Abstract
Interleukin-5 (IL-5) is a hematopoietic differentiation factor that promotes the development of mature eosinophils from progenitors in bone marrow. We present a multifactorial microarray study documenting the transcriptional events in bone marrow of wild-type and IL-5–deficient mice at baseline and in response to infection with Schistosoma mansoni. The microarray data were analyzed by a 4-way subtractive algorithm that eliminated confounding non-IL-5–related sequelae of schistosome infection as well as alterations in gene expression among uninfected mice. Among the most prominent findings, we observed 7- to 40-fold increased expression of transcripts encoding the classic eosinophil granule proteins (eosinophil peroxidase, major basic protein, the ribonucleases) together with arachidonate-15-lipoxygenase and protease inhibitor plasminogen activator inhibitor 2 (PAI-2), in the IL-5–producing, infected wild-type mice only. This was accompanied by increased transcription of genes involved in secretory protein biosynthesis and granule-vesicle formation. Interestingly, we did not detect increased expression of genes encoding eosinophil-related chemokine receptors (CCR1, CCR3) or members of the GATA or CCAAT/enhancer binding protein (C/EBP) transcription factor families. These data suggest that the IL-5–responsive progenitors in the mouse bone marrow are already significantly committed to the eosinophil lineage and that IL-5 promotes differentiation of these committed progenitors into cells with recognizable and characteristic cytoplasmic granules and granule proteins.
Introduction
The eosinophilic leukocyte remains among the most poorly understood of all cells in mammalian physiology. Although we have a reasonably clear (albeit controversial) understanding of the detrimental sequelae of eosinophil activities as they relate to the pathogenesis of disease (allergic, helminthic, idiopathic), the beneficial end points of eosinophil differentiation, recruitment, and activation have not been clearly defined.1-5
The cytokine, interleukin-5 (IL-5), a 45-kDa homodimeric protein produced primarily by T helper 2 (Th2) lymphocytes, has been linked almost inextricably to the biology of the eosinophilic leukocyte. IL-5 promotes eosinophil differentiation in bone marrow, mediates eosinophil activation, and delays eosinophil apoptosis in target tissues.5-8 Although IL-5 is a prominent participant in numerous eosinophil-mediated and eosinophil-related disease processes,2,9-12 its role in the development and differentiation of eosinophils under physiologic, unstressed conditions remains unclear. For example, although IL-5 transgenic mice demonstrate profound blood and tissue eosinophilia,13,14 the converse—that IL-5 gene-deleted mice should have no eosinophils—is not the case. Both the IL-5 gene-deleted mouse described by Kopf et al15 and the IL-5 receptor gene-deleted mouse described by Yoshida et al16 maintain normal or near-normal eosinophil numbers in bone marrow and peripheral blood. Furthermore, eosinophil recruitment into the pregnant uterus proceeds normally in IL-5 gene-deleted mice,17 as does the recruitment observed in response to respiratory virus infection,18 and, although blunted, one can observe eosinophil recruitment to the lungs of allergen-challenged, interleukin-5–deficient mice.5,19 Although these results suggest that there may be unique IL-5–dependent and IL-5–independent subsets of what has until now been presumed to be a homogeneous leukocyte population,18,20 the results of Foster et al5 suggest that all eosinophils maintain IL-5 responsiveness and that IL-5 is necessary not so much for baseline eosinophil production, but for lineage expansion (ie, eosinophilopoiesis in response to pathophysiologic provocation).
Although recent results have improved our understanding of IL-5–mediated signal transduction,21,22 at current writing, we have only a limited understanding of the way in which these findings translate into IL-5–dependent eosinophilopoiesis. Although several groups have studied the transcription of individual eosinophil-specific genes,23-26 and most recently, Yu et al27 have identified a promoter element in the GATA-1 gene that is clearly crucial to creation of this lineage overall, no one has identified any IL-5–specific, lineage-defining transcription factors or events.
The experiments described here address the question of IL-5–dependent hematopoiesis in vivo via a 4-way gene microarray approach. We present here a subtractive analysis of gene transcripts detected in wild-type and IL-5–deficient mice, both at baseline and in the setting of increased serum IL-5 (in wild-type, but not IL-5–deficient mice) as part of the overall late-stage Th2 response to infection with the helminthic parasite, Schistosoma mansoni. Among the most interesting findings, the microarray data suggest that the target cells responding to IL-5 are already highly committed to the eosinophil lineage (ie, already contain eosinophil-related and eosinophil-specific transcripts) and that IL-5–mediated eosinophil expansion involves transcriptional activation of genes related to granule and granule protein synthesis.
Materials and methods
Infection of mice with S mansoni
Age-matched C57BL/6 wild-type (WT) and IL-5–deficient (IL-5–/–) mice were used in all experiments described. Infected mice were exposed to 25 to 40 cercariae of S mansoni by percutaneous exposure as described.28 Serum IL-5 was monitored on a weekly basis by enzyme-linked immunosorbent assay (ELISA; R&D Systems, Minneapolis, MN). The mice were killed by cervical dislocation at week 8. Experimental protocols were reviewed by the Animal Care and Use Committee, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), protocol numbers LPD-16 and LHD-8E.
Collection and analysis of bone marrow cells
Mouse bone marrow was collected from femurs and tibiae of the hind legs of S mansoni–infected and uninfected mice by flushing the opened bones with sterile phosphate-buffered saline (PBS) through opened bones. Cells were washed in PBS, and red blood cells were lysed by addition of ACK lysis buffer (Biowhittaker, Gaithersburg, MD) to the cells, incubation for 5 minutes, and subsequent washing in PBS + 1% bovine serum albumin (BSA). The bone marrow cells were counted in a hemocytometer, and 1 × 105 cells/mL were subjected to cytospin. The cytospins were fixed in methanol and stained using Diff Quik (Dade Behring AG, Dudingen, Switzerland). The percentage of eosinophils was determined by counting 1000 cells on 3 slides from each of the 4 conditions. Mature eosinophils were identified by their deep-red staining cytoplasm and bilobed nucleus. Furthermore, 1 × 106 bone marrow cells from S mansoni-infected wild-type and IL-5–/– mice (t = 8 weeks) were treated with ACK lysis as described earlier and fixed with 200 μL 1.5% paraformaldehyde for flow cytometric analysis (FACSort; Becton Dickinson, San Jose, CA). The different cell populations were identified by their forward/side scatter pattern.
Preparation of RNA from bone marrow cells
Bone marrow was harvested from both wild-type and IL-5–deficient mice, both infected with S mansoni and uninfected (ie, 4 distinct conditions). RNazol B (1 mL; Teltest, Friendswood, TX), was added to each 2 × 106 cells (15-25 × 106 cells total), and extraction proceeded as per manufacturer's instructions. The precipitated RNA was harvested by centrifugation, washed in 70% ethanol, dried, and resuspended in diethyl-pyrocarbonate (DEPC)–treated sterile water. RNA concentration was measured spectrophotometrically at optical density (OD) 260, with typical yields of 60 μg total RNA at OD 260/OD 280 ratios of 2.0. Equal amounts of bone marrow RNA were pooled from 3 mice per condition to be used for Northern blotting and complementary (c)RNA and cDNA synthesis.
Gene microarray
The gene array procedures were performed at the Functional Genomics Center, University of Rochester Medical Center using the protocol described in our previous publication.29 Briefly, 5 μg from each of the pooled RNAs representing the 4 conditions was first-strand transcribed using T7-oligo deoxythymidine (dT). Second-strand DNA synthesis was performed with primer recognizing the T7 promoter sequence. Subsequently, the synthesized DNA was cleaned by phenol/chloroform extraction and ethanol precipitation. The second-strand DNA was mixed with T7 RNA polymerase and was in vitro transcribed to cRNA by addition of ribonucleotides of which deoxyuridine triphosphate (dUTP) was biotin labeled (Affymetrix, Santa Clara, CA). The in vitro transcription step increased the RNA copy number by a thousand-fold. Unincorporated nucleotides were removed by RNeasy Mini kit (Qiagen, Valencia, CA), and 15 μg cRNA was fragmented to 35 to 200 bp by metal-induced hydrolysis (Affemetrix). As a control of the hybridization efficiency, known concentrations of biotinylated cRNAs of 4 noneukaryotic genes (bioB, bioC, bioD, and cre) were included in each of the 4 reactions. The 4 biotin-labeled cRNA pools were hybridized to one each of U74Av2 gene arrays in morpholinoethanesulfonic acid (MES) hybridization buffer. The array was washed and subsequently incubated with streptavidin-phycoerythrin in Fluidics Station. Finally, the 4 arrays were scanned with Agilent Gene Array scanner. TheU74Av2 array contains 6000 known genes from the Mouse UNI gene database (Build 74) and 6000 expressed sequence tagged (EST) sequences. As normalization of each array was performed by comparing the signals of the spiked nonmammalian genes, of known concentrations, it was possible to compare the signals between the 4 arrays of the 4 different conditions.
Evaluation of array data
Primary data from the gene microarray experiments were analyzed by using algorithms within the GeneSpring software package (Silicon Genetics, Redwood City, CA), which was also used to define the IL-5–responsive profiles to be described in the text. The fold-change was calculated by the change in expression level of wild-type infected versus IL-5–deficient infected for those transcripts following the IL-5-–responsive profiles.
Northern blotting
RNA (10 μg) from uninfected and infected mouse bone marrow cells was subjected to electrophoresis on a 1.2% agarose gel containing 18% formaldehyde and subjected to Northern blotting by standard procedures. Blots were probed sequentially with a 471–base pair (bp) 32P deoxycytidine triphosphate (dCTP)–labeled cDNA probe encoding mouse eosinophilassociated ribonuclease (mEar) 2 and a 60-bp oligo encoding mouse eosinophil peroxidase (mEPO) 5′-TCGGTACTTGTTGCTGCACCTCTCAGC CTGGTCCTGGAGA GCACAGCCACTGGCCTGGGA-3′. The control probe was a cDNA encoding β-actin.
RT-PCR–based identification of mEars
For identification of specific mEars expressed in bone marrow, 2 μg total bone marrow RNA, from C56Black/6 mice, uninfected and infected with S mansoni, were treated with DNase I (Invitrogen, Gaithersburg, MD) to remove contaminating genomic DNA. DNase I-treated RNA (0.7 μg) was reverse transcribed by using the First-strand cDNA synthesis kit (Roche Molecular Biochemicals, Indianapolis, IN) following the manufacturer's instructions. All reactions were accompanied by one identical control reaction devoid of the avian myeloblastosis virus (AMV) reverse-transcriptase (RT) enzyme. The RT reactions were subjected to polymerase chain reaction (PCR) amplification with Pfx proofreading polymerase by addition of 5 μL cDNA to a total volume of 50 μL containing standard components. The primers, 5′-ATGGGTCCGAAGCTGCTTGAGTC-3′ and 5′-CTAAAATGTCCCATCCAAGTGAAC-3′, correspond to bases 72 to 94 and 542 to 519 of mEar 2 (GenBank AF3056664) and were chosen so as to amplify the open reading frames of all known mEars. The approximate 470-bp amplicons were gel purified (Geneclean Spin Kit; Q-Bio Gene, Carlsbad, CA) and ligated into the PCR-Blunt vector (Invitrogen) for subsequent restriction analysis and/or sequencing.
Identifying unique restriction sites among the mouse eosinophil-associated ribonucleases (mEars)
Accession numbers containing the open reading frames of all mEars30 were loaded into the web tool provided by New England Biolabs NEBcutter version 1.0 (http://tools.neb.com/NEBcutter/index.php3). Restriction sites determined for each Ear were compared so as to find enzymes that might cut 1 or at most 2 or 3 genes specifically. Criteria for choosing a restriction enzyme included (1) that it cuts the sequence into products that can be distinguished from the starting material and (2) that the optimal buffer could accommodate one or more additional restriction enzymes if necessary. Inserts were PCR-amplified from bacterial colonies resuspended in sterile PBS, using primers designed to be equidistant from the cloning site to avoid multiple restriction patterns resulting from variant orientations. Combinations of restriction enzymes used to identify specific mEars include 2.5 U SnaBI and 5 U BtsI in New England Biolabs (NEB) buffer 4 for mEar 2; 1 U SfaNI and 10 U BsaBI for mEar 1; 5 U BsmBI in NEB buffer 3 for mEar 6. All reactions were digested at 37°C for 2 hours and subsequently analyzed by electrophoresis on 20% polyacrylamide gels (Invitrogen) with digestion products visualized by ethidium bromide staining.
RT–PCR confirmation of gene microarray data
For confirmation of expression patterns indicated by microarray analysis, 0.1 μg total RNA was subjected to first strand cDNA synthesis as described “Gene microarray.” RT-minus controls were included for each RT reaction. Primers used to amplify specific transcripts were as follows: CCR1, 5′-GCCCTGAGGGCCCGAACTGTTACT and 3′-CAGACGCACGGCTTTGACCTTCTT; CCR2, 5′-CTTGGCCATCTCTGACCTGCTCTT and 3′-ACTAGACTTGTTATCACCCCAAAG; CCR3, 5′-GTGGGGTTTTGGCCACTACATGTG and 3′-GTGGGGTTTTGGCCACTACATGTG; CCR5, 5′-AAGTCAGAACGGTCAACTTTGGGG and 3′-TCAGGAGGAGGACAATGTTGTAGG; IL-5R, 5′-AAAGCAAATGTGTGACCCCCCTTC and 3′-CAGCACGCTTGCTTGAGCCATTAATGT; PAI-2, 5′-AACAAAGGTGAAATCCCAAACCTG and 3′-GCTGATCCACTTGTTGAAGTTGGC. Cycling conditions were denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, extension at 72°C for 1 minute, 25 or 30 cycles as indicated.
Results
Induction of IL-5 and bone marrow eosinophilia
Wild-type and interleukin-5–deficient (IL-5–/–) mice were exposed to cercariae of S mansoni on day 0 as described in “Materials and methods.” Serum samples were obtained weekly and evaluated for IL-5 by ELISA assay. As anticipated, no IL-5 could be detected in serum from the IL-5–deficient mice at any point during the experiment (ie, < 7 pg/mL, lower limit of detection). Consistent with earlier studies,31 IL-5 was first detected in serum at 6 weeks after exposure and at week 8 reached 160 ± 15 pg/mL in the wild-type mice (Figure 1). We also observed marked hepatosplenomegaly among the S mansoni–infected mice, with classic granulomata and eosinophil infiltration noted on microscopic examination of liver tissue, with eosinophils diminished approximately 10- to 20-fold in the liver granulomata of infected IL-5–/– mice (data not shown).
Serum IL-5 (pg/mL) in wild-type (WT) and interleukin-5–deficient (IL-5–/–) C57Black/6 mice detected at 1 and 8 weeks after percutaneous inoculation with cercariae (40/mL) S mansoni versus uninfected controls. The detectable limit of the ELISA assay is indicated by the horizontal dashed line (7 pg/mL). The bars represent average ± standard error (SE) of values from 7 mice.
Serum IL-5 (pg/mL) in wild-type (WT) and interleukin-5–deficient (IL-5–/–) C57Black/6 mice detected at 1 and 8 weeks after percutaneous inoculation with cercariae (40/mL) S mansoni versus uninfected controls. The detectable limit of the ELISA assay is indicated by the horizontal dashed line (7 pg/mL). The bars represent average ± standard error (SE) of values from 7 mice.
As anticipated, the increase in serum IL-5 correlated with an increase in eosinophil number and percentage of eosinophils in the bone marrow of the wild-type mice. Shown in Figure 2A-B are samples of bone marrow from both wild-type and interleukin-5–deficient mice, demonstrating the morphology and staining properties of mature eosinophils, scored quantitatively in Figure 2C. Consistent with the initial description of Kopf et al,15 the fraction of eosinophils present in the bone marrow of IL-5–deficient mice did not differ substantially from that of the wild-type mice at baseline, both detected at approximately 3% to 4%.At week 8, corresponding to the rise in serum interleukin-5, we observed a marked increase in eosinophils in the bone marrow of the wild-type mice, reaching 20% ± 3%. The interleukin-5–deficient mice do not respond in this fashion; eosinophils in the bone marrow remain at baseline (3.7% ± 0.3%). Bone marrow cells from infected wild-type and IL-5–/– mice were analyzed by flow cytometry. As anticipated from the results in Figure 2C, the population containing myeloid cells, including eosinophils and their progenitors, was 4.5-fold larger in the infected wild-type mice than in the IL-5–/– mice. Statistically insignificant increases were observed among monocyte and lymphocyte populations. Mice were killed at week 8 after inoculation, and the bone marrow was collected for the gene array studies described in “Identification of IL-5–dependent transcriptional targets by gene microarray.”
Cytospin preparations of bone marrow cells. Diff-Quik–stained cells from wild-type (A) and IL-5–deficient (B) mice evaluated 8 weeks after inoculation with S mansoni. Eosinophils are indicated at the arrows. (C) Percentage of mature eosinophils detected in bone marrow from S mansoni–infected wild-type and IL-5–deficient mice at zero and 8 weeks after inoculation. Each bar represents average ± SE of absolute counts per 1000 cells from 3 mice of each of the conditions; statistical significance, *P < .01, S mansoni-infected wild-type mice versus any of the other 3 conditions.
Cytospin preparations of bone marrow cells. Diff-Quik–stained cells from wild-type (A) and IL-5–deficient (B) mice evaluated 8 weeks after inoculation with S mansoni. Eosinophils are indicated at the arrows. (C) Percentage of mature eosinophils detected in bone marrow from S mansoni–infected wild-type and IL-5–deficient mice at zero and 8 weeks after inoculation. Each bar represents average ± SE of absolute counts per 1000 cells from 3 mice of each of the conditions; statistical significance, *P < .01, S mansoni-infected wild-type mice versus any of the other 3 conditions.
Identification of IL-5–dependent transcriptional targets by gene microarray
Gene expression data generated for all 4 physiologic conditions— uninfected and infected IL-5–deficient mice and uninfected and infected wild-type mice—were compared quantitatively to one another as shown in Figure 3. Figure 3A includes the complete set of 12 000 tracings. The overdrawn lines represent specific expression profiles: profiles 1 and 2 demarcating genes whose expression is increased predominantly (profile 1) or only (profile 2) in infected, wild-type mice. By this experimental design, we have limited the data set to responses to IL-5 alone and have not included responses to S mansoni infection in general, which would be represented as more or less equal expression in bone marrow from both wild-type and IL-5–/– mice. Nor are we considering any innate differences present in bone marrow from wild-type versus IL-5–/– mice at baseline. Profile 3 is the inverse of profile 1 and represents genes whose expression decreases predominantly in infected, wild-type mice (ie, in response to IL-5). Genes whose expression patterns followed the profiles 1, 2, and 3 at a correlation coefficient of at least 0.95 were selected for further analysis. Figure 3B-D represents the tracings of gene groups corresponding to profiles 1, 2, and 3, respectively. There were no transcripts detected within the 0.95 correlation coefficient that followed the profile represented by the inverse of line 2.
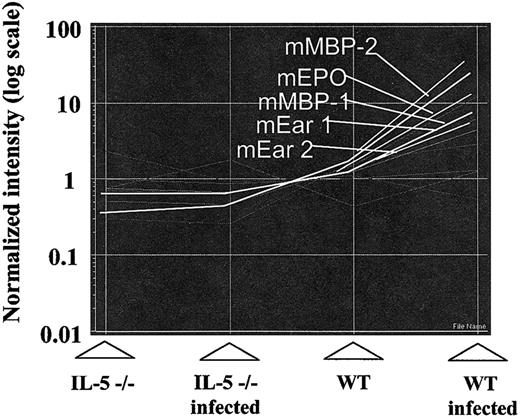
Tracking the expression patterns of 12 000 mouse genes within the U74Av.2 gene chip. The images are generated using algorithms of the GeneSpring analysis program (described in “Materials and methods”). (A) The S mansoni–infected wild-type pattern (WT Infected) is set as default, with the high values (approaching 10) indicating genes that are highly expressed in bone marrow from these mice, with gradations approaching 0.1 indicating genes that are expressed minimally. The “overdrawn” lines indicate selected patterns (correlation coefficient ≥ 0.95); profile 1, at or near baseline (= 1.0) in all but IL-5–expressing, infected wild-type mice; profile 2, at baseline for all but the infected wild-type mice. Profiles 1 and 2 represent gene expression correlating directly with levels of serum IL-5 and bone marrow eosinophilia. Profile 3, is the inverse of profile 1, as shown. There were no transcripts corresponding to the inverse of profile 2. (B-D) Graphic representations of transcripts following expression patterns delineated by profiles 1, 2, and 3 respectively.
Tracking the expression patterns of 12 000 mouse genes within the U74Av.2 gene chip. The images are generated using algorithms of the GeneSpring analysis program (described in “Materials and methods”). (A) The S mansoni–infected wild-type pattern (WT Infected) is set as default, with the high values (approaching 10) indicating genes that are highly expressed in bone marrow from these mice, with gradations approaching 0.1 indicating genes that are expressed minimally. The “overdrawn” lines indicate selected patterns (correlation coefficient ≥ 0.95); profile 1, at or near baseline (= 1.0) in all but IL-5–expressing, infected wild-type mice; profile 2, at baseline for all but the infected wild-type mice. Profiles 1 and 2 represent gene expression correlating directly with levels of serum IL-5 and bone marrow eosinophilia. Profile 3, is the inverse of profile 1, as shown. There were no transcripts corresponding to the inverse of profile 2. (B-D) Graphic representations of transcripts following expression patterns delineated by profiles 1, 2, and 3 respectively.
We identified 122 genes expressed in mouse bone marrow within profiles 1 and 2, demonstrating increased expression (2-fold or higher) in response to physiologic production of IL-5. Interestingly, of these genes, only 9 genes demonstrated increases of 5-fold or more (maximum 40-fold). We detected a more-or-less equal number of genes whose expression is diminished in response to IL-5, although with 47 genes demonstrating decreases of 5-fold or more (maximum decrease, 34-fold).
In terms of specifics, we demonstrated increased expression of transcripts encoding all of the eosinophil granule proteins including mouse eosinophil peroxidase (mEPO), major basic proteins 1 and 2 (mMBP-1 and -2), and the mouse eosinophil-associated ribonucleases (mEars). All follow the pattern delineated by profile 1 (Figure 4). Although the microarray readout vis à vis the eosinophils ribonucleases lists mEars 1 and 2, the 15 genes of this cluster are highly homologous and cross-hybridize with one another.32 The actual distribution of mEars in mouse bone marrow under the 4 conditions depicted is discussed in “Identification of mEars expressed in bone marrow at baseline and in response to interleukin-5.”
As inFigure 3, the relative expression patterns of eosinophil granule protein genes mEPO (mouse eosinophil peroxidase), mMBP-1 and mMBP-2 (mouse major basic protein), and mEar-1 and mEar-2 (mouse eosinophil-associated ribonucleases 1 and 2). NB indicates the probes used on the array will also cross-react with all mEars save for 4, 5, 11, and 14.
As inFigure 3, the relative expression patterns of eosinophil granule protein genes mEPO (mouse eosinophil peroxidase), mMBP-1 and mMBP-2 (mouse major basic protein), and mEar-1 and mEar-2 (mouse eosinophil-associated ribonucleases 1 and 2). NB indicates the probes used on the array will also cross-react with all mEars save for 4, 5, 11, and 14.
Results of the microarray analysis are shown in Table 1. We have grouped the genes as increased expression (left-hand column) versus decreased expression (right-hand column) and have delineated several specific but not mutually exclusive categories. As noted earlier, several of the transcripts detected most prominently in RNA from wild-type, infected mice alone include those expressed uniquely or primarily in eosinophils, including mouse eosinophil peroxidase, mouse eosinophil major basic protein, and arachidonate-15-lipoxygenase. Other categories of altered gene expression include transcripts associated with signal transduction, vesicular transport, gene transcription, and cell cycling. These findings will be considered at greater length in “Discussion.” Among the genes undergoing the most prominent alterations in expression are Syntaxin 3 (+8-fold), which has been identified as a component of the eosinophil exocytotic apparatus,33 the signal transduction elements phospholipase D1 (–34-/–2.1-fold), syk (–27-fold) which is intriguing, given the role of syk in IL-5–mediated eosinophil activation.34 Also undergoing transcriptional alteration is the retinoic acid receptor (–14-fold), an interesting parallel given that its ligand, retinoic acid, has been shown to modulate expression of the receptor for IL-5.35
Detection of transcripts correlating directly (profiles 1 and 2) and inversely (profile 3) with increased levels of serum IL-5
Direct correlation . | . | . | Indirect correlation . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. . | Name . | Fold . | Accession no. . | Name . | Fold . | |||||
| Eosinophil-granulocyte—inflammation associated | ||||||||||
| AI326147* | MBP-2 | 40 | Y18101 | Macrophage actin-associated protein | -6.9 | |||||
| D78353 | EPO | 26 | U43384 | gp91 phox | -5.1 | |||||
| L34570 | Arachidonate 15-lipoxygenase | 18 | X66084 | CD44 | -4.1 | |||||
| L46768 | MBP-1 | 11 | AB002663 | gp47 phox | -3.8 | |||||
| U72032 | mEar-1 | 9.4 | M60778 | LFA-1 | -2.2 | |||||
| X16490 | Plasminogen activator inhibitor 2 | 7.6 | ||||||||
| AF017258 | mEar-2 | 6.7 | ||||||||
| U12884 | VCAM-1 | 2.4 | ||||||||
| D16432 | CD63 | 2.2 | ||||||||
| X94353 | Cathelicidin | 2.2 | ||||||||
| Lymphocyte-related genes | ||||||||||
| D86232 | Lymphocyte antigen 6 complex | 2.4 | AF101164 | CEA-related CAM 2 | -18 | |||||
| M21332 | MHC class III RD gene | 2.0 | X88902 | Immunoglobulin heavy chain 4 | -10 | |||||
| L33954 | Ig variable heavy chain precursor | -8.2 | ||||||||
| L28060 | B-cell antigen receptor | -5.0 | ||||||||
| L43568 | B-cell receptor gene | -4.3 | ||||||||
| M23158 | Ly-5 | -3.1 | ||||||||
| D37797 | Lymphocyte antigen 78 | -2.6 | ||||||||
| X07640 | Mac-1 | -2.2 | ||||||||
| Nuclear and nucleolar proteins | ||||||||||
| AV306387* | Squamous cell carcinoma antigen | 2.9 | X04435 | Nuclear receptor subfamily 3 | -13.7 | |||||
| AI838366* | Brd4 | 2.6 | A1325802*† | NLVP.1 homolog | -5.3 | |||||
| AW121424*† | DNA-directed RNA pol II | 2.4 | U39074 | Thymopoietin | -3.9 | |||||
| U31758 | Histone deacetylase 2 | 2.4 | AB009275 | Major nuclear matrix protein | -3.4 | |||||
| AI854006* | SET | 2.3 | AW045530* | Inner centromere protein | -2.9 | |||||
| AV252495* | Smarcd 2 | 2.3 | AB026432 | Damage-specific DNA bind protein 1 | -2.0 | |||||
| AW120628*† | Nuclear LIM interacting factor | 2.3 | ||||||||
| M96823 | Nucleobindin | 2.0 | ||||||||
| AI849356* | Chromobox homolog 3 | 2.0 | ||||||||
| Receptors/signal transduction/cell cycle | ||||||||||
| AI563854* | TROP 2 | 3.5 | U87868‡ | Phospholipase D1b (mPLD1b) | -34/-2.1 | |||||
| AF059583 | Dok-R | 2.8 | U36776 | Syk, tyrosine kinase | -27 | |||||
| U69491 | IL-11 receptor α | 2.8 | M60909 | Retinoic receptor α | -14 | |||||
| X64534 | IL-3 receptor α | 2.4 | X83106 | MAD | -5.1 | |||||
| AW045910*† | HPK/CGK kinase | 2.2 | U29056 | Src-like adapter | -4.6 | |||||
| M80739 | Cytoplasmic protein tyrosine phosphate | 2.0 | D49393 | Protein tyrosine phosphatase | -4.3 | |||||
| AW122170* | Raf-1 | 2.0 | U21110 | STAT 5B | -3.5 | |||||
| U50413 | PI3K, regulatory subunit | -3.5 | ||||||||
| AF073997 | Myotubularin-related protein 1 | -3.5 | ||||||||
| AI747899* | Phosphatidylinositol transfer protein | -3.5 | ||||||||
| U86587 | PI3K, delta polypeptide | -3.3 | ||||||||
| AF054819 | NKG2D | -3.3 | ||||||||
| X76850 | MAPKAP kinase2 | -3.0 | ||||||||
| X62646 | IL-6 signal transducer | -2.8 | ||||||||
| L31532 | Bcl-2 | -2.7 | ||||||||
| AF005423 | CDC-like kinase 4 | -2.4 | ||||||||
| AF035646 | RAB 10 | -2.3 | ||||||||
| AB005664 | MAPK9 | -2.2 | ||||||||
| Vesicular transport/Golgi/endosome | ||||||||||
| D29797§ | Syntaxin 3 | 7.8 | AF011379 | ADAM 10 | -23 | |||||
| AJ222586 | NEFA protein | 3.3 | X54424 | Adaptor protein complex AP-1 | -12 | |||||
| AI425990* | Contain RabGAP/TBC domain | 2.9 | AW227650*† | Translocon-associated protein γ | -8.1 | |||||
| AW121185* | Dynein, heavy chain 1 | 2.8 | AB015790 | Sortilin-related receptor | -2.7 | |||||
| AI849207* | Golgi reassembly stacking protein 2 | 2.6 | AA763937* | Translocating chain ass membrane protein | -2.6 | |||||
| U06119 | Cathepsin H | 2.5 | AW259199* | Sorting nexin 2 | -2.5 | |||||
| AF085809 | Synapsin 1 | 2.3 | U52461 | Lysosomal trafficking regulator | -2.1 | |||||
| AI846182* | Huntington interacting protein 1 | 2.3 | D50032 | Trans Golgi network protein 2 | -2.0 | |||||
| W65964 | VAMP-8 | 2.2 | ||||||||
| AW125311* | ARF binding protein 1 | 2.0 | ||||||||
| AV347947*† | Ankyrin repeat/SOCS box protein | 2.0 | ||||||||
| Transcription and proliferation factors | ||||||||||
| AI854029* | PERP, p53 apoptosis effector | 6.5 | AF020772 | Karyopherin (importin) alpha 3 | -29 | |||||
| AV055186* | Ribosomal protein, large P1 | 4.0 | Y12783 | Ring finger protein 1B | -20 | |||||
| D63707 | Hepatoma-derived growth factor | 3.1 | L03547 | IKAROS | -10 | |||||
| AW208513* | TSSC6 | 2.3 | AV380793*† | Eukaryotic initiation factor 4G | -7.5 | |||||
| M36146 | Zfp-35 | 2.3 | X66091 | Alternate splicing factor | -6.4 | |||||
| U93862 | Ribosomal protein L41 | 2.1 | X53250 | Zfa gene | -6.0 | |||||
| X15962 | Ribosomal protein S12 | 2.1 | AA674798*† | TATA-BP-interacting protein | -5.3 | |||||
| AV105022* | Ribosomal protein 10 | 2.1 | AI131735* | Poly A polymerase α | -4.9 | |||||
| AI836082* | TF IIIC 1 | 2.1 | AF099974 | Schlafen 3 | -3.9 | |||||
| L21973 | E2F transcription factor 1 | 2.1 | U80821 | C1 transcription factor gene | -3.6 | |||||
| AI844751*† | Eukaryotic translation terminal factor 1 | 2.1 | AF043939 | DP-1 | -3.4 | |||||
| X57687 | Lyl | 2.0 | Y15907 | Myc-intron binding protein 1 | -3.1 | |||||
| U35141 | Retinoblastoma binding protein 4 | 2.0 | U21855 | CCR4-NOT transcription complex | -3.1 | |||||
| M57999 | NF kappa B | 2.0 | AF037206 | Ring finger protein 13 | -3.0 | |||||
| AB000096¶ | GATA-2 | 1.6 | AF077658 | Homeodomain interact protein kinase | -2.9 | |||||
| X15763¶ | GATA-1 | 1.5 | AF084480 | Bromodomain adj zing finger domain | -2.8 | |||||
| U97572 | Excision repair cross-complementary | -2.8 | ||||||||
| D86527 | Werner syndrome homolog | -2.7 | ||||||||
| AI844532*† | Similar to splicing factor 3b | -2.5 | ||||||||
| AF061270 | Pbx/knotted 1 homeobox | -2.4 | ||||||||
| AW209414* | Zfp422 | -2.4 | ||||||||
| M13945 | Pim-1 | -2.2 | ||||||||
| U67321 | Caspase 7 | -2.3 | ||||||||
| U10325 | Aryl hydrocarbon rec nucl transloc | -2.3 | ||||||||
| Y11245 | Forkhead box M1 | -2.1 | ||||||||
| U09928 | Eukaryot TIF 2-alpha kinase | -2.1 | ||||||||
| AF052506 | Adenosine deaminase, RNA spec | -2.0 | ||||||||
| AF026524 | Inhibitor of kappa B kinase β | -2.0 | ||||||||
Direct correlation . | . | . | Indirect correlation . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Accession no. . | Name . | Fold . | Accession no. . | Name . | Fold . | |||||
| Eosinophil-granulocyte—inflammation associated | ||||||||||
| AI326147* | MBP-2 | 40 | Y18101 | Macrophage actin-associated protein | -6.9 | |||||
| D78353 | EPO | 26 | U43384 | gp91 phox | -5.1 | |||||
| L34570 | Arachidonate 15-lipoxygenase | 18 | X66084 | CD44 | -4.1 | |||||
| L46768 | MBP-1 | 11 | AB002663 | gp47 phox | -3.8 | |||||
| U72032 | mEar-1 | 9.4 | M60778 | LFA-1 | -2.2 | |||||
| X16490 | Plasminogen activator inhibitor 2 | 7.6 | ||||||||
| AF017258 | mEar-2 | 6.7 | ||||||||
| U12884 | VCAM-1 | 2.4 | ||||||||
| D16432 | CD63 | 2.2 | ||||||||
| X94353 | Cathelicidin | 2.2 | ||||||||
| Lymphocyte-related genes | ||||||||||
| D86232 | Lymphocyte antigen 6 complex | 2.4 | AF101164 | CEA-related CAM 2 | -18 | |||||
| M21332 | MHC class III RD gene | 2.0 | X88902 | Immunoglobulin heavy chain 4 | -10 | |||||
| L33954 | Ig variable heavy chain precursor | -8.2 | ||||||||
| L28060 | B-cell antigen receptor | -5.0 | ||||||||
| L43568 | B-cell receptor gene | -4.3 | ||||||||
| M23158 | Ly-5 | -3.1 | ||||||||
| D37797 | Lymphocyte antigen 78 | -2.6 | ||||||||
| X07640 | Mac-1 | -2.2 | ||||||||
| Nuclear and nucleolar proteins | ||||||||||
| AV306387* | Squamous cell carcinoma antigen | 2.9 | X04435 | Nuclear receptor subfamily 3 | -13.7 | |||||
| AI838366* | Brd4 | 2.6 | A1325802*† | NLVP.1 homolog | -5.3 | |||||
| AW121424*† | DNA-directed RNA pol II | 2.4 | U39074 | Thymopoietin | -3.9 | |||||
| U31758 | Histone deacetylase 2 | 2.4 | AB009275 | Major nuclear matrix protein | -3.4 | |||||
| AI854006* | SET | 2.3 | AW045530* | Inner centromere protein | -2.9 | |||||
| AV252495* | Smarcd 2 | 2.3 | AB026432 | Damage-specific DNA bind protein 1 | -2.0 | |||||
| AW120628*† | Nuclear LIM interacting factor | 2.3 | ||||||||
| M96823 | Nucleobindin | 2.0 | ||||||||
| AI849356* | Chromobox homolog 3 | 2.0 | ||||||||
| Receptors/signal transduction/cell cycle | ||||||||||
| AI563854* | TROP 2 | 3.5 | U87868‡ | Phospholipase D1b (mPLD1b) | -34/-2.1 | |||||
| AF059583 | Dok-R | 2.8 | U36776 | Syk, tyrosine kinase | -27 | |||||
| U69491 | IL-11 receptor α | 2.8 | M60909 | Retinoic receptor α | -14 | |||||
| X64534 | IL-3 receptor α | 2.4 | X83106 | MAD | -5.1 | |||||
| AW045910*† | HPK/CGK kinase | 2.2 | U29056 | Src-like adapter | -4.6 | |||||
| M80739 | Cytoplasmic protein tyrosine phosphate | 2.0 | D49393 | Protein tyrosine phosphatase | -4.3 | |||||
| AW122170* | Raf-1 | 2.0 | U21110 | STAT 5B | -3.5 | |||||
| U50413 | PI3K, regulatory subunit | -3.5 | ||||||||
| AF073997 | Myotubularin-related protein 1 | -3.5 | ||||||||
| AI747899* | Phosphatidylinositol transfer protein | -3.5 | ||||||||
| U86587 | PI3K, delta polypeptide | -3.3 | ||||||||
| AF054819 | NKG2D | -3.3 | ||||||||
| X76850 | MAPKAP kinase2 | -3.0 | ||||||||
| X62646 | IL-6 signal transducer | -2.8 | ||||||||
| L31532 | Bcl-2 | -2.7 | ||||||||
| AF005423 | CDC-like kinase 4 | -2.4 | ||||||||
| AF035646 | RAB 10 | -2.3 | ||||||||
| AB005664 | MAPK9 | -2.2 | ||||||||
| Vesicular transport/Golgi/endosome | ||||||||||
| D29797§ | Syntaxin 3 | 7.8 | AF011379 | ADAM 10 | -23 | |||||
| AJ222586 | NEFA protein | 3.3 | X54424 | Adaptor protein complex AP-1 | -12 | |||||
| AI425990* | Contain RabGAP/TBC domain | 2.9 | AW227650*† | Translocon-associated protein γ | -8.1 | |||||
| AW121185* | Dynein, heavy chain 1 | 2.8 | AB015790 | Sortilin-related receptor | -2.7 | |||||
| AI849207* | Golgi reassembly stacking protein 2 | 2.6 | AA763937* | Translocating chain ass membrane protein | -2.6 | |||||
| U06119 | Cathepsin H | 2.5 | AW259199* | Sorting nexin 2 | -2.5 | |||||
| AF085809 | Synapsin 1 | 2.3 | U52461 | Lysosomal trafficking regulator | -2.1 | |||||
| AI846182* | Huntington interacting protein 1 | 2.3 | D50032 | Trans Golgi network protein 2 | -2.0 | |||||
| W65964 | VAMP-8 | 2.2 | ||||||||
| AW125311* | ARF binding protein 1 | 2.0 | ||||||||
| AV347947*† | Ankyrin repeat/SOCS box protein | 2.0 | ||||||||
| Transcription and proliferation factors | ||||||||||
| AI854029* | PERP, p53 apoptosis effector | 6.5 | AF020772 | Karyopherin (importin) alpha 3 | -29 | |||||
| AV055186* | Ribosomal protein, large P1 | 4.0 | Y12783 | Ring finger protein 1B | -20 | |||||
| D63707 | Hepatoma-derived growth factor | 3.1 | L03547 | IKAROS | -10 | |||||
| AW208513* | TSSC6 | 2.3 | AV380793*† | Eukaryotic initiation factor 4G | -7.5 | |||||
| M36146 | Zfp-35 | 2.3 | X66091 | Alternate splicing factor | -6.4 | |||||
| U93862 | Ribosomal protein L41 | 2.1 | X53250 | Zfa gene | -6.0 | |||||
| X15962 | Ribosomal protein S12 | 2.1 | AA674798*† | TATA-BP-interacting protein | -5.3 | |||||
| AV105022* | Ribosomal protein 10 | 2.1 | AI131735* | Poly A polymerase α | -4.9 | |||||
| AI836082* | TF IIIC 1 | 2.1 | AF099974 | Schlafen 3 | -3.9 | |||||
| L21973 | E2F transcription factor 1 | 2.1 | U80821 | C1 transcription factor gene | -3.6 | |||||
| AI844751*† | Eukaryotic translation terminal factor 1 | 2.1 | AF043939 | DP-1 | -3.4 | |||||
| X57687 | Lyl | 2.0 | Y15907 | Myc-intron binding protein 1 | -3.1 | |||||
| U35141 | Retinoblastoma binding protein 4 | 2.0 | U21855 | CCR4-NOT transcription complex | -3.1 | |||||
| M57999 | NF kappa B | 2.0 | AF037206 | Ring finger protein 13 | -3.0 | |||||
| AB000096¶ | GATA-2 | 1.6 | AF077658 | Homeodomain interact protein kinase | -2.9 | |||||
| X15763¶ | GATA-1 | 1.5 | AF084480 | Bromodomain adj zing finger domain | -2.8 | |||||
| U97572 | Excision repair cross-complementary | -2.8 | ||||||||
| D86527 | Werner syndrome homolog | -2.7 | ||||||||
| AI844532*† | Similar to splicing factor 3b | -2.5 | ||||||||
| AF061270 | Pbx/knotted 1 homeobox | -2.4 | ||||||||
| AW209414* | Zfp422 | -2.4 | ||||||||
| M13945 | Pim-1 | -2.2 | ||||||||
| U67321 | Caspase 7 | -2.3 | ||||||||
| U10325 | Aryl hydrocarbon rec nucl transloc | -2.3 | ||||||||
| Y11245 | Forkhead box M1 | -2.1 | ||||||||
| U09928 | Eukaryot TIF 2-alpha kinase | -2.1 | ||||||||
| AF052506 | Adenosine deaminase, RNA spec | -2.0 | ||||||||
| AF026524 | Inhibitor of kappa B kinase β | -2.0 | ||||||||
See Figure 3 and text for profile descriptions.
LFA-1 indicates lymphocyte function antigen-1; VCAM-1, vascular cell adhesion molecule-1; CEA, carcinoembryonic antigen; CAM 2, cellular adhesion molecule 2; MHC, major histocompatibility complex; Ig, immunoglobulin; SET, leukemia-associated protein; TROP 2, tropisetron 2; Dok-R, downstream-of-kinase—related; MAD, myoadenylate deaminase; HPK/CGK, histidine protein kinase/complex glycerol kinase; STAT 5B, signal transducer and activator of transcription 5B; PI3K, phosphatidyl inositol-3 kinase; NKG2D, natural killer—activating receptor G2D; MAPKAP, mitogen-activated protein kinase-activated protein; CDC, complement-dependent cytotoxicity; RAB 10, rac-activating and binding protein 10; MAPK9 mitogen-activated protein kinase 9; ADAM 10, adamalysin family protein 10; NEFA, nonesterified fatty acid; VAMP-8, vesicle-associated membrane protein 8; SOCS, suppressor of cytokine signaling family of proteins; TSSC6, tetraspanin 6; TATA-BP, tumor-associated transplantation antigen basic protein; TF, tissue factor; DP-1, differentiation polypeptide-1; CCR4-NOT, regulator of gene transcription in yeast; NF, nuclear factor; TIF 2-alpha, tumor-inhibiting factor 2-alpha.
EST sequence, identity identified by BLAST search.
Identified by homology to genes in Homo sapiens or Rattus norvegicus.
Duplicate readouts with significant divergence.
Profile 2.
Not ≥2.0-fold increase, included for specific interest related to eosinophilopoiesis.
Many genes that are traditionally associated with eosinophil development and differentiation were either not detected or did not respond within the patterns defined by profiles 1 or 2 in Figure 3 (Table 2). These findings will be considered and interpreted in “Discussion.”
Genes implicated in eosinophilopoiesis but with expression patterns that do not correlate with levels of serum IL-5
Cytokines/chemokines and their receptors . | Transcription factors . | Signal transduction . |
|---|---|---|
| GM-CSF | GATA-3, 4, 5, 6 | JAK2 |
| IL-5R_ | C/EBP-α | VAV |
| IL-3 | C/EBP-β | Src |
| Eotaxin | C/EBP-γ | Lyn |
| CCR1 | C/EBP-ϵ | Shc |
| CCR2 | myb | |
| CCR3 | ||
| CCR5 |
Cytokines/chemokines and their receptors . | Transcription factors . | Signal transduction . |
|---|---|---|
| GM-CSF | GATA-3, 4, 5, 6 | JAK2 |
| IL-5R_ | C/EBP-α | VAV |
| IL-3 | C/EBP-β | Src |
| Eotaxin | C/EBP-γ | Lyn |
| CCR1 | C/EBP-ϵ | Shc |
| CCR2 | myb | |
| CCR3 | ||
| CCR5 |
Confirmation of gene microarray findings by Northern blotting
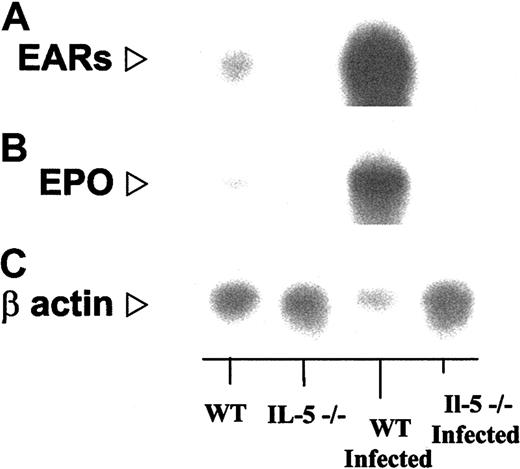
We have confirmed the increased expression predicted by gene microarray using more traditional Northern blotting methodology. In Figure 5 we demonstrate the relative expression of transcripts encoding the mouse eosinophil-ribonucleases, as noted earlier, cross-hybridizing cluster of 15 genes that cannot be distinguished from one another by Northern blotting or for that matter by gene microarray36 (“Confirmation of additional gene microarray findings by RT-PCR”) and mouse eosinophil peroxidase. Densitometry scanning (ratio of signal to actin) confirms not only the increased expression of these transcripts in wild-type infected mice but recapitulates the pattern defined by line 1 in Figure 3. The relative ratios, in order, IL-5–deficient/IL-5–deficient infected/wild-type/wild-type infected, with wild type arbitrarily normalized to 1.0, are for mouse eosinophil-associated ribonucleases, 0.13:0.19:1.0:16; mouse eosinophil peroxidase, 0.08:0.12:1.0:15, data points that are completely consistent with the values are presented in Table 2.
Northern blot analysis confirming findings of gene array. In each lane, total bone marrow RNA (approximately 10 μg) from S mansoni–infected and uninfected wild-type and IL-5–/– mice as indicated, probed with (A) a 86-bp oligonucleotide probe encoding mEar-2, which will cross-react with most of the 15 known genes of the mEar cluster; (B) an 60-bp oligonucleotide probe encoding eosinophil peroxidase (EPO; see “Materials and methods”), and (C) human β-actin. Relative signals (probe/actin) determined by scanning densitometry are as follows: IL-5–/–/WT/:IL-5–/– infected/WT/infected, mEar/β-actin, 0.13:0.19:1.0:16 (16-fold over WT, 86-fold over IL-5–/– infected); mEPO/β-actin, 0.08:0.12:1.0:15 (15-fold over WT, 122-fold over IL-5–/– infected).
Northern blot analysis confirming findings of gene array. In each lane, total bone marrow RNA (approximately 10 μg) from S mansoni–infected and uninfected wild-type and IL-5–/– mice as indicated, probed with (A) a 86-bp oligonucleotide probe encoding mEar-2, which will cross-react with most of the 15 known genes of the mEar cluster; (B) an 60-bp oligonucleotide probe encoding eosinophil peroxidase (EPO; see “Materials and methods”), and (C) human β-actin. Relative signals (probe/actin) determined by scanning densitometry are as follows: IL-5–/–/WT/:IL-5–/– infected/WT/infected, mEar/β-actin, 0.13:0.19:1.0:16 (16-fold over WT, 86-fold over IL-5–/– infected); mEPO/β-actin, 0.08:0.12:1.0:15 (15-fold over WT, 122-fold over IL-5–/– infected).
Identification of mEars expressed in bone marrow at baseline and in response to interleukin-5
As noted earlier, the mouse eosinophil-ribonuclease (mEar) genes exist in an unusually large, species-limited cluster, with significant homology (70%-95%) among individual gene pairs. As such, methods used to detect overall homology, such as gene microarray or Northern blotting, are not sufficiently precise to distinguish among individual members of this gene cluster. As part of an analysis of the IL-5–dependent transcriptional targets, we have isolated and characterized individual mEars amplified by RT-PCR from bone marrow from the 4 conditions under study. The results of the differential restriction analysis described in “Materials and methods” are shown in Figure 6. In all cases, the predominant expression pattern was more or less equal levels of mRNAs encoding mEars 1 and 2, consistent with that described by Cormier et al.37 Although other mEars were detected in all but the IL-5–/–, uninfected mice, the overall pattern was of no significant change. In other words, although there was a substantial increase in expression of mEar mRNA in the bone marrow S mansoni–infected wild-type mice, there was no significant change in distribution among the 15 or more mEars known to exist in the mouse genome. This finding is particularly intriguing, given evidence suggesting that eosinophils undergo differential transcription of mEars in response to IL-5 once they leave the bone marrow and undergo activation in the periphery (Nitto et al, manuscript submitted, 2003).
Quantitative analysis of mouse eosinophil-associated ribonucleases expressed in mouse bone marrow by way of restriction enzyme analysis of RT-PCR amplicons. Relative expression of different mouse eosinophil-associated ribonuclease genes in bone marrow in mouse bone marrow under the 4 conditions shown. Despite increased transcription of mRNAs encoding the mEars in the wild-type mice, no significant differences in distribution were observed (chi square test, P > .05).
Quantitative analysis of mouse eosinophil-associated ribonucleases expressed in mouse bone marrow by way of restriction enzyme analysis of RT-PCR amplicons. Relative expression of different mouse eosinophil-associated ribonuclease genes in bone marrow in mouse bone marrow under the 4 conditions shown. Despite increased transcription of mRNAs encoding the mEars in the wild-type mice, no significant differences in distribution were observed (chi square test, P > .05).
Confirmation of additional gene microarray findings by RT-PCR
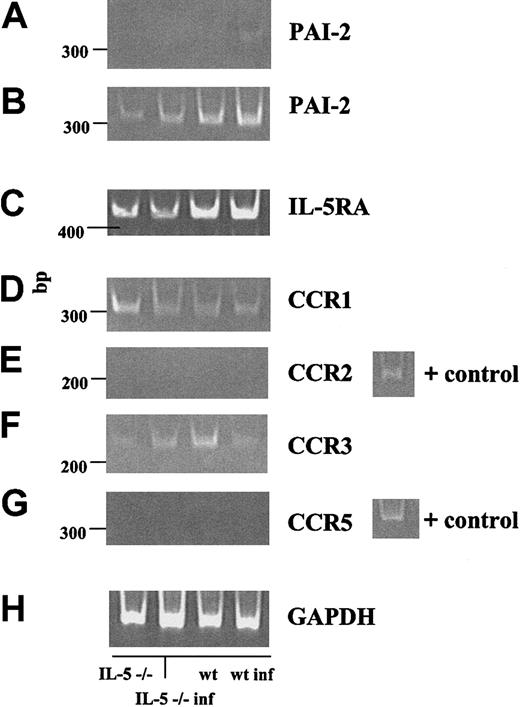
RT-PCR analysis of specific gene transcripts in bone marrow RNA from wild-type and IL-5–/– mice, both at baseline and at 8 weeks after inoculation is shown in Figure 7. Figure 7A demonstrates the anticipated IL-5–dependent increase of mRNA encoding plasminogen activator inhibitor 2 (PAI-2) in infected wild-type mice as compared with the other 3 conditions, consistent with the 7-fold increased detection documented by gene microarray (Table 1). In Figure 7B-F, we confirm the IL-5–independent expression (or lack thereof) of several specific receptors characteristic of mature peripheral blood eosinophils. We detected transcripts encoding IL-5 receptor-α, and CC chemokine receptors 1 and 3 (CCR1 and CCR3), although, consistent with findings on gene microarray, the expression of these receptors did not correlate with the presence of serum IL-5 or with the level of mature, granulated eosinophils present in the bone marrow at this time point. We were unable to detect transcripts encoding chemokine receptors CCR2 or CCR5 in these samples. RT-minus control reactions were performed with each set of primer pairs and were uniformly negative (data not shown).
RT-PCR confirmation of specific microarray results. RT-PCR was performed on 0.5-μg samples of total bone marrow RNA. (A-B) The serpin-protease inhibitor PAI-2, at 25 and 30 cycles, respectively. (B-G) Receptors IL-5Rα, CCR1, CCR2, CCR3, and CCR5. (H) glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control. All RT-minus control reactions were negative. Far right lane, positive controls as indicated.
RT-PCR confirmation of specific microarray results. RT-PCR was performed on 0.5-μg samples of total bone marrow RNA. (A-B) The serpin-protease inhibitor PAI-2, at 25 and 30 cycles, respectively. (B-G) Receptors IL-5Rα, CCR1, CCR2, CCR3, and CCR5. (H) glyceraldehyde-3-phosphate dehydrogenase (GAPDH) control. All RT-minus control reactions were negative. Far right lane, positive controls as indicated.
Discussion
Here, we present the results of a subtractive 4-way gene microarray, data that provide a virtual “snapshot” of transcriptional alterations observed in mouse bone marrow in response to IL-5 in vivo. To model a natural hematopoietic response, we infected mice with the helminthic parasite, S mansoni, and harvested bone marrow during the initial stages of the secondary Th2 response characteristic of this infection.38,39 By way of the 4-way approach (IL-5–/– uninfected versus wild-type uninfected versus IL-5–/– infected versus wild-type infected), which is the equivalent of 6 pairwise primary comparisons, we subtract out the non-IL-5–dependent effects of S mansoni infection as well as any alterations in bone marrow transcription because of the absence of IL-5 at baseline, in the uninfected state.
Most prominent among our findings was the profoundly increased transcription of genes encoding the “classic” eosinophil granule proteins. We observed 7- to 40-fold IL-5–dependent increases in transcription of the genes encoding mouse MBP, mouse EPO, and mouse eosinophil associated ribonucleases, with the identity and distribution of the latter confirmed by RT-PCR and sequencing. In addition, we observed similarly increased transcription of the gene encoding arachidonate 15-lipoxygenase, a prominent eosinophil component and crucial enzyme for leukotriene biosynthesis and lipid body formation.40,41
Among our findings were components of eosinophils that are not undergoing transcriptional activation in response to IL-5 in the bone marrow of the S mansoni–infected mice. Among this latter group are the chemoattractant receptors CCR1 and CCR3, the interleukin-5 receptor itself, interleukin-13 and its receptors, most of the GATA and all of CCAAT/enhancer binding protein (C/EBP) transcription factors, and several crucial components known to participate in IL-5–dependent signal transduction (Table 2). Although this was unexpected, and at first glance, somewhat counter-intuitive, the fact that certain eosinophil-genes are not undergoing transcriptional up-regulation led us to speculate on this nature of the IL-5–responsive eosinophil progenitor cell in this particular setting. The true nature of the IL-5–responsive eosinophil progenitor—what are its components? what is its potential?—these matters remain largely undefined. In efforts to characterize the IL-5–responsive progenitor cell(s), Denburg and colleagues42-45 demonstrated that a small proportion of human CD34+ bone marrow progenitors are also IL-5R+, and, when examined in allergic individuals, the population of IL-5R+ cells expands in response to allergen challenge. This has also been observed in allergen-sensitized and -challenged mice.46,47 However, other than concomitant CD34 and IL-5R positivity, it is not clear what eosinophil components likewise exist in these and/or whether there are in fact additional IL-5R–positive eosinophil progenitor cells. Interestingly, and consistent with the findings on our array, CCR3 was detected in “primitive” progenitors (CD34+, IL-5R–) prior to the onset of IL-5 responsiveness.45 Of course, the IL-5 responsiveness in atopic and/or asthmatic individuals may differ significantly from “wild-type” humans. Bone marrow responses to IL-5 in the setting schistosome infection might likewise have features distinct from those present after allergen sensitization.
Our data might indicate that the IL-5–responsive eosinophil progenitors in mouse bone marrow are highly committed and that IL-5 induces eosinophil proliferation in this setting by promoting the conversion of highly committed eosinophil precursor cells into ones with the more recognizable, granulated phenotype. As noted earlier, we speculate that the IL-5–responsive eosinophil precursors might contain most, if not all, of the receptors necessary for eosinophil function in the periphery and that IL-5 would promote the final steps of the differentiation pathway, involving the formation of eosinophil granules—including genes encoding elements of the Golgi and other granule-protein synthesis components—as well as the synthesis of eosinophil granule proteins themselves.
Another feature of IL-5 biology, observed in mice, but not in humans, is the relationship of this cytokine to the development of CD5+ B-1a lymphocytes, a minor subset of B lymphocytes that react to natural autoantigens.48,49 Among the findings relating to IL-5, Kopf et al15 demonstrated (temporarily) reduced numbers of B1-a lymphocytes in the peritoneal cavities of 4- to 8-week-old IL-5–/– mice, and Lee et al50 demonstrated the increased abundance of these lymphocytes in the spleens of IL-5 transgenics. Although B-1a lymphocytes arise from the fetal liver during prenatal life, reconstitution experiments in adult animals demonstrated that the bone marrow cannot reconstitute this population of cells after birth,51 and current models suggest that they arise from repopulating peritoneal stem cells.52,53 Our results are consistent with these latter findings, as we observe little to no transcriptional modulation of B lymphocyte–specific transcripts (and no modulation of the identifying cell surface marker, CD5/Ly-1) in mouse bone marrow in response to IL-5.
Several genes of specific interest have emerged from the microarray data. Among these is the 7-fold increase in transcript encoding plasminogen activator inhibitor 2 (PAI-2) a spontaneously polymerizing serpin expressed in macrophages and lymphoma cells and has been recognized as a marker for metastatic invasion.54 Although there is no direct evidence for expression of PAI-2 in eosinophils, Hara et al55 have shown that cells of the EoL-1 line inhibit the production of plasmin from the plasminogen produced by bronchial epithelial cells in culture, a function that might easily be attributed to PAI-2. PAI-2 is also prominent on the list of “asthma signature genes”56 which includes genes present in the eosinophils that are recruited to the airways in response to presensitization and challenge with both ovalbumin and aspergillus. The presence of an efficient protease inhibitor would stand in support of the hypothesis presented by Lee57 regarding an innate and essential role for eosinophils in postinflammatory “clean up” and tissue remodeling.
Although not prominent on the list, increased expression of the transcription factor, E2F-1, is worthy of some comment. E2F-1 is a member of a larger family of E2F factors, and together with E2F-2, it has been characterized as an activator of transcription and plays a central role in regulating cell cycle and cellular apoptosis.58,59 Relevant to this study, E2F-1 interacts with a number of other proteins, many of which are also subject to transcriptional regulation by interleukin-5 in bone marrow cells, including DP-1 (–3-fold), Rb binding proteins 4 (+2-fold) and 7 (–3-fold), histone deacetylase 2 (+2-fold), and, most prominently, PERP (+7-fold), a target of the p53 gene that promotes fibroblast apoptosis60 but that has not been explored at all in the context of hematopoiesis or granulocyte development and function. E2F-1–/– mice are viable, fertile, appear normal, but show deficiencies in thymocyte development and have increased susceptibility to neoplasia.61
There are 2 papers that touch on a role for E2F-1 in granulocyte development. Costa et al,62 demonstrate that, although there was no direct physical interaction with retinoic acid response elements, E2F-1 inhibited expression of transcripts controlled by retinoic acid in P19 embryonal carcinoma cells in vitro. In that light, it is interesting to note the 14-fold decreased expression of retinoic acid receptor transcript (Table 1) observed in mouse bone marrow in response to IL-5. A second article, by Porse et al,63 demonstrated an interplay between C/EBPα and E2F-dependent transcription in the process of granulocyte differentiation. It is impossible at this point to make complete sense of these findings; indeed, La Thangue64 recently reviewed the entire E2F-related field and remarked on the somewhat inscrutable “yin-yang” quality of the oft-contradictory scenarios. Nonetheless, our results suggest that IL-5 may use the E2F system in some fashion to activate quiescent precursors and/or to induce differentiation of eosinophils in vivo.
Of the 3 prominently down-regulated transcription factors, there have been no reports documenting any relationship between karyopherin (–29-fold) and granulocyte development. Regarding the ring finger factors, the single publication relating to granulocyte biology cites the retinoic acid receptor (RAR):PML—a protein of unknown function that is nonetheless structurally related to ring finger proteins—fusion created in the t(15;17) translocation characteristic of acute promyelocytic leukemia.65 Finally, and perhaps most interesting is IKAROS, a zinc finger transcription factor characterized in lymphocyte differentiation and just recently shown to regulate early stages of neutrophil differentiation in a mouse model66 ; although those researchers did not comment on the presence and/or function of eosinophils in gene-disrupted (IkL/L) mice, the pronounced down-regulation of the IKAROS transcript (–10-fold) as eosinophils are induced to mature with IL-5 would be consistent with the aforementioned studies.
In summary, the 4-way subtractive gene microarray has suggested several novel and completely test-able hypotheses regarding eosinophil components and eosinophil-related transcription, and it has provided us with specific insight into the nature of the interleukin-5–responsive eosinophil progenitor cell.
Prepublished online as Blood First Edition Paper, October 2, 2003; DOI 10.1182/blood-2003-08-2778.
Supported by the Intramural NIH Program (T.A.W. and H.F.R.) and an American Heart Association Scientist Development Grant (J.B.D.). J.B. is the recipient of a fellowship from the Fulbright Foundation of Sweden.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Andrew Brooks of the Functional Genomics Center, Rochester, NY, for running the microarray chip hybridization, and the members of the Eosinophil Pathophysiology Section, LHD, NIAID, NIH for their helpful discussions and careful consideration of this manuscript. We also thank Howard Adams and the staff of the 14BS animal facility for their care of the mice used in these experiments.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal