Abstract
Gene expression patterns of CD34+CD38- cells derived from human embryonic stem cells (ESCs) were compared with those of cells isolated from adult human bone marrow (BM) using microarrays; 1692 and 1494 genes were expressed at levels at least 3-fold above background in cells from BM and ESCs, respectively. Of these, 494 showed similar levels of expression in cells from both sources, 791 genes were overexpressed in cells from BM (BM versus ESCs, at least 2-fold), and 803 genes were preferentially expressed in cells from ESCs (ESCs versus BM, at least 2-fold). The message of the flt-3 gene was markedly decreased in cells from ESCs, whereas there was substantial flt-3 expression in cells from BM. High levels of embryonic ϵ-globin expression were observed—but no adult β-globin message—in CD34+CD38- cells from ESCs, whereas high levels of β-globin expression—but no embryonic ϵ-globin message—could be detected in cells from BM. Furthermore, high levels of major histocompatibility complex (MHC) gene expression were demonstrated in cells from BM but very low levels of MHC message in corresponding cells from ESCs. These observations demonstrate that CD34+CD38- cells derived from ESCs correspond consistently to an early developmental stage at which the yolk sac and fetal liver are the primary sites of hematopoiesis.
Introduction
Embryonic stem cells (ESCs), pleuripotent cells that can differentiate to form all of the various cell types of the body, have been established from a variety of mammalian species, including nonhuman primates and man.1-4 The hematopoietic differentiation of ESCs in vitro has been investigated extensively, and hematopoietic precursors as well as differentiated progeny representing erythroid, myeloid, megakaryocytic, and lymphoid lineages have been demonstrated in differentiation cultures of ESCs from several species including rhesus monkey and man.5-7 The hematopoietic character of these precursors is further supported by the demonstration that they express genes associated with early hematopoietic differentiation.5
Among the distinctive characteristics of hematopoietic stem cells (HSCs) is their ability to repopulate bone marrow (BM)–ablated animals. Murine HSCs derived from ESC differentiation in vitro, however, generally lack long-term reconstitution potential when transplanted into adult recipient mice.8,9 The failure of ESC-derived HSCs to engraft could be explained by 2 broad hypotheses. First, ESC differentiation under culture conditions could have resulted in altered regulation of certain genes in the hematopoietic progenitor/stem cells that might have impaired their function as HSCs. Alternatively, ESC differentiation conditions could have resulted in the generation of hematopoietic precursors that retained critical properties of embryonic cells, and did not undergo complete maturation to adult HSCs, and consequently lacked the ability to engraft in adult BM.10-12 The objective of the present study was to compare gene expression profiles between CD34+CD38- hematopoietic cells with known BM repopulation potential—that is, cells harvested from adult BM—against CD34+CD38- cells from human ESC differentiation cultures.
We previously compared the expression patterns of multiple genes associated with hematopoietic differentiation, HSC homing and engraftment, and cell cycle control in hematopoietic precursors derived from rhesus monkey ESCs, and in those isolated from adult rhesus monkey BM, by semiquantitative reverse transcriptase–polymerase chain reaction (RT-PCR). These studies, encompassing about 60 genes, demonstrated a remarkable degree of similarity in the expression patterns of these genes in cells from both sources, with only few exceptions.13,14 The approach we used, however, could only examine a limited number of genes. To expand this study, we took advantage of the availability of high-density microarrays, which we applied to characterize the expression of more than 10 000 genes in CD34+CD38- cells from cultures of human ESCs. An initial obstacle to applying this approach was the limited number of cells that could be generated from ESC differentiation in vitro. The quantity of RNA required for high-density microarray analysis is normally in the microgram range and at the least is on the order of 100 ng with the most advanced array technology. In this study, however, we were able to utilize SMART technology (Clontech, Palo Alto, CA) successfully for amplifying minute amounts of RNA to examine gene expression patterns in CD34+CD38- cells derived from human ESC cultures and from adult BM with high-density microarrays.
Materials and methods
Hematopoietic differentiation of human ESCs
Human ESC line H1 was obtained from the WiCell Research Institute (Madison, WI). Undifferentiated H1 cells were maintained according to the culture protocol provided by WiCell. Briefly, cells were cultured on mouse embryonic fibroblast feeder cells irradiated with 3000 cGy in complete ESC medium consisting of 20% KNOCK-OUT serum replacement (Gibco, Grand Island, NY), 80% Dulbecco modified Eagle medium (DMEM)/F12 medium (Invitrogen, Carlsbad, CA), 1 mM l-glutamine, 0.1 mM β-mercaptoethanol, 1% nonessential amino acids, and 4 ng/mL human fibroblast growth factor (hb-FGF). ESCs from passage no. 29 were used for these analyses. Subconfluent layers of murine S17 BM stromal cells (kindly provided by Dr Kenneth Dorshkind, UCLA Medical Center, Los Angeles, CA) were grown in 6-well culture plates, and collagenase-dissociated H1 ESCs were seeded onto the S17 cell layers. The hematopoietic differentiation medium consisted of Iscove modified Dulbecco medium (IMDM) supplemented with 8% horse serum (Gibco), 8% fetal bovine serum, 5 × 10-6 M hydrocortisone, 15 ng/mL bone morphogenetic protein 4 (BMP-4), 5 ng/mL BMP-2, and 5 ng/mL BMP-7 (R&D Systems, Minneapolis, MN), levels previously determined in dose-response studies using rhesus monkey ESCs.15 At day 6 of differentiation, recombinant human stem cell factor (SCF), interleukin-3 (IL-3), vascular endothelial growth factor (VEGF), granulocyte colony-stimulating factor (G-CSF; 20 ng/mL of each), IL-6, Flt-3 ligand (10 ng/mL of each), and erythropoietin (Epo; 2 U/mL; R&D Systems) were added to the cultures. Differentiation cultures were fed periodically with fresh medium including cytokines. At day 20, differentiated cells were rinsed off the culture plates for flow cytometry analysis and CD34+CD38- cell purification.
Purification of human CD34+CD38- cells
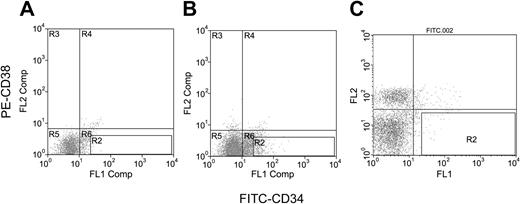
Day-20 H1 ESC-derived hematopoietic precursors were rinsed from the culture wells by gentle pipetting. The cells were passed through a 40-μm nylon cell strainer (Falcon, Deerfield, IL), washed twice with phosphate-buffered saline (PBS; Life Technologies, Bethesda, MD) supplemented with 1% bovine albumin, and stained with biotinylated antihuman CD34 (clone 12.8; Baxter) plus streptavidin–fluorescein isothiocyanate (FITC) and phycoerythrin (PE)–antihuman CD38 (clone HIT2; PharMingen, San Diego, CA) antibodies. The CD34+38- cells were collected by a Moflow high-speed sorter (DakoCytomation, Fort Collins, CO). CD34+CD38- cells comprised about 0.3% of the total population, and a total of 2840 CD34+CD38- cells were obtained. Normal human BM was purchased from AllCells (Berkeley, CA); CD34+CD38- cells were purified as described previously in this paragraph. Figure 1 shows the fluorescence-activated cell sorter (FACS) profiles that define the gates for purification of the CD34+CD38- cells.
Immunofluorescence flow cytometric gate profiles for CD34+CD38- cells. (A) Isotype antibody controls for cells derived from ESC differentiation. (B-C) FITC-CD34 and PE-CD38 antibodies for cells derived from ESC differentiation (B) and for cells of human bone marrow (C). Cells in gate R2 are defined as CD34+CD38- cells.
Immunofluorescence flow cytometric gate profiles for CD34+CD38- cells. (A) Isotype antibody controls for cells derived from ESC differentiation. (B-C) FITC-CD34 and PE-CD38 antibodies for cells derived from ESC differentiation (B) and for cells of human bone marrow (C). Cells in gate R2 are defined as CD34+CD38- cells.
Cytoplasmic RNA isolation and cDNA pool construction
Cytoplasmic RNA was isolated from purified CD34+CD38- cells derived from ESC cultures and from BM using an RNAeasy Mini Kit (Qiagen, Valencia, CA) following the procedure recommended by the supplier. RNA was subjected to first-strand cDNA synthesis with SMART II and CDS primers (Clontech), using Superscript II reverse transcriptase (Invitrogen), and cDNA pools were constructed using the SMART cDNA synthesis kit (Clontech) as described previously.13 Complementary DNA pools generated by the SMART procedure have been shown to preserve the relative abundance relationship of the original mRNA populations,16-19 and this procedure has successfully been employed in the construction of cDNA pools using fewer than 1000 cells.20
Hybridization of cDNA probes to Atlas cDNA arrays
SMART-generated double-strand cDNAs were purified with the Qiagen PCR purification kit (Qiagen), and the quantities of DNA were determined by UV 260 nm absorption. About 500 ng cDNA was labeled with Random Primer mix plus cDNA Synthesis Control primers (Clontech) using [α-33P]deoxyadenosine monophosphate ([α-33P]dATP) (Amersham, Arlington Heights, IL) and Klenow polymerase as suggested by the supplier. The probes were purified with the NucleoSpin Extraction Kit supplied by Clontech. Equal amounts of labeled probes from the ESCs and BM cells were hybridized to the BD Atlas Plastic Human 12K microarrays (Clontech), according to the manufacturer's instructions. After completing the washing procedure, the arrays were exposed with a low-energy phosphorimaging screen (Molecular Dynamics, Sunnyvale, CA) for 2 to 3 days. The signal intensity was detected using a Storm Phosphor imaging system (Molecular Dynamics) at 50 μ resolution and analyzed using AtlasImage 2.7 software (Clontech) following the manufacturer's guidelines. The intensities of the cDNA double spots from the ESC and the BM arrays were compared, and the data were exported as Excel files (Microsoft, Seattle, WA) for further analysis. To confirm the initial data, the probes were stripped from the arrays following recommendations of the supplier, and they were rehybridized with newly labeled probes as described above. In the repeat assays, the BM array was rehybridized with ESC cDNA, and the ESC array was reprobed with BM cDNA. The resulting expression profiles were virtually identical to those from the first hybridization but with only 60% to 70% of the original signal intensity. Consequently, only data from the first experiment are presented.
Gene expression quantification by semiquantitative PCR
The DNA templates in cDNA pools from the CD34+CD38- cells were equalized based on their relative expression of the α-tubulin gene as described previously.13 The PCR conditions for the CD14, myeloperoxidase, flt-3, and α-tubulin genes were as described13 ; 10 μL PCR products were separated on 1.5% agarose gel and visualized by ethidium bromide staining. For quantitative comparisons of the expression of the non–α-globin genes, a single set of primers with specificity for sequences common to the β-, δ-, γ-, and ϵ-globin genes were employed. They were sense, 5′-GTY-TAC-CCH-TGG-ACC-CAG-A-3′, and antisense, 5′-GCA-GCT-TGT-CAC-AGT-GCA-G-3′, encompassing nucleotides 103 to 292, to generate a PCR product of 190 bp. PCR conditions were 95° C, 1 minute; 55° C, 1.5 minutes; 72° C, 2 minutes with Mg++ concentration of 2.0 mM. The PCR products were digested with restriction enzymes specific for each globin gene. Restriction enzymes were DraIII for the β-globin gene, BfaI for the δ-globin gene, XcmI for the γ-globin gene, and NcoI for the ϵ-globin gene. The digested PCR products were separated on a 10% native polyacrylamide gel electrophoresis (PAGE) gel, stained with ethidium bromide, and visualized with a UV transilluminator.
Results
Hematopoietic differentiation of human ESCs in vitro has previously been demonstrated by 2 groups, from studies that employed different culture conditions.6,7 We have previously identified combinations of BMPs as having high efficiency in the induction of hematopoietic differentiation of rhesus monkey ESCs and in enhancing the formation of clonogenic hematopoietic-like precursors in differentiated monkey ESC colonies.15 In the present study, we observed that combinations of BMP-4, BMP-2, and BMP-7 plus cytokines efficiently promoted the development of hematopoietic clusters from human ESCs when cultured on S17 stromal cell layers; the hematopoietic progeny cells from these cultures exhibited characteristic morphologic features of erythroid cells, granulocytes, megakaryocytes, macrophages, and monocytes (data not shown).
To compensate for the limited numbers of early hematopoietic precursors in the human ESC differentiation cultures, cytoplasmic RNA isolated from CD34+CD38- cells was reverse transcripted and amplified utilizing SMART technology. SMART-generated cDNAs have been successfully used in gene expression analysis utilizing low-density (nylon membrane–based filters) cDNA arrays,21,22 but no such study has been reported using high-density oligonucleotide microarrays. To validate the hybridization results between SMART-generated cDNAs and plastic high-density arrays, we first examined several genes that were included repeatedly at different locations in the BD Atlas Plastic Human 12K microarrays. As shown in Table 1, the signal intensities for each of these genes were very similar at 3 different locations in the array, with only minor exceptions. These findings were consistent for genes with different expression levels, including those of low, intermediate, and high abundance, further supporting the validity of this approach.
Signal intensities of control genes
Array location . | BM intensity . | ESC intensity . | ESC BM ratio . | Gene-protein description . | GenBank no. . |
|---|---|---|---|---|---|
| H12cd2 | 747 | 44 | 0.0589 | Major histocompatibility complex, class I, C | M11886 |
| A01cd2 | 667 | 42 | 0.063 | Major histocompatibility complex, class I, C | M11886 |
| P12cd2 | 711 | 48 | 0.0675 | Major histocompatibility complex, class I, C | M11886 |
| A01cd3 | 3 | 0 | ↓ | Ubiquitin C | M26880 |
| H12cd3 | 5 | 0 | ↓ | Ubiquitin C | M26880 |
| P12cd3 | 18 | 6 | 0.3333 | Ubiquitin C | M26880 |
| A01cd1 | 692 | 406 | 0.5867 | α-tubulin, ubiquitous | K00558 |
| H12cd1 | 602 | 472 | 0.7841 | α-tubulin, ubiquitous | K00558 |
| P12cd1 | 954 | 755 | 0.7914 | α-tubulin, ubiquitous | K00558 |
| P12cd7 | 308 | 307 | 0.9968 | β-actin | X00351 |
| A01cd7 | 284 | 289 | 1.0176 | β-actin | X00351 |
| H12cd7 | 286 | 396 | 1.3846 | β-actin | X00351 |
| A01ef1 | 180 | 72 | 0.4 | Ribosomal protein L13a | X56932 |
| P12ef1 | 377 | 219 | 0.5809 | Ribosomal protein L13a | X56932 |
| H12ef1 | 165 | 126 | 0.7636 | Ribosomal protein L13a | X56932 |
| A01cd5 | 27 | 42 | 1.5556 | Ribosomal protein S9 | U14971 |
| P12cd5 | 20 | 34 | 1.7 | Ribosomal protein S9 | U14971 |
| H12cd5 | 18 | 50 | 2.7778 | Ribosomal protein S9 | U14971 |
| A01cd8 | 161 | 788 | 4.8944 | Glyceraldehyde-3-phosphate dehydrogenase | X01677 |
| P12cd8 | 142 | 800 | 5.6338 | Glyceraldehyde-3-phosphate dehydrogenase | X01677 |
| H12cd8 | 116 | 747 | 6.4397 | Glyceraldehyde-3-phosphate dehydrogenase | X01677 |
Array location . | BM intensity . | ESC intensity . | ESC BM ratio . | Gene-protein description . | GenBank no. . |
|---|---|---|---|---|---|
| H12cd2 | 747 | 44 | 0.0589 | Major histocompatibility complex, class I, C | M11886 |
| A01cd2 | 667 | 42 | 0.063 | Major histocompatibility complex, class I, C | M11886 |
| P12cd2 | 711 | 48 | 0.0675 | Major histocompatibility complex, class I, C | M11886 |
| A01cd3 | 3 | 0 | ↓ | Ubiquitin C | M26880 |
| H12cd3 | 5 | 0 | ↓ | Ubiquitin C | M26880 |
| P12cd3 | 18 | 6 | 0.3333 | Ubiquitin C | M26880 |
| A01cd1 | 692 | 406 | 0.5867 | α-tubulin, ubiquitous | K00558 |
| H12cd1 | 602 | 472 | 0.7841 | α-tubulin, ubiquitous | K00558 |
| P12cd1 | 954 | 755 | 0.7914 | α-tubulin, ubiquitous | K00558 |
| P12cd7 | 308 | 307 | 0.9968 | β-actin | X00351 |
| A01cd7 | 284 | 289 | 1.0176 | β-actin | X00351 |
| H12cd7 | 286 | 396 | 1.3846 | β-actin | X00351 |
| A01ef1 | 180 | 72 | 0.4 | Ribosomal protein L13a | X56932 |
| P12ef1 | 377 | 219 | 0.5809 | Ribosomal protein L13a | X56932 |
| H12ef1 | 165 | 126 | 0.7636 | Ribosomal protein L13a | X56932 |
| A01cd5 | 27 | 42 | 1.5556 | Ribosomal protein S9 | U14971 |
| P12cd5 | 20 | 34 | 1.7 | Ribosomal protein S9 | U14971 |
| H12cd5 | 18 | 50 | 2.7778 | Ribosomal protein S9 | U14971 |
| A01cd8 | 161 | 788 | 4.8944 | Glyceraldehyde-3-phosphate dehydrogenase | X01677 |
| P12cd8 | 142 | 800 | 5.6338 | Glyceraldehyde-3-phosphate dehydrogenase | X01677 |
| H12cd8 | 116 | 747 | 6.4397 | Glyceraldehyde-3-phosphate dehydrogenase | X01677 |
↓ indicates genes with expression levels undetectable in cells from ESCs.
Using AtlasImage 2.7 software, the array was aligned with the AtlasImage Grid Template (both from Clontech) automatically and fine-tuned for each individual grid manually. The background was calculated based on the median intensity of the blank spaces between the different panels of the array (default method), and the raw signal intensity of each spot was measured. A raw intensity (before normalization) of 3-fold over background was taken as an indication that a gene was expressed at a significant level. By this criterion, we determined that 1692 and 1494 genes were expressed in CD34+CD38- cells from BM and ESCs, respectively. The list of all of the expressed genes is presented in the Supplemental Data Set (see the Supplemental Data Set link at the top of the online article on the Blood website).
For comparing the expression patterns of cells derived from BM and from ESCs, the signal intensities were normalized by the global normalization-sum method, which is best suited for the comparison of 2 similar samples. Signal values in arrays hybridized with ESC cDNA were normalized with respect to those from arrays probed with cDNA of BM origin, and the signal intensities in the ESC array were adjusted accordingly. The adjusted signal intensities for each of the individual cDNA spots in arrays hybridized with cDNA derived from BM and ESCs were compared, and the results were exported as an Excel file for further analysis. These analyses revealed that 494 genes showed similar levels of expression in CD34+CD38- cells from both sources, 791 genes were relatively overexpressed in cells from BM (BM versus ESCs, at least 2-fold), and 803 genes were preferentially expressed in cells from ES cell cultures (ESCs versus BM, at least 2-fold). These genes comprise all categories of function (Table 2), and a large number of genes associated with transcription (177), membrane channels and transporters (101), trafficking/targeting proteins (133), metabolism (308), protein translation (121), cell receptors (109), and intracellular transducers/effectors/modulators (246) were detected. A total of 409 genes that have not been functionally classified were also expressed. Further analysis showed that most of the genes were of low abundance, and less than 20% of them (387 of 2088 genes) were expressed at levels at least 10-fold over the background. Less than 10% of cell adhesion proteins (5 of 65), extracellular transport/carrier proteins (5 of 57), and cell receptors (9 of 109) were expressed at these high levels. Whereas 55% of genes associated with translation were expressed at levels at least 10-fold over background, most of these were ribosomal proteins, and most of them were expressed at higher levels in cells derived from BM than in those from ESCs. We also found that a relatively high percentage of genes for RNA processing/turnover/transport (28%), DNA binding/chromatin proteins (33%), and stress response proteins (22%) were expressed at high levels. These observations are consonant with recent findings that these genes are associated with the “stemness” of multiple types of stem cells.23-25 These comparative analyses also demonstrated that CD34+CD38- cells derived from ESCs expressed more high-activity genes (more than 10-fold over background) associated with intracellular signal transduction than did cells from BM (ESCs 25 versus BM 8), even though cells from both sources showed similar numbers of genes of relatively low activity (ESCs 85 versus BM 77) (Table 2). A similar pattern of expression was also observed for genes associated with cellular trafficking/targeting proteins.
Gene expression profiles of CD34+CD38- cells derived from human BM and ESCs
. | ESC/BM ratio* . | . | . | ESC/BM ratio† . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene functional classification . | 0.5 or less . | 0.5 to less than 2.0 . | 2.0 or more . | 0.5 or less . | 0.5 to less than 2.0 . | 2.0 or more . | ||||
| All genes | 791 | 494 | 803 | 132 | 128 | 127 | ||||
| Cell surface antigens | 14 | 5 | 33 | 1 | 2 | 3 | ||||
| Transcription | 77 | 39 | 61 | 6 | 10 | 12 | ||||
| Cell cycle | 15 | 3 | 20 | 2 | 2 | 4 | ||||
| Cell adhesion receptors/proteins | 21 | 13 | 31 | 1 | 1 | 3 | ||||
| Immune system proteins | 17 | 4 | 11 | 5 | 1 | 3 | ||||
| Extracellular transport/carrier proteins | 24 | 14 | 19 | 2 | 0 | 3 | ||||
| Oncogenes/tumor suppressor | 13 | 10 | 20 | 3 | 2 | 4 | ||||
| Stress response proteins | 39 | 18 | 26 | 9 | 5 | 4 | ||||
| Membrane channels and transporters | 29 | 32 | 40 | 2 | 9 | 8 | ||||
| Extracellular matrix proteins | 4 | 1 | 7 | 0 | 0 | 1 | ||||
| Trafficking/targeting proteins | 44 | 39 | 50 | 2 | 2 | 14 | ||||
| Metabolism | 97 | 91 | 120 | 15 | 27 | 16 | ||||
| Posttranslation/protein folding | 22 | 26 | 24 | 3 | 7 | 2 | ||||
| Translation | 58 | 40 | 23 | 41 | 21 | 4 | ||||
| Apoptosis-associated proteins | 14 | 4 | 9 | 4 | 0 | 1 | ||||
| RNA processing/turnover/transport | 29 | 22 | 27 | 6 | 11 | 5 | ||||
| DNA binding and chromatin proteins | 22 | 13 | 13 | 6 | 6 | 4 | ||||
| Cell receptors | 41 | 15 | 53 | 2 | 2 | 5 | ||||
| Cell signaling, extracellular communication proteins | 31 | 15 | 26 | 4 | 3 | 3 | ||||
| Intracellular transducers/effectors/modulators | 85 | 51 | 110 | 8 | 7 | 25 | ||||
| Protein turnover | 30 | 27 | 32 | 3 | 8 | 3 | ||||
| Cytoskeleton/mobility proteins | 23 | 27 | 28 | 1 | 6 | 3 | ||||
| DNA synthesis/recombination/repair | 11 | 5 | 19 | 1 | 0 | 4 | ||||
| Hematopoiesis | 79 | 33 | 94 | 12 | 5 | 17 | ||||
| Functionally unclassified | 176 | 67 | 166 | 22 | 12 | 21 | ||||
. | ESC/BM ratio* . | . | . | ESC/BM ratio† . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene functional classification . | 0.5 or less . | 0.5 to less than 2.0 . | 2.0 or more . | 0.5 or less . | 0.5 to less than 2.0 . | 2.0 or more . | ||||
| All genes | 791 | 494 | 803 | 132 | 128 | 127 | ||||
| Cell surface antigens | 14 | 5 | 33 | 1 | 2 | 3 | ||||
| Transcription | 77 | 39 | 61 | 6 | 10 | 12 | ||||
| Cell cycle | 15 | 3 | 20 | 2 | 2 | 4 | ||||
| Cell adhesion receptors/proteins | 21 | 13 | 31 | 1 | 1 | 3 | ||||
| Immune system proteins | 17 | 4 | 11 | 5 | 1 | 3 | ||||
| Extracellular transport/carrier proteins | 24 | 14 | 19 | 2 | 0 | 3 | ||||
| Oncogenes/tumor suppressor | 13 | 10 | 20 | 3 | 2 | 4 | ||||
| Stress response proteins | 39 | 18 | 26 | 9 | 5 | 4 | ||||
| Membrane channels and transporters | 29 | 32 | 40 | 2 | 9 | 8 | ||||
| Extracellular matrix proteins | 4 | 1 | 7 | 0 | 0 | 1 | ||||
| Trafficking/targeting proteins | 44 | 39 | 50 | 2 | 2 | 14 | ||||
| Metabolism | 97 | 91 | 120 | 15 | 27 | 16 | ||||
| Posttranslation/protein folding | 22 | 26 | 24 | 3 | 7 | 2 | ||||
| Translation | 58 | 40 | 23 | 41 | 21 | 4 | ||||
| Apoptosis-associated proteins | 14 | 4 | 9 | 4 | 0 | 1 | ||||
| RNA processing/turnover/transport | 29 | 22 | 27 | 6 | 11 | 5 | ||||
| DNA binding and chromatin proteins | 22 | 13 | 13 | 6 | 6 | 4 | ||||
| Cell receptors | 41 | 15 | 53 | 2 | 2 | 5 | ||||
| Cell signaling, extracellular communication proteins | 31 | 15 | 26 | 4 | 3 | 3 | ||||
| Intracellular transducers/effectors/modulators | 85 | 51 | 110 | 8 | 7 | 25 | ||||
| Protein turnover | 30 | 27 | 32 | 3 | 8 | 3 | ||||
| Cytoskeleton/mobility proteins | 23 | 27 | 28 | 1 | 6 | 3 | ||||
| DNA synthesis/recombination/repair | 11 | 5 | 19 | 1 | 0 | 4 | ||||
| Hematopoiesis | 79 | 33 | 94 | 12 | 5 | 17 | ||||
| Functionally unclassified | 176 | 67 | 166 | 22 | 12 | 21 | ||||
Threshold = 3; BM, 1692; ES, 1494. Due to genes having been classified in multiple categories, the total number of genes listed in this table is not equal to the number of expressed genes (2086).
Genes with expression levels at least 3-fold of background
Genes with expression levels at least 10-fold of background
Six mitogen-activated protein kinases (MAPK3, MAPK4, MAPK7, MAPK8, MAPKK3, and MAP4K1) were expressed at relatively high levels in CD34+CD38- cells derived from ESC cultures, whereas only 3 such genes (MAPK7, MAPK8, and MAPKK3), at expression levels just over background, were detected in cells from BM. Both the CDK5 and cdc27 genes were also found to be up-regulated in ESC-derived hematopoietic precursors, consistent with the notion that HSCs from BM are in the G0 resting state. The ESC-derived hematopoietic precursors, which were exposed to multiple hematopoietic growth factors, were in active growth, and that might have resulted in the activation of signal transduction pathways. However, only 3 cyclin messages were detected: Cyclin M4 and D3 were expressed in BM-derived cells, and cyclin F was observed in ESC-derived CD34+CD38- cells.
We also analyzed the expression patterns of genes associated with hematopoietic cells. As shown in Tables 2 and 3, most of these genes were expressed at relatively low levels in CD34+CD38- cells derived from both adult BM and ESCs, with only a few exceptions. A substantial level of laminin receptor 1 message was expressed in CD34+CD38- cells from BM, and a much lower expression of this gene was observed in the corresponding cells from ESCs (ratio of BM versus ESCs, about 10). Another finding of the comparative gene expression was that low, but clearly above background, levels of multiple cytokines were observed in BM cells. The genes of inhibin βA and βC, granulocyte-macrophage colony-stimulating factor (GM-CSF), hepatocyte growth factor (HGF)–like factor, interferon-α14, IL-8, IL-13, IL-15, IL-25, CCL2, monocyte chemotactic protein 3 (MCP-3), CCL27, CXCL6, and small inducible cytokines A1 and A5 were only detected in CD34+CD38- cells from BM, whereas 4 such factors, IL-1δ, FGF-20, BMP-1, and epidermal growth factor-β (EGF-β), were expressed exclusively in cells from ESC cultures. Interferon-α2 and chemokine-like factor 2 were preferentially expressed in BM-derived cells, whereas higher levels of platelet factor 4, platelet-derived growth factor (PDGF) α chain, and stromal cell-derived factor 2 (SCDF2) expression were detected in cells derived from ESC cultures. No difference in angiopoietin-1, BMP-4, BMP-7, CCL-11, SCDF2-like 1, angiopoietin-like 1, and chemokine-like factor 1 expression was seen (Table 3).
Genes associated with hematopoietic cells
Array location . | BM intensity . | ESC intensity . | ESC/BM ratio . | Gene-protein description/name . | GenBank no. . |
|---|---|---|---|---|---|
| D05ef5 | 2 | 0 | ↓ | fms-related tyrosine kinase 3, flt3/flk2 | U02687 |
| F11cd2 | 2 | 0 | ↓ | Tyrosine kinase with immunoglobulin and EGF homology domains, Tie-1 | NM_005424 |
| J21ab6 | 2 | 0 | ↓ | Interleukin-12 receptor, β2 | NM_001559 |
| O11ef7 | 2 | 0 | ↓ | Interleukin-7 receptor | M29696 |
| L20ab4 | 3 | 0 | ↓ | GATA binding protein 2, GATA-2 | NM_002050 |
| M01ab5 | 6 | 0 | ↓ | v-ets erythroblastosis virus E26 oncogene-like (avian), Erg-2 | NM_004449 |
| C01ef6 | 2 | 0 | ↓ | Ras-related C3 botulinum toxin substrate 1, rac | M29870 |
| H06ef5 | 3 | 0 | ↓ | Growth factor receptor-bound protein 2, Grb2 | L29511 |
| J09ef1 | 2 | 0 | ↓ | Phosphoinositide-3-kinase, class 2, β polypeptide, Pl3K | Y11312 |
| E06ef6 | 2 | 0 | ↓ | B-cell CLL/lymphoma 2, BCL2 | M14745 |
| K24ef1 | 47 | 0 | ↓ | BCL2-antagonist of cell death | U66879 |
| B09gh1 | 4 | 0 | ↓ | Bcl-2-associated transcription factor | NM_014739 |
| N04ab6 | 2 | 0 | ↓ | Laminin, α 4 | NM_002290 |
| D15ef7 | 2 | 0 | ↓ | Inhibin, βA (activin A, activin AB alpha polypeptide) | J03634 |
| O24ab7 | 2 | 0 | ↓ | Inhibin, βC | NM_005538 |
| G22gh6 | 2 | 0 | ↓ | Colony-stimulating factor 2 (granulocyte-macrophage), GM-CSF | NM_000758 |
| M24ef7 | 2 | 0 | ↓ | Macrophage stimulating 1 (hepatocyte growth factor-like) | M74178 |
| H02ab7 | 3 | 0 | ↓ | Interferon, α14 | NM_002172 |
| H03ef7 | 4 | 0 | ↓ | Interleukin-13 | L06801 |
| H13ef7 | 5 | 0 | ↓ | Interleukin-15 | U14407 |
| K09ef4 | 2 | 0 | ↓ | Interleukin-26 | NM_018402 |
| O22ef7 | 2 | 0 | ↓ | Interleukin-8 | Y00787 |
| C15cd3 | 2 | 0 | ↓ | Small inducible cytokine A2 (monocyte chemotactic protein 1), CCL2 | NM_002982 |
| E23ef5 | 6 | 0 | ↓ | Small inducible cytokine A7 (monocyte chemotactic protein 3), MCP3 | X72308 |
| F14cd1 | 2 | 0 | ↓ | Small inducible cytokine subfamily B (Cys-Xaa-Cys), member 6, CXCL6 | NM_002993 |
| K22ef7 | 13 | 0 | ↓ | Small inducible cytokine A5 (RANTES) | M21121 |
| M08ef7 | 16 | 0 | ↓ | Small inducible cytokine A1, I-309 | M57502 |
| N08cd6 | 2 | 0 | ↓ | Small inducible cytokine subfamily A (Cys-Cys), member 27, CCL27 | NM_006664 |
| N01ab7 | 7 | 0 | ↓ | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila), MLL | NM_005933 |
| G21ef7 | 77 | 8 | 0.104 | Laminin receptor 1 (67 kDa, ribosomal protein SA) | U43901 |
| G14gh6 | 6 | 2 | 0.333 | Interferon, α2 | NM_000605 |
| N05ef4 | 24 | 10 | 0.417 | Chemokine-like factor 2 | NM_016326 |
| G13ab2 | 4 | 2 | 0.5 | Activin A receptor, type IIB | NM_001106 |
| J01ab7 | 4 | 2 | 0.5 | MAD, mothers against decapentaplegic homolog 5 (Drosophila), SMAD5 | NM_005903 |
| M13cd2 | 3 | 2 | 0.667 | T-cell acute lymphocytic leukemia 1, SCL | NM_003189 |
| G14ab2 | 3 | 2 | 0.667 | Angiopoietin-1 | NM_001146 |
| G16ef5 | 3 | 2 | 0.667 | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, c-kit | X06182 |
| C23ab3 | 50 | 38 | 0.76 | β2-microglobulin | NM_004048 |
| I23ef4 | 43 | 34 | 0.791 | Chemokine-like factor 1 | NM_016951 |
| K23cd4 | 9 | 8 | 0.889 | Mitogen-activated protein kinase-activated protein kinase 3, MAPAPK3 | NM_004635 |
| A10ab3 | 2 | 2 | 1 | Bone morphogenetic protein 7 (osteogenic protein 1), BMP-7 | NM_001719 |
| F02cd1 | 2 | 2 | 1 | Small inducible cytokine subfamily A (Cys-Cys), member 11 (eotaxin), CCL 11 | NM_002986 |
| L23ab2 | 2 | 2 | 1 | Angiopoietin-like 1 | NM_004673 |
| N12ab3 | 2 | 2 | 1 | Bone morphogenetic protein 4, BMP-4 | NM_001202 |
| A05gh1 | 2 | 2 | 1 | Stromal cell—derived factor 2-like 1, SCDF2L1 | NM_022044 |
| G11ab2 | 3 | 4 | 1.333 | Activin A receptor, type II | NM_001616 |
| M01cd1 | 8 | 12 | 1.5 | Nuclear factor (erythroid-derived 2), 45 kDa, Nf-E2 | NM_006163 |
| C18ab6 | 0 | 16 | ↑ | Glycophorin B (includes Ss blood group) | NM_002100 |
| L03ab3 | 0 | 4 | ↑ | CD14 antigen | NM_000591 |
| E21ef7 | 0 | 8 | ↑ | Integrin, β6 | M35198 |
| E21gh1 | 0 | 4 | ↑ | Signal transducer and activator of transcription 5A, Stat5a | NM_003152 |
| G21cd2 | 0 | 4 | ↑ | Signal transducer and activator of transcription 4, Stat4 | NM_003151 |
| B13ef5 | 0 | 6 | ↑ | Mitogen-activated protein kinase 3, MAPK3 | X60188 |
| B19ef5 | 0 | 8 | ↑ | Mitogen-activated protein kinase 4, MAPK4 | X59727 |
| L09ab6 | 0 | 4 | ↑ | Inositol polyphosphate phosphatase-like 1, SHIP-2 | NM_001567 |
| L16ef5 | 0 | 4 | ↑ | Mitogen-activated protein kinase 8, MAPK8 | L26318 |
| N04ef5 | 0 | 4 | ↑ | Janus kinase 2 (a protein tyrosine kinase), JAK2 | AF005216 |
| A08ef7 | 0 | 12 | ↑ | Interleukin-1 receptor, type II | X59770 |
| K08ab5 | 0 | 6 | ↑ | Chemokine (C motif) XC receptor 1 | NM_005283 |
| C03cd8 | 0 | 4 | ↑ | Interleukin-1, δ | NM_012275 |
| L04ef8 | 0 | 4 | ↑ | Fibroblast growth factor 20, FGF-20 | NM_019851 |
| N08ab3 | 0 | 4 | ↑ | Bone morphogenetic protein 1, BMP-1 | NM_001199 |
| P20ab5 | 0 | 4 | ↑ | Epidermal growth factor (β-urogastrone), EGF | NM_001963 |
| P24cd2 | 2 | 4 | 2 | Platelet factor 4 | NM_002619 |
| G09ab2 | 4 | 8 | 2 | Activin A receptor, type IB | NM_004302 |
| F14cd2 | 2 | 6 | 3 | Stromal cell-derived factor 2, SCDF2 | NM_006923 |
| I14ab6 | 2 | 6 | 3 | Integrin, β2 (antigen CD18 [p95]) | NM_000211 |
| M03cd1 | 1 | 4 | 4 | Nuclear factor (erythroid-derived 2)-like 2, Nrf-2 | NM_006164 |
| L18ef4 | 1 | 4 | 4 | Putative leukocyte platelet-activating factor receptor | M76676 |
| O16ef7 | 1 | 4 | 4 | Platelet-derived growth factor alpha polypeptide, PDGF-a | X06374 |
| G24cd7 | 1 | 4 | 4 | Mitogen-activated protein kinase kinase kinase kinase 1, MAP4K1 | NM_007181 |
| B21ef5 | 1 | 6 | 6 | Mitogen-activated protein kinase 7, MAPK7 | U25278 |
| J05ab7 | 1 | 10 | 10 | MAD, mothers against decapentapiegic homolog 9 (Drosophila), SMAD9 | NM_005905 |
| M02ab6 | 11 | 116 | 10.55 | Myeloperoxidase, MPO | NM_000250 |
Array location . | BM intensity . | ESC intensity . | ESC/BM ratio . | Gene-protein description/name . | GenBank no. . |
|---|---|---|---|---|---|
| D05ef5 | 2 | 0 | ↓ | fms-related tyrosine kinase 3, flt3/flk2 | U02687 |
| F11cd2 | 2 | 0 | ↓ | Tyrosine kinase with immunoglobulin and EGF homology domains, Tie-1 | NM_005424 |
| J21ab6 | 2 | 0 | ↓ | Interleukin-12 receptor, β2 | NM_001559 |
| O11ef7 | 2 | 0 | ↓ | Interleukin-7 receptor | M29696 |
| L20ab4 | 3 | 0 | ↓ | GATA binding protein 2, GATA-2 | NM_002050 |
| M01ab5 | 6 | 0 | ↓ | v-ets erythroblastosis virus E26 oncogene-like (avian), Erg-2 | NM_004449 |
| C01ef6 | 2 | 0 | ↓ | Ras-related C3 botulinum toxin substrate 1, rac | M29870 |
| H06ef5 | 3 | 0 | ↓ | Growth factor receptor-bound protein 2, Grb2 | L29511 |
| J09ef1 | 2 | 0 | ↓ | Phosphoinositide-3-kinase, class 2, β polypeptide, Pl3K | Y11312 |
| E06ef6 | 2 | 0 | ↓ | B-cell CLL/lymphoma 2, BCL2 | M14745 |
| K24ef1 | 47 | 0 | ↓ | BCL2-antagonist of cell death | U66879 |
| B09gh1 | 4 | 0 | ↓ | Bcl-2-associated transcription factor | NM_014739 |
| N04ab6 | 2 | 0 | ↓ | Laminin, α 4 | NM_002290 |
| D15ef7 | 2 | 0 | ↓ | Inhibin, βA (activin A, activin AB alpha polypeptide) | J03634 |
| O24ab7 | 2 | 0 | ↓ | Inhibin, βC | NM_005538 |
| G22gh6 | 2 | 0 | ↓ | Colony-stimulating factor 2 (granulocyte-macrophage), GM-CSF | NM_000758 |
| M24ef7 | 2 | 0 | ↓ | Macrophage stimulating 1 (hepatocyte growth factor-like) | M74178 |
| H02ab7 | 3 | 0 | ↓ | Interferon, α14 | NM_002172 |
| H03ef7 | 4 | 0 | ↓ | Interleukin-13 | L06801 |
| H13ef7 | 5 | 0 | ↓ | Interleukin-15 | U14407 |
| K09ef4 | 2 | 0 | ↓ | Interleukin-26 | NM_018402 |
| O22ef7 | 2 | 0 | ↓ | Interleukin-8 | Y00787 |
| C15cd3 | 2 | 0 | ↓ | Small inducible cytokine A2 (monocyte chemotactic protein 1), CCL2 | NM_002982 |
| E23ef5 | 6 | 0 | ↓ | Small inducible cytokine A7 (monocyte chemotactic protein 3), MCP3 | X72308 |
| F14cd1 | 2 | 0 | ↓ | Small inducible cytokine subfamily B (Cys-Xaa-Cys), member 6, CXCL6 | NM_002993 |
| K22ef7 | 13 | 0 | ↓ | Small inducible cytokine A5 (RANTES) | M21121 |
| M08ef7 | 16 | 0 | ↓ | Small inducible cytokine A1, I-309 | M57502 |
| N08cd6 | 2 | 0 | ↓ | Small inducible cytokine subfamily A (Cys-Cys), member 27, CCL27 | NM_006664 |
| N01ab7 | 7 | 0 | ↓ | Myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila), MLL | NM_005933 |
| G21ef7 | 77 | 8 | 0.104 | Laminin receptor 1 (67 kDa, ribosomal protein SA) | U43901 |
| G14gh6 | 6 | 2 | 0.333 | Interferon, α2 | NM_000605 |
| N05ef4 | 24 | 10 | 0.417 | Chemokine-like factor 2 | NM_016326 |
| G13ab2 | 4 | 2 | 0.5 | Activin A receptor, type IIB | NM_001106 |
| J01ab7 | 4 | 2 | 0.5 | MAD, mothers against decapentaplegic homolog 5 (Drosophila), SMAD5 | NM_005903 |
| M13cd2 | 3 | 2 | 0.667 | T-cell acute lymphocytic leukemia 1, SCL | NM_003189 |
| G14ab2 | 3 | 2 | 0.667 | Angiopoietin-1 | NM_001146 |
| G16ef5 | 3 | 2 | 0.667 | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog, c-kit | X06182 |
| C23ab3 | 50 | 38 | 0.76 | β2-microglobulin | NM_004048 |
| I23ef4 | 43 | 34 | 0.791 | Chemokine-like factor 1 | NM_016951 |
| K23cd4 | 9 | 8 | 0.889 | Mitogen-activated protein kinase-activated protein kinase 3, MAPAPK3 | NM_004635 |
| A10ab3 | 2 | 2 | 1 | Bone morphogenetic protein 7 (osteogenic protein 1), BMP-7 | NM_001719 |
| F02cd1 | 2 | 2 | 1 | Small inducible cytokine subfamily A (Cys-Cys), member 11 (eotaxin), CCL 11 | NM_002986 |
| L23ab2 | 2 | 2 | 1 | Angiopoietin-like 1 | NM_004673 |
| N12ab3 | 2 | 2 | 1 | Bone morphogenetic protein 4, BMP-4 | NM_001202 |
| A05gh1 | 2 | 2 | 1 | Stromal cell—derived factor 2-like 1, SCDF2L1 | NM_022044 |
| G11ab2 | 3 | 4 | 1.333 | Activin A receptor, type II | NM_001616 |
| M01cd1 | 8 | 12 | 1.5 | Nuclear factor (erythroid-derived 2), 45 kDa, Nf-E2 | NM_006163 |
| C18ab6 | 0 | 16 | ↑ | Glycophorin B (includes Ss blood group) | NM_002100 |
| L03ab3 | 0 | 4 | ↑ | CD14 antigen | NM_000591 |
| E21ef7 | 0 | 8 | ↑ | Integrin, β6 | M35198 |
| E21gh1 | 0 | 4 | ↑ | Signal transducer and activator of transcription 5A, Stat5a | NM_003152 |
| G21cd2 | 0 | 4 | ↑ | Signal transducer and activator of transcription 4, Stat4 | NM_003151 |
| B13ef5 | 0 | 6 | ↑ | Mitogen-activated protein kinase 3, MAPK3 | X60188 |
| B19ef5 | 0 | 8 | ↑ | Mitogen-activated protein kinase 4, MAPK4 | X59727 |
| L09ab6 | 0 | 4 | ↑ | Inositol polyphosphate phosphatase-like 1, SHIP-2 | NM_001567 |
| L16ef5 | 0 | 4 | ↑ | Mitogen-activated protein kinase 8, MAPK8 | L26318 |
| N04ef5 | 0 | 4 | ↑ | Janus kinase 2 (a protein tyrosine kinase), JAK2 | AF005216 |
| A08ef7 | 0 | 12 | ↑ | Interleukin-1 receptor, type II | X59770 |
| K08ab5 | 0 | 6 | ↑ | Chemokine (C motif) XC receptor 1 | NM_005283 |
| C03cd8 | 0 | 4 | ↑ | Interleukin-1, δ | NM_012275 |
| L04ef8 | 0 | 4 | ↑ | Fibroblast growth factor 20, FGF-20 | NM_019851 |
| N08ab3 | 0 | 4 | ↑ | Bone morphogenetic protein 1, BMP-1 | NM_001199 |
| P20ab5 | 0 | 4 | ↑ | Epidermal growth factor (β-urogastrone), EGF | NM_001963 |
| P24cd2 | 2 | 4 | 2 | Platelet factor 4 | NM_002619 |
| G09ab2 | 4 | 8 | 2 | Activin A receptor, type IB | NM_004302 |
| F14cd2 | 2 | 6 | 3 | Stromal cell-derived factor 2, SCDF2 | NM_006923 |
| I14ab6 | 2 | 6 | 3 | Integrin, β2 (antigen CD18 [p95]) | NM_000211 |
| M03cd1 | 1 | 4 | 4 | Nuclear factor (erythroid-derived 2)-like 2, Nrf-2 | NM_006164 |
| L18ef4 | 1 | 4 | 4 | Putative leukocyte platelet-activating factor receptor | M76676 |
| O16ef7 | 1 | 4 | 4 | Platelet-derived growth factor alpha polypeptide, PDGF-a | X06374 |
| G24cd7 | 1 | 4 | 4 | Mitogen-activated protein kinase kinase kinase kinase 1, MAP4K1 | NM_007181 |
| B21ef5 | 1 | 6 | 6 | Mitogen-activated protein kinase 7, MAPK7 | U25278 |
| J05ab7 | 1 | 10 | 10 | MAD, mothers against decapentapiegic homolog 9 (Drosophila), SMAD9 | NM_005905 |
| M02ab6 | 11 | 116 | 10.55 | Myeloperoxidase, MPO | NM_000250 |
↓ indicates genes with expression levels undetectable in cells from ESCs; and ↑, genes with expression levels undetectable in cells from BM.
We also examined the expression of genes that are known to be expressed in HSCs. Similar expression levels of the c-kit and scl genes were detected in cells from both sources; however, low but clearly over background levels of Tie-1, flt-3, bcl-2, GATA-2, and Erg-2 were observed in CD34+CD38- cells derived from BM but not from ESCs. We previously determined that hematopoietic precursors derived from rhesus monkey and mouse ES cells also failed to express the flt-3 gene, consistent with the observations from this study. Genes involved in downstream signal transduction of FLT-3, and of other hematopoietic receptors such as c-KIT and IL-6 receptor, including phosphoinositide-3-kinase (PI3K) and growth factor receptor-bound protein 2 (GRB2), were detected only in CD34+CD38- cells from BM. However, several MAP kinases and signal transducer genes, such as Janus kinase 2 (JAK2), signal transducer and activator of transcription 4 (STAT4), and STAT5a, were preferentially expressed in cells from ESC cultures. We also determined that the CD14, glycophorin B, and myeloperoxidase genes, all of which are associated with lineage commitment, were expressed at higher levels in cells of ESC origin than in those from BM (Table 3).
Several of the major histocompatibility complex (MHC) genes were expressed at much higher levels in CD34+CD38- cells of BM origin than in those derived from ESCs. Very high levels (more than 100-fold over background) of MHC class IC and II DRβ5 genes were detected in BM-derived cells, which were 15 and 20 times higher than those in cells of ESC origin, respectively. However, similar levels of the β2-microglobulin gene, the common molecule of the MHC I complex, were observed in the cells of both sources. A high level of the adult β-globin message, but none of the embryonic ϵ-globin mRNA, was expressed in CD34+CD38- cells derived from BM. In contrast, the message of embryonic ϵ-globin gene was detected in CD34+CD38- cells derived from ESCs, and no β-globin mRNA was detectable (Table 4).
MHC and hemoglobin gene expression profiles
Array location . | BM intensity . | ESC intensity . | ESC/BM ratio . | Gene-protein description . | GenBank no. . |
|---|---|---|---|---|---|
| D08ab5 | 10 | 0 | ↓ | Major histocompatibility complex, class II, DMα | NM_006120 |
| F06ab7 | 124 | 6 | 0.0484 | Major histocompatibility complex, class II, DRβ5 | NM_002125 |
| H12cd2 | 747 | 44 | 0.0589 | Major histocompatibility complex, class I, C | M11886 |
| F04ab7 | 66 | 4 | 0.0606 | Major histocompatibility complex, class II, DRβ1 | NM_002124 |
| A01cd2 | 667 | 42 | 0.063 | Major histocompatibility complex, class I, C | M11886 |
| P12cd2 | 711 | 48 | 0.0675 | Major histocompatibility complex, class I, C | M11886 |
| M24ab7 | 20 | 2 | 0.1 | Major histocompatibility complex, class I, E | NM_005516 |
| F08ab7 | 42 | 6 | 0.1429 | HLA-G histocompatibility antigen, class I, G | NM_002127 |
| C23ab3 | 50 | 38 | 0.76 | β2-microglobulin | NM_004048 |
| K21ef7 | 2 | 2 | 1 | Major histocompatibility complex, class II, DRα | K01171 |
| K01ab6 | 37 | 0 | ↓ | Hemoglobin, β | NM_000518 |
| I23ab6 | 6 | 2 | 0.3333 | Hemoglobin, α2 | NM_000517 |
| K03ab6 | 6 | 10 | 1.6667 | Hemoglobin, δ | NM_000519 |
| G17ab6 | 0 | 1 | ↑ | Hemoglobin, ϵ | NM_005330 |
| K07ab6 | 0 | 18 | ↑ | Hemoglobin, γA | NM_000559 |
Array location . | BM intensity . | ESC intensity . | ESC/BM ratio . | Gene-protein description . | GenBank no. . |
|---|---|---|---|---|---|
| D08ab5 | 10 | 0 | ↓ | Major histocompatibility complex, class II, DMα | NM_006120 |
| F06ab7 | 124 | 6 | 0.0484 | Major histocompatibility complex, class II, DRβ5 | NM_002125 |
| H12cd2 | 747 | 44 | 0.0589 | Major histocompatibility complex, class I, C | M11886 |
| F04ab7 | 66 | 4 | 0.0606 | Major histocompatibility complex, class II, DRβ1 | NM_002124 |
| A01cd2 | 667 | 42 | 0.063 | Major histocompatibility complex, class I, C | M11886 |
| P12cd2 | 711 | 48 | 0.0675 | Major histocompatibility complex, class I, C | M11886 |
| M24ab7 | 20 | 2 | 0.1 | Major histocompatibility complex, class I, E | NM_005516 |
| F08ab7 | 42 | 6 | 0.1429 | HLA-G histocompatibility antigen, class I, G | NM_002127 |
| C23ab3 | 50 | 38 | 0.76 | β2-microglobulin | NM_004048 |
| K21ef7 | 2 | 2 | 1 | Major histocompatibility complex, class II, DRα | K01171 |
| K01ab6 | 37 | 0 | ↓ | Hemoglobin, β | NM_000518 |
| I23ab6 | 6 | 2 | 0.3333 | Hemoglobin, α2 | NM_000517 |
| K03ab6 | 6 | 10 | 1.6667 | Hemoglobin, δ | NM_000519 |
| G17ab6 | 0 | 1 | ↑ | Hemoglobin, ϵ | NM_005330 |
| K07ab6 | 0 | 18 | ↑ | Hemoglobin, γA | NM_000559 |
↓ indicates genes with expression levels undetectable in cells from ESCs; ↑, genes with expression levels undetectable in cells from BM.
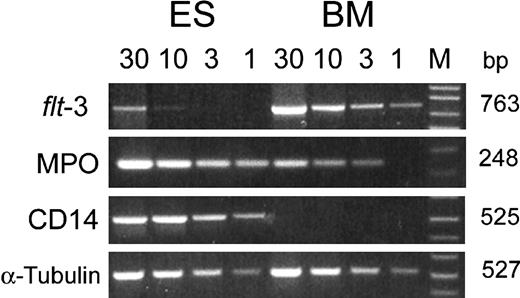
To validate further the results from the array analysis, we performed semiquantitative PCR analyses for several genes that showed different levels of expression in the array experiments. As shown in Figure 2, there was a close correlation between the 2 methods. A markedly decreased expression of flt-3 gene was observed in CD34+CD38- cells derived from ESCs as compared with the substantial level of expression in the corresponding cell preparations from BM (Figure 2). Similarly, no CD14 expression was detected in CD34+CD38- cells from BM, but a substantial level of CD14 message was found in cells from ESC cultures, consistent with the results obtained from the array analysis. For the myeloperoxidase gene, the variance in expression between the 2 sources of cells was again demonstrated, although a somewhat lesser difference was seen by the PCR method (Figure 2).
Analysis of gene expression in CD34+CD38- cells derived from human ESCs and adult BM. Cytoplasmic RNA from CD34+ CD38- cells was used to construct cDNA pools, and the expression of genes was examined by semiquantitative PCR. The number at the top of each lane indicates the amount (microliters) of cDNA used in the 50-μL PCR reaction. M = 1 kb plus DNA ladder.
Analysis of gene expression in CD34+CD38- cells derived from human ESCs and adult BM. Cytoplasmic RNA from CD34+ CD38- cells was used to construct cDNA pools, and the expression of genes was examined by semiquantitative PCR. The number at the top of each lane indicates the amount (microliters) of cDNA used in the 50-μL PCR reaction. M = 1 kb plus DNA ladder.
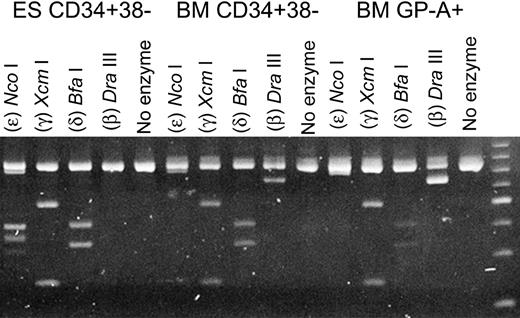
To permit a more quantitative comparison of the expression of the non–α-globin genes in the PCR analysis, we utilized a group of restriction enzymes with specificity for the β-, δ-, γ-, and ϵ-globin genes and a single set of primers with specificity for sequences common to each of these 4 genes. The PCR products were then digested with restriction enzymes specific for each globin gene. The comparative expression of the β-, δ-, γ-, and ϵ-globin genes of CD34+CD38- cells derived from human ESCs, and of those harvested from adult BM, is shown in Figure 3. In CD34+CD38- cells from BM, the messages of the non–α-globin genes were those of the adult β- and δ-globins and fetal γ-globin, and none could be detected for the embryonic ϵ-globin gene. These results were also very similar to those we observed in glycophorin A–positive erythroid cells, also derived from human adult BM (Figure 3). With the CD34+CD38- cells derived from human ESCs, no β-globin gene expression was apparent, whereas the ϵ-globin gene was prominently expressed (Figure 3). Again, these results are entirely consistent with those obtained from the array analyses.
Semiquantitative expression analysis of the ϵ-, γ-, δ-, and β-globin genes. For the RT-PCR, a single set of primers was used. The products were then digested with restriction enzymes specific for each specific globin gene. The individual enzymes are indicated at the top of each lane.
Semiquantitative expression analysis of the ϵ-, γ-, δ-, and β-globin genes. For the RT-PCR, a single set of primers was used. The products were then digested with restriction enzymes specific for each specific globin gene. The individual enzymes are indicated at the top of each lane.
Discussion
Complementary DNA microarray technology, which can detect and quantify the expression of thousands of genes simultaneously, represents one of the most powerful approaches in the field of gene expression profiling and has greatly improved our understanding of the complex patterns of gene expression in cells. Expression studies on cDNA arrays, especially oligonucelotide-based high-density arrays, normally require substantial quantities of RNA samples for probe preparation, which limits its application when only small samples are available. Although hematopoietic differentiation of human ESCs has been successfully achieved, the generation of large numbers of early hematopoietic precursors/ stem cells such as CD34+CD38- cells remains a major obstacle to this type of analysis. In the present study, we were able to take advantage of SMART technology and cDNA microarrays to compare systematically the expression patterns of genes of CD34+CD38- cells derived from adult BM and from ESCs. The consistent signal intensities we observed for reference genes placed at different locations in the arrays provided additional confirmation of the observed results. Moreover, the results from the array assay were entirely consistent with those from PCR analyses using the same cDNA libraries and served further to validate this strategy.
Our analyses showed that similar numbers of genes (1494 versus 1692, ESCs versus BM, respectively) were expressed in CD34+CD38- cells derived from ESCs and from BM. However, the expression patterns showed numerous differences, involving all of the functional gene categories; within these categories, some genes were expressed primarily or exclusively in cells from BM, whereas others were only detected in the corresponding cell populations derived from ESCs. In general, the numbers of up- and down-regulated genes in cells from BM and ESCs were similar in almost all of the functional categories. A possible explanation for this disparity may be that one or another alternative equivalent pathway within these individual functional categories may be active in these different cell types and therefore may not necessarily be reflective of an overall difference in cell function activity. Alternatively, the CD34+CD38- cells purified from BM and ESC differentiation could contain dissimilar subpopulations of hematopoietic precursors, possibly at varying stages of differentiation. Simultaneous characterization for the content of functionally distinct precursors and analysis of gene expression in the purified cell populations26 might serve to resolve this question.
On the other hand, these analyses provide a clear indication that CD34+CD38- cells derived from ESCs and those from BM differ in their developmental phenotypes, with the hematopoietic precursors derived from ESCs corresponding consistently to the embryonic stage of yolk sac/hepatic hematopoiesis. Normal expression of the various globin genes, with their distinct expression patterns corresponding to the embryonic, fetal, and adult developmental stages in man and other primates, provides a particularly useful measure of the maturational stage.27 We observed that the globin gene expression pattern of CD34+CD38- cells derived from human ESCs was typical of the embryonic pattern; in contrast, the CD34+CD38- cells from BM expressed a substantial level of the β-globin gene, with absent expression of the ϵ-globin gene, corresponding to the adult phenotype.
In human adult hematopoiesis, MHC I genes are expressed on both undifferentiated progenitors and morphologically recognizable precursors. MHC II molecules are highly expressed on early precursors but are decreased on late progenitors and differentiated precursors.28,29 An absence of MHC II and of classical MHC I expression has been reported in early human embryos, although low levels of nonclassical MHC I (HLA-G) molecules were demonstrated in some preimplantation embryos.30-33 These genes are also not expressed in primitive erythoid lineages (ie, in yolk sac–derived megaloblasts circulating in the peripheral blood at 5 weeks to 6 weeks) but have been observed from 6 weeks outward,31,32 when “definitive” macrocytic erythoblasts start differentiating in the fetal liver. Our array analyses demonstrate high levels of both MHC I and II gene expression in CD34+CD38- cells from BM, whereas there were low or negligible levels of most MHC I and II molecules in the cells derived from ESC cultures. These results are also consistent with observations that low levels of MHC I proteins and undetectable levels of MHC II molecules were present in human ESCs and in their differentiated derivatives.34
The results from these analyses are consonant with the hypothesis that ESC-derived HSCs may retain functionally important properties of embryonic cells, which in turn may require specific elements of a fetal environment for BM engraftment to take place.10-12 Supporting this idea are observations showing that when mouse yolk sac progenitor cells were transplanted into livers of newborn pups, the presence of long-term repopulating stem cells could be demonstrated, whereas the same cells transplanted into adult animals exhibited no such repopulating potential.10-12 Culture conditions or factors that could enhance the maturation of ESC-derived HSCs from the embryonic/fetal phenotype to an adult/definitive phenotype might therefore function to generate HSCs with the potential for engraftment in adult recipients.
In the present and previous studies, we have consistently demonstrated that there was little or no expression of flt-3 in hematopoietic progenitor cells derived from human, rhesus monkey, and mouse ESCs.13,15 FLT-3 is a member of the class III receptor tyrosine kinase (RTK-III) family and shares considerable structural homology with other members of this family, which include c-KIT, FMS, and platelet-derived growth factor receptor (PDGF-R).35-37 The signal transduction activity of FLT-3, in response to stimulation by its ligand, FL, results in phosphorylation with activation of a number of cellular pathways, including phospholipase C gamma (PLCγ), Ras guanosine triphosphatase (GTPase)–activating protein (Ras-GAP), phosphatidylinositol 3′-kinase (PI3K) SHC, growth factor receptor-bound protein 2 (GRB2), VAV, FYN, SRC, mitogen-activated protein (MAP) kinases, and signal transducer and activator of transcription (Stat5a),38 several of which have a well-defined role in the regulation of embryonic cell differentiation and hematopoiesis.39-42 In the mouse, the HSC population has been shown to consist entirely of the Flt-3- cells.43-47 However, recent studies by Sitnicka et al48 and Ebihara et al49 have shown that virtually all human BM and umbilical cord blood lymphomyeloid stem cells capable of reconstituting nonobese diabetic/severe combined immunodeficiency (NOD-SCID) mice expressed the flt-3 gene. Moreover, the Flt-3 ligand, FL, efficiently supports the viability of human HSCs48 but has no such effect on HSCs from the mouse.43 Although the understanding of the function of flt-3 in hematopoiesis is as yet incomplete, these various observations suggest that its role in this process may be a significant one. Our findings of diminished or absent expression of the flt-3 gene in ESC-derived hematopoietic precursors from 3 different mammalian species, suggesting that this may be a consistent and general difference between hematopoietic precursors obtained from BM and those from the maturation of ESCs, could represent a fruitful avenue for further study.
Prepublished online as Blood First Edition Paper, February 12, 2004; DOI 10.1182/blood-2003-10-3575.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal