOD)/SCID mice appeared to be very promising, results obtained in the nonhuman primate model have been rather disappointing overall. Gene transfer levels were highly variable, and not higher than what had previously been achieved with the use of oncoretroviral vectors.5,11,12 One report suggested that gene transfer with lentiviral vectors may be superior when hematopoietic stem cells from mobilized peripheral blood are used rather than bone marrow.12 A potential explanation for the overall low marking efficiency in the nonhuman primate model may have been the presence of lentivirus-specific inhibitors, as recently described by 2 research groups.13,14
Here we used the dog as a clinically relevant large animal model to study lentiviral gene transfer into hematopoietic repopulating cells. No lentiviral inhibitors have been described for dog cells, and studies in the dog have been highly predictive of studies in humans.
Materials and methods
Animals
Dogs were raised and housed at the Fred Hutchinson Cancer Research Center (FHCRC) (Seattle, WA) under conditions approved by the American Association for Accreditation of Laboratory Animal Care. All animals were provided with commercial chow and chlorinated tap water ad libitum. In preparation for the harvest of stem/progenitor cells, the dogs received recombinant canine granulocyte-colony stimulating factor (cG-CSF) (5 μg/kg body weight subcutaneously, twice daily) and canine stem cell factor (cSCF) (25 μg/kg body weight subcutaneously, once daily) for 5 consecutive days. Marrow draws were performed after animals had been anesthetized with ketamine. Immediately after the marrow draw, a leukapheresis was performed by means of the Spectra Apheresis System (COBE BCT, Lakewood, CO). The machine was primed with autologous blood. A dual-lumen venous catheter was inserted and connected to the apheresis machine. During the procedure, the dogs were constantly monitored for level of sedation or signs of distress, and a slow infusion of 10% calcium gluconate was given to prevent cramping.
As preparation for transplantation, all animals received a single myeloablative dose of 920 cGy total body irradiation (TBI) administered from a linear accelerator at 7 cGy/min. The animals received broad-spectrum antibiotics and cG-CSF after transplantation until absolute neutrophil count (ANC) exceeded 1 × 109/L (1000/μL). The animals also received cyclosporine from the day before transplantation to 35 days after transplantation.
Lentivirus vectors and production of virus-containing medium
The lentiviral transfer vectors RRLsin.cPPT.hPGK.GFP.Wpre and RRLsin. cPPT.hPGK.YFP.Wpre (kindly provided by Dr L. Naldini, Torino, Italy) are self-inactivating HIV-derived vectors expressing the enhanced green fluorescent protein (GFP) or its yellow variant (YFP) from the internal human phosphoglycerate kinase promoter (hPGK) containing a woodchuck hepatitis pre-element as well as a central polypurine tract, and have been described previously.15
Vector stocks of vesicular stomatitis virus glycoprotein (VSV-G)–pseudotype lentiviral vectors were prepared by calcium phosphate–mediated 3-plasmid transfection of 293T cells essentially as described.16 Briefly, 27 μg transfer vector construct, 17.5 μg second-generation gag-pol packaging construct pCMV.ΔR8.74, and 9.5 μg VSV-G expression construct pMD.G were used for transfection of 12 × 106 293T cells overnight in 25 mL Dulbecco modified Eagle medium (DMEM) with 10% heat-inactivated fetal bovine serum. The cells were treated with 10 mM sodium butyrate during the first of three 12-hour vector supernatant collections. The supernatant was filtered through 0.22-μm-pore-size filters (Nalge Nunc International, Rochester, NY) and concentrated 100-fold by ultracentrifugation before freezing and storing at –70° C. All vector stocks were titered by transducing HT1080 cells with the use of limiting dilutions of the stock with analysis for GFP or YFP expression by flow cytometry. The titers of the concentrated VSV-G–pseudotype vector preparations were between 3.7 × 108 and 6.4 × 108 infectious units per milliliter.
CD34 enrichment
The method has been described previously.17,18 Briefly, cells were labeled with biotinylated monoclonal antibody (MoAb) 1H6 (immunoglobulin G1 [IgG1] anticanine CD34) at 4° C for 30 minutes. The cells were washed twice, then incubated with streptavidin-conjugated microbeads for 30 minutes at 4° C, washed again, and then separated by means of an immunomagnetic column technique (Miltenyi Biotec, Auburn, CA) according to the manufacturer's instructions.
Transduction of CD34-enriched cells
Similar numbers of CD34-enriched cells from peripheral blood and bone marrow were exposed directly (without prior cryopreservation) to the corresponding lentiviral vectors at a multiplicity of infection (MOI) of 100 for 18 hours in 75 cm2 canted-neck flasks (Corning, Corning, NY) coated with CH-296 (RetroNectin; Takara Shuzo, Otsu, Japan) at a concentration of 2 μg/cm2 in Iscove modified Dulbecco medium (IMDM) supplemented with 10% fetal bovine serum (FBS) (GIBCO BRL, Gaithersburg, MD), 1% sodium pyruvate, 1% l-glutamine, 1% penicillin/streptomycin (GIBCO BRL) in the presence of fms-like tyrosine kinase 3 ligand (Flt3-L), cSCF, and cG-CSF at a concentration of 50 ng/mL each, and protamine sulfate (8 μg/mL). The vector titer used during transduction was kept constant between the 2 experimental arms compared within one animal (G115, 1.5 × 108 infectious units [iu]/mL; G136, 9 × 107 iu/mL; G206, 8 × 107 iu/mL). After transduction, nonadherent and adherent cells were pooled, counted, and infused intravenously into the animal.
Analysis of gene expression in colony-forming cells (CFCs)
CD34-enriched cells were cultured in a double-layer agar culture system. Isolated cells were cultured in alpha minimal essential medium supplemented with FBS (Hyclone, Logan, UT), bovine serum albumin (BSA) (fraction V) (Sigma, St Louis, MO), 1% (wt/vol) agar (Difco, Detroit, MI), overlaid on medium with 0.6% agar (wt/vol) containing 100 ng/mL cSCF, cG-CSF, canine granulocyte-macrophage colony-stimulating factor (cGM-CSF), and 4 U/mL erythropoietin. Cultures were incubated at 37° C in 5% CO2 and 95% air in a humidified incubator. CD34 cells after transduction were plated at a density of 2000 cells prior to transduction per plate. Nontransduced control cells were plated at the same time. All cultures were performed at least in triplicate. The total number as well as the number of GFP+/YFP+ colonies were enumerated at day 14 of culture by fluorescence microscopy.
Flow-cytometric analysis
GFP/YFP-expressing white blood cells were quantitated by flow-cytometric analysis of at least 250 000 events (propidium iodide [1 μg/mL]–excluding, forward and right-angle light scatter–gated) on a FACS Vantage (Becton Dickinson, San Jose, CA). For analysis of red blood cells and platelets, a FACS Calibur was used (Becton Dickinson). Flow-cytometric data were analyzed by CELLQuest v3.1f software (Becton Dickinson) with gating to exclude fewer than 0.1% control cells in the relevant region. The results were then plotted over time in an MS Excel (Microsoft, Redmond, WA) chart. Murine antihuman monoclonal antibodies conjugated to phycoerythrin (PE), which had been shown to bind to canine CD antigens included CD21 (clone CA2.1D6; Serotec, Raleigh, NC) for B cells, and CD14 (clone TÜK4; DAKO, Carpinteria, CA) for monocytes. The monoclonal antibody DM5 as a marker for granulocytes and anti-CD3 (clone 17.6B3) for T cells were kindly provided by Drs Peter Moore and Brenda Sandmaier (University of California, Davis, and Fred Hutchinson Cancer Research Center, Seattle, WA, respectively).
Fluorescent probe PCR assay (TaqMan)
Relative marking levels with EGFP and EYFP were analyzed with the TaqMan 5′ nuclease quantitative real-time polymerase chain reaction (PCR) assay.19 Calculated gene marking percentages assume that the marrow and peripheral blood cells contain one copy of the corresponding vector per cell. We amplified 300 ng DNA in at least duplicate with an EGFP-specific primer/probe combination (5′-CTG CAC CAC CGG CAA-3′ and 5′-GTA GCG GCT GAA GCA CTG-3′; probe, 5′-FAM-CCA CCC TGA CCT ACG GCG TG -TAMRA-3′) (Synthegen, Houston, TX), and with an EYFP-specific primer/probe combination (5′-CTG CAC CAC CGG CAA-3′ and 5′-GTA GCG GGC GAA GCA CT-3′; probe, 5′-FAM-CCA CCT TCG GCT ACG GCC TG -TAMRA-3′). A canine interleukin 3 (IL-3)–specific primer/probe combination (5′-ATG AGC AGC TTC CCC ATC C-3′, 5′-GTC GAA AAA GGC CTC CCC-3′; probe, 5′-FAM-TCC TGC TTG GAT GCC AAG TCC CAC -TAMRA-3′) was used to adjust for equal loading of genomic DNA per reaction. Standards consisted of dilutions of DNA extracted from cell lines transduced with a single copy of the GFP or YFP vector. Negative controls consisted of DNA extracted from peripheral blood mononuclear cells (PBMCs) obtained before transplantation from control animals or water. Reactions were run by means of the ABI master mix (Applied Biosystems, Branchburg, NJ) on the ABI Prism 7700 sequence detection system (Applied Biosystems) under the following thermal cycling conditions: 50° C for 2 minutes and 95° C for 10 minutes, then 40 cycles of 95° C for 15 seconds and 60° C for 1 minute.
Linear amplification–mediated PCR
Integration site analysis by linear amplification–mediated PCR (LAM-PCR) was performed on canine DNA isolated from either total bone marrow (BM) or peripheral blood leukocytes (PBLs). We used 100 ng DNA from peripheral blood samples or crude lysate from plucked colonies to serve as a template for LAM-PCR that was performed as previously described.20,21
Detection of replication-competent virus
Serum of all 3 animals was tested for the presence of recombinant replication-competent lentiviral vectors by means of the p24 Antigen Assay (Beckman Coulter, Miami, FL) according to the manufacturer's recommendations. The p24 antigen was not detectable at any time point, indicating the absence of helper-virus production.
Results
Experimental design
We used a competitive repopulation assay in dogs to study lentiviral gene transfer into hematopoietic repopulating cells using a short, 18-hour transduction protocol. Since we had observed immune responses against lentivirally transferred gene products, even after myeloablative conditioning,22 cyclosporine was given to all 3 dogs after transplantation to avoid the development of immune responses. Animals have been followed for a mean of 51 (range, 39-63) weeks. All animals were analyzed for replication-competent virus by testing for the p24 antigen. The p24 antigen was not detectable at any time point, indicating the absence of helper-virus production.
Rapid engraftment after transplantation of lentivirally transduced cells
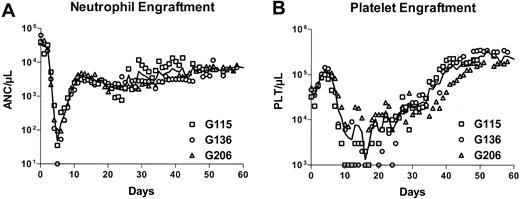
In the 3 animals, a stable ANC exceeding 0.5 × 109/L (500/μL) was reached within 8 to 9 days, and a platelet count exceeding 50 ×109/L (50 000/μL) was reached at a median of 35 days (Table 1). Figure 1 displays ANCs (panel A) and platelet counts (panel B) after transplantation for all 3 animals in this study. Engraftment was remarkably fast in comparison with historic controls, which received hematopoietic stem cells transduced with oncoretroviral vectors in a 3-day transduction protocol.17,23
Engraftment of lentivirally transduced dog peripheral blood and marrow cells
Dog . | Days to ANC exceeding 500/μL . | Days to platelet count exceeding 50 000/μL . |
|---|---|---|
| G115 | 8 | 35 |
| G136 | 9 | 35 |
| G206 | 8 | 41 |
Dog . | Days to ANC exceeding 500/μL . | Days to platelet count exceeding 50 000/μL . |
|---|---|---|
| G115 | 8 | 35 |
| G136 | 9 | 35 |
| G206 | 8 | 41 |
Rapid hematopoietic recovery in dogs receiving transplants of lentivirally transduced CD34-enriched cells in an overnight transduction protocol. For all 3 dogs, the absolute neutrophil counts (A) and the platelet counts (B) after transplantation are displayed. The solid line marks the time course of average cell numbers of all 3 dogs. All animals engrafted without delay and faster than historic controls receiving transplants of cells transduced in a 3-day transduction protocol.
Rapid hematopoietic recovery in dogs receiving transplants of lentivirally transduced CD34-enriched cells in an overnight transduction protocol. For all 3 dogs, the absolute neutrophil counts (A) and the platelet counts (B) after transplantation are displayed. The solid line marks the time course of average cell numbers of all 3 dogs. All animals engrafted without delay and faster than historic controls receiving transplants of cells transduced in a 3-day transduction protocol.
Gene transfer efficiency in progenitor cells before transplantation
Transduction efficiency before transplantation was assessed by flow-cytometric determination of GFP+ or YFP+ CD34-enriched cells or by scoring GFP+/YFP+ colony-forming cells by fluorescence microscopy on day 14. Gene transfer levels were very high overall, with about 50% to 80% GFP/YFP-expressing colonies. Table 2 summarizes the results of the pretransplantation analysis of hematopoietic progenitor cells. Gene transfer efficiency was overall slightly higher in CD34-enriched cells from marrow than from mobilized peripheral blood (43.0% ± 3.6% versus 30.3% ± 6.5%) although this difference was not statistically significant (P = .11; 1-sided t test).
Lentiviral transduction of dog peripheral blood and marrow cells
Dog and source . | No. CD34-enriched cells × 106/kg before culture . | Purity of CD34-enriched cells, % . | No. infused cells per kg × 106 . | Transduced CFCs, %* . |
|---|---|---|---|---|
| G115 | ||||
| PB, GFP | 3.3 | 86 | 3.6 | 67 |
| BM, YFP | 3.4 | 84 | 4.9 | 81 |
| G136 | ||||
| PB, YFP | 1.5 | 83 | 0.8 | 60 |
| BM, GFP | 1.8 | 69 | 0.9 | 73 |
| G206 | ||||
| PB, GFP | 2.8 | 87 | 2.5 | 49 |
| BM, YFP | 2.3 | 86 | 1.7 | 65 |
Dog and source . | No. CD34-enriched cells × 106/kg before culture . | Purity of CD34-enriched cells, % . | No. infused cells per kg × 106 . | Transduced CFCs, %* . |
|---|---|---|---|---|
| G115 | ||||
| PB, GFP | 3.3 | 86 | 3.6 | 67 |
| BM, YFP | 3.4 | 84 | 4.9 | 81 |
| G136 | ||||
| PB, YFP | 1.5 | 83 | 0.8 | 60 |
| BM, GFP | 1.8 | 69 | 0.9 | 73 |
| G206 | ||||
| PB, GFP | 2.8 | 87 | 2.5 | 49 |
| BM, YFP | 2.3 | 86 | 1.7 | 65 |
CFC indicates colony-forming cell; PB, peripheral blood; BM, bone marrow.
Percentage of fluorescence-positive colonies plated immediately after transduction and assessed on day 14 by fluorescence microscopy.
Efficient gene transfer into canine repopulating cells
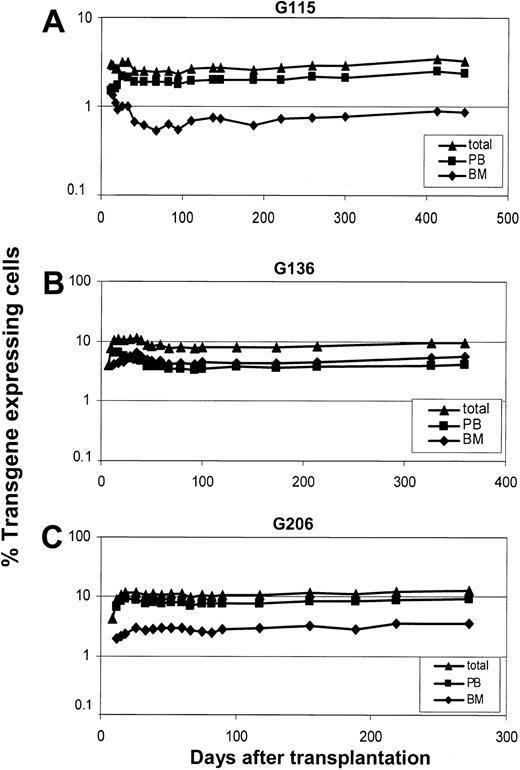
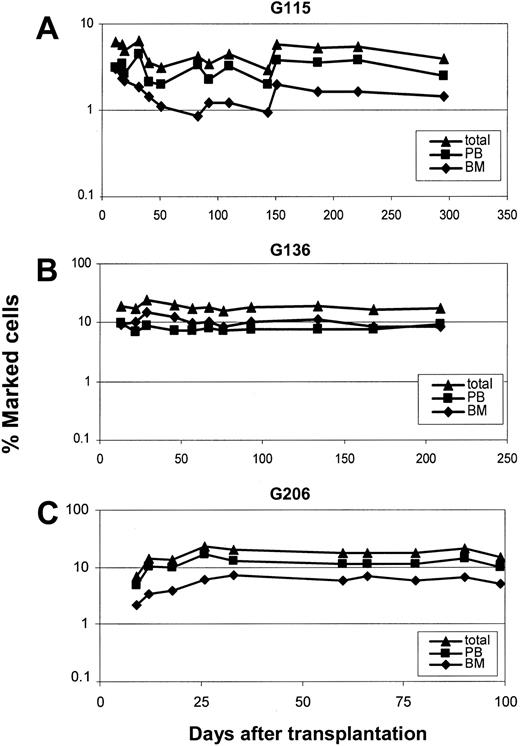
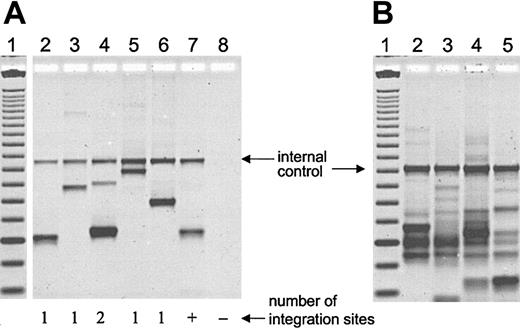
Transduction efficiency of hematopoietic repopulating cells after transplantation was measured by simultaneous flow-cytometric detection of GFP and YFP in peripheral blood cells (Figure 2). The overall percentage of transgene-expressing cells was high, with up to 12% showing long-term marking (Figure 2B-C). Two different cell sources, primed marrow and mobilized peripheral blood, were used for transduction of CD34-enriched cells. In 2 animals (G115 and G206), mobilized peripheral blood resulted in 3- to 4-fold higher gene transfer levels than primed marrow. In one animal (G136) marking levels were slightly higher with the use of primed marrow as a stem cell source. Follow-up of longer than 9 months is available for all 3 animals, and 2 animals have been followed for more than 1 year. The level of transgene-expressing cells was very stable over time; this is in contrast to the marking with oncoretroviral vectors, which typically declines over time. Gene transfer rates after transplantation were also measured by real-time PCR (Figure 3). The difference between the 2 experimental arms (peripheral blood versus marrow) determined by flow cytometry could be confirmed by quantitative PCR in all cases. There was also a very good correlation between gene marking levels determined by flow cytometry and real-time PCR, indicating that gene expression from the vectors used was stable over time. On average, there was less than a 2-fold difference between flow cytometry (Figure 2) and real-time PCR (Figure 3), suggesting that, on average, repopulating cells contained fewer than 2 vector copies. To directly assess the number of transgene copies per cell, bone marrow from animal G115 was plated in semisolid medium, and individual colonies were plucked and subjected to LAM-PCR. A total of 16 colonies were evaluable, 14 of which contained only 1 vector copy while 2 had 2 copies. A representative analysis is shown in Figure 4A. In addition, LAM-PCR on peripheral blood from this animal obtained at different time points after transplantation demonstrated a polyclonal repopulation pattern (Figure 4B).
Gene expression levels in peripheral blood cells of dogs that received lentivirally transduced stem cells from mobilized peripheral blood and primed bone marrow. Displayed are the percentages of transgene-expressing leukocytes detected by flow-cytometry in all 3 dogs: G115 (A), G136 (B), G206 (C).
Gene expression levels in peripheral blood cells of dogs that received lentivirally transduced stem cells from mobilized peripheral blood and primed bone marrow. Displayed are the percentages of transgene-expressing leukocytes detected by flow-cytometry in all 3 dogs: G115 (A), G136 (B), G206 (C).
Gene marking levels in peripheral blood cells of dogs that received lentivirally transduced stem cells from mobilized peripheral blood and primed bone marrow. Displayed are the percentages of transgene-positive leukocytes as determined by quantitative PCR in all 3 dogs: G115 (A), G136 (B), G206 (C).
Gene marking levels in peripheral blood cells of dogs that received lentivirally transduced stem cells from mobilized peripheral blood and primed bone marrow. Displayed are the percentages of transgene-positive leukocytes as determined by quantitative PCR in all 3 dogs: G115 (A), G136 (B), G206 (C).
Polyclonal repopulation with hematopoietic cells containing only 1 or 2 vector integrations. (A) LAM-PCR was performed on cell lysate from CFCs plucked 1 year after transplantation of dog G115. Lane 1, 25 base pair (bp) DNA ladder; lanes 2 to 6, individual colonies; lane 7, positive control (+); lane 8, negative control (–). Of a total of 16 evaluable colonies, 14 contained 1 integration site, and only 2 contained 2 integration sites. (B) Peripheral blood samples from dog G115 at different time points after transplantation were analyzed by LAM-PCR, revealing that multiple different clones persist in the long term. Lane 1, 25 bp DNA ladder; lane 2, day 83; lane 3, day 110; lane 4, day 418; lane 5, day 453.
Polyclonal repopulation with hematopoietic cells containing only 1 or 2 vector integrations. (A) LAM-PCR was performed on cell lysate from CFCs plucked 1 year after transplantation of dog G115. Lane 1, 25 base pair (bp) DNA ladder; lanes 2 to 6, individual colonies; lane 7, positive control (+); lane 8, negative control (–). Of a total of 16 evaluable colonies, 14 contained 1 integration site, and only 2 contained 2 integration sites. (B) Peripheral blood samples from dog G115 at different time points after transplantation were analyzed by LAM-PCR, revealing that multiple different clones persist in the long term. Lane 1, 25 bp DNA ladder; lane 2, day 83; lane 3, day 110; lane 4, day 418; lane 5, day 453.
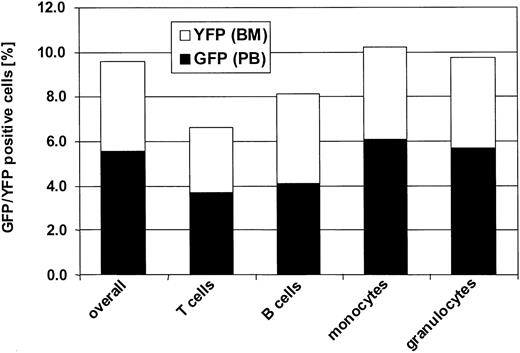
GFP and YFP expression is detectable in all peripheral blood subsets
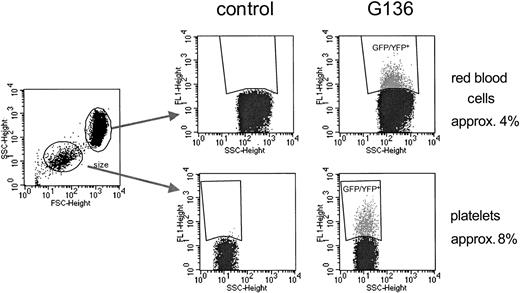
To assess gene expression in different hematopoietic lineages, peripheral blood cells in all 3 dogs were labeled with antibodies against granulocytes (DM-5), T lymphocytes (CD3), and monocytes (CD14), and analyzed by flow cytometry at different time points. Figure 5 shows a representative result obtained in 1 of the 3 dogs at 1-year after transplantation. Sustained GFP expression could be detected in all subsets examined in the 3 dogs. In all 3 dogs, we were also able to detect fluorescence-positive platelets and fluorescence-positive erythrocytes (Figure 6). Since fluorescence intensity in these cell populations is significantly lower than in white blood cells, a different filter setup that did not allow differentiation between GFP and YFP had to be used for detection (see “Materials and methods”).
Flow-cytometric analysis of transgene-expressing cells in peripheral blood subpopulations. Displayed is the percentage of transgene-positive cells in different leukocyte subpopulations in the peripheral blood of dog G136 at 12 months after transplantation. In all dogs, GFP- and YFP-expressing cells were found in all lineages examined.
Flow-cytometric analysis of transgene-expressing cells in peripheral blood subpopulations. Displayed is the percentage of transgene-positive cells in different leukocyte subpopulations in the peripheral blood of dog G136 at 12 months after transplantation. In all dogs, GFP- and YFP-expressing cells were found in all lineages examined.
Flow-cytometric detection of transgene expression in red blood cells and platelets. Gating on red blood cells and platelets was based on scatter characteristics (left panel). Transgene-expressing red blood cells (upper panels) and platelets (lower panels) were detected in all 3 dogs. Because of the overlapping positive and negative populations due to low fluorescence intensity, especially in red blood cells, the percentages displayed most likely underestimate the actual percentage of GFP/YFP-expressing cells.
Flow-cytometric detection of transgene expression in red blood cells and platelets. Gating on red blood cells and platelets was based on scatter characteristics (left panel). Transgene-expressing red blood cells (upper panels) and platelets (lower panels) were detected in all 3 dogs. Because of the overlapping positive and negative populations due to low fluorescence intensity, especially in red blood cells, the percentages displayed most likely underestimate the actual percentage of GFP/YFP-expressing cells.
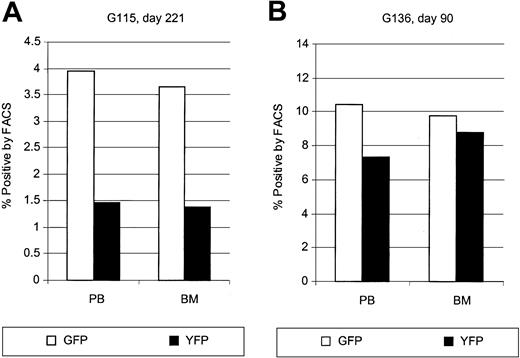
Similar marking levels in bone marrow and peripheral blood
To compare marking/expression levels in marrow and peripheral blood cells, marrow samples were obtained at various time points after transplantation. The percentage of transgene-expressing cells was determined by flow cytometry, and the percentage of gene-marked cells by real-time PCR. Representative examples are shown in Figure 7. There was no obvious difference in the percentage of gene-marked cells in marrow and peripheral blood. Additionally, marrow cells were plated in agar cultures to assess transgene marking in a functionally defined precursor population. Owing to the overlapping emission spectra of GFP and YFP, it was not possible to differentiate between GFP- and YFP-expressing colonies. However, the overall percentage of fluorescent colonies correlated closely to the flow-cytometric results, with up to 12% colonies expressing GFP or YFP.
Comparison of gene transfer in peripheral blood and bone marrow leukocytes after transplantation. Displayed are representative examples of the percentages of gene-expressing cells detected in the peripheral blood and bone marrow at 221 days after transplantation in dog G115 (A) and at 90 days after transplantation in dog G136 (B).
Comparison of gene transfer in peripheral blood and bone marrow leukocytes after transplantation. Displayed are representative examples of the percentages of gene-expressing cells detected in the peripheral blood and bone marrow at 221 days after transplantation in dog G115 (A) and at 90 days after transplantation in dog G136 (B).
Discussion
In the current study, we compared lentiviral transduction efficiency of hematopoietic repopulating cells from mobilized PB and primed BM cells using a very short, 18-hour transduction protocol. Stable gene transfer into hematopoietic repopulating cells was up to 12%, which is a remarkable level considering the short transduction period. In addition, the close correlation between gene expression and gene marking, and the analysis of single CFCs by LAM-PCR, suggest that the number of vector copies per cell in long-term repopulating cells is low, even when high MOIs are used for lentiviral transduction.
Over the past years, oncoretroviral transduction of hematopoietic stem cells has significantly improved.17,24-26 However, relatively long ex vivo culture periods of 3 to 4 days have been required for efficient oncoretroviral gene transfer. Unfortunately, extended culture periods have been associated with a decline in hematopoietic repopulating cells and thus increased risk of delayed engraftment or even graft failure.6 Short transduction cultures are therefore preferred for clinical stem cell gene therapy applications, especially for diseases such as Fanconi anemia, where maintenance of stem cells has been a significant obstacle to successful gene therapy. Because of their ability to stably transduce nondividing cells, lentiviral vectors have been considered as a potential alternative vector system for stem cell transduction.
Hematopoietic recovery in the dogs presented in this study was considerably more rapid than in previous autologous transplantation studies using oncoretrovirally transduced hematopoietic stem cells.17,23 In our previous studies, mean neutrophil engraftment (ANC greater than 0.5 × 109/L [500/μL]) occurred at day 18 as compared with day 8 in the current study. The difference was, however, not statistically significant owing to the small number of animals in the current study. In addition, it is not clear how much the combination of 2 stem cell sources (PB and BM) may have contributed to a faster hematopoietic recovery; our data suggest that the repopulation capability of the cultured cells did not decline within this brief ex vivo time.
Lentiviral marking was very stable over time and did not decline as is commonly observed with the use of oncoretroviral vectors. In contrast to previously reported highly variable results with lentiviral vectors in nonhuman primates,5,11,12,24 we observed very reproducible marking levels in the dogs. Potential explanations for this variable and for rather low overall marking efficiency may be the presence of lentivirus-specific inhibitors or immune responses. The presence of such inhibitors in nonhuman primates has recently been reported.13,14 Furthermore, immune responses against the transfer vector or the transgene itself have been a long-standing concern in gene therapy and may have contributed to the highly variable results previously reported for lentiviral transduction of hematopoietic stem cells.5,11,12 Antibody and cytotoxic T lymphocyte (CTL) responses to gene-modified cells have been reported after transplantation of GFP-transduced CD34+ cells following a nonmyeloablative conditioning regimen.27 More recently, we have encountered immune responses against GFP/YFP in baboons after a fully myeloablative conditioning regimen.22 Additionally, we have previously observed a very sudden decrease and even loss of marking in some dogs, suggestive of an immune response against genetically marked cells. In the current study, we therefore included cyclosporine as an immunosuppressive drug after transplantation to prevent potential immune responses to the gene-modified cells. While we have not formally investigated immune responses in the current study, it appears that the administration of immunosuppressive drugs has contributed to the stable and consistent marking described in this study.
Previous studies by Dunbar and colleagues28 in a human autologous transplantation study suggested that mobilized PB cells may be preferable to BM for gene therapy applications. Although a number of publications have since dealt with this important issue of the optimal stem cell source for oncoretroviral and lentiviral gene transfer into hematopoietic stem cells,12,25,26,29 no formal direct comparison of primed BM and mobilized PB in a large animal model has been performed with the use of lentiviral vectors. We therefore directly compared these 2 stem cell sources in a competitive repopulation assay in the canine autologous transplant model. In all 3 animals, gene transfer into the bulk cell population and into colony-forming cells was higher in primed BM cells than in mobilized PB cells. The most obvious explanation for this finding seems to be that, in contrast to mobilized PB cells, a significant proportion of progenitor cells from BM are actively proliferating30,31 and that gene transfer efficiency is related to the activation status of the target cells.10
In contrast to the in vitro results obtained before transplantation, in vivo gene marking was 3- to 4-fold higher with the use of mobilized PB product as a stem cell source in 2 animals, while in 1 animal primed BM was slightly superior. This finding is consistent with previous reports that suggested higher gene transfer rates with the use of mobilized PB.12 However, this difference was not consistently observed in all 3 animals. In addition, an influence of the vector on transduction efficiency, although unlikely, cannot be completely ruled out, since it was the GFP-expressing vector in all the dogs that led to higher long-term marking. However, our data clearly demonstrate that efficient transduction with lentiviral vectors is possible into cells of either stem cell source.
The most accepted assay for the transduction of stem cells is the stable generation of transgene-carrying progeny that will differentiate into all blood lineages and persist for the lifetime of the recipient. We were able to follow the dogs that received lentivirally transduced primed BM and mobilized PB cells for more than 1 year and observed stable marking in the peripheral blood of all 3 dogs. We furthermore detected transgene-expressing cells of all hematopoietic lineages, including red blood cells and platelets. These data strongly suggest that long-term, multipotent repopulating cells were transduced with lentiviral vectors.
Gene transfer levels detected in peripheral blood by both flow cytometry and quantitative real-time PCR were very close to those determined in bone marrow leukocytes (Figure 7). This observation is consistent with reports of a similar finding in nonhuman primates by our group5 and other investigators.32 To assess gene transfer levels in functionally defined precursor cells, we determined the percentage of transduced CFCs at various time points after transplantation. Although it was technically not possible to differentiate between GFP and YFP expression by fluorescence microscopy, the overall percentage of transduced colonies correlated well with the marking levels in peripheral blood and bone marrow leukocytes. These data support the interpretation that there is no block in differentiation or specific elimination of mature genemodified cells as had previously been suggested by other investigators.33,34
The risk of malignant transformation of stably transduced hematopoietic stem cells by insertional mutagenesis has been of concern because 2 children in Hacein-Bey-Abina et al's gene therapy study for X-linked SCID have developed leukemia.35 The number of proviral copies per cell is thus a very important aspect in gene transfer studies because the risk of insertional mutagenesis increases with the number of proviral copies per cell,36 although it is currently not clear whether this increase is exponential or linear.37,38 In the current study, we demonstrated by LAM-PCR that only 1 or 2 proviral copies were present in colony-forming cells after transplantation, even though a high MOI of 100 was used for transduction. This finding is in contrast to studies in the NOD/SCID mouse where transductions with an MOI of 100 (as used in the current study) have resulted in an average of 5.6 vector copies in transduced NOD/SCID repopulating cells.39 These data suggest that the safety of lentiviral vectors, with regard to the number of insertion sites in repopulating cells, may be similar to oncoretroviral vectors in large animals and humans.
In conclusion, we report efficient and reproducible transduction of long-term, multipotent canine repopulating cells in a short overnight transduction protocol with lentiviral vectors. The transduction protocol described here should be particularly important for diseases like Fanconi anemia in which maintenance of stem cells in transduction cultures has been a major obstacle to successful gene therapy.
Prepublished online as Blood First Edition Paper, January 22, 2004; DOI 10.1182/blood-2003-07-2414.
Supported in part by grants HL36444, DK47754, HL074162, and DK56465 from the National Institutes of Health, Bethesda, MD; H.P.K. is a Markey Molecular Medicine Investigator.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We would like to thank Michele Spector, DVM; the technicians in the canine facilities of the Fred Hutchinson Cancer Research Center; and the investigators of the Program in Transplantation Biology who participated in the weekend treatments. We wish to thank Drs Rainer Storb, Peter Moore, and Brenda Sandmaier for providing antibodies for subset analyses; Amgen for providing canine-specific growth factors; and the technicians of the hematology and pathology laboratories of the Fred Hutchinson Cancer Research Center. We also acknowledge the assistance of Bonnie Larson and Helen Crawford in preparing the manuscript.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal