Abstract
Identification of the targets of mixed lineage leukemia (MLL) fusion genes will assist in understanding the biology of MLL fusion gene leukemias and in development of better therapies. Numerous studies have implicated HOXA9 as one of the possible targets of MLL fusion proteins. To determine if HOXA9 was required for leukemia development by MLL fusion genes, we compared the effects of the Mll-AF9 knock-in mutation in mice in the presence or absence of Hoxa9. Both groups of mice showed myeloid expansion at 8 weeks and then developed myeloid leukemia with a similar incidence and time course. The leukemia in the mice lacking Hoxa9 generally displayed a more immature myeloid phenotype than that in the mice that were wild-type for Hoxa9. Gene expression profiling revealed that expression of Mll-AF9 led to overexpression of Hoxa5, Hoxa6, Hoxa7, Hoxa9, and Hoxa10. Thus, genes of the Hox-a cluster are important in defining the phenotype but not the incidence of Mll-AF9 leukemia. These results demonstrate that the Mll-AF9 fusion gene disrupts the expression of several Hox genes, none of which as a single gene is likely to be necessary for development of leukemia. Instead, we propose that the “Hox code” minimally defined by the Hoxa5-a9 cluster is central to MLL leukemogenesis.
Introduction
Translocations involving the mixed lineage leukemia (MLL, ALL-1, HRX) gene are encountered in both myeloid and lymphoid leukemias. These MLL leukemias are often found in infants and also in adults previously treated with chemotherapy for other cancers.1 The mechanisms by which the translocations cause leukemia remain unknown. Gene expression profile studies demonstrate that lymphoid leukemias with MLL rearrangements exhibit an increase in expression of certain homeobox (HOX) genes compared with phenotype-matched leukemias without MLL rearrangements.2-5 The HOXA9 gene may hold an important key to the MLL leukemias because it is the one homeobox gene most frequently overexpressed in these leukemias.5 Recent evidence also indicates that MLL is part of a multiprotein complex that regulates the transcription of HOXA9 by directly binding to promoter sequences.6,7 Overexpression of Hoxa9 is also known to transform primary myeloid bone marrow cells.8-10 HOXA9 is directly involved in human leukemia caused by the NUP98-HOXA9 fusion gene11 and in the BXH-2 mouse model of leukemia.12 This encouraged us to study the relationship between Hoxa9 and Mll fusion genes and to ask whether Hoxa9 is necessary for the development of Mll leukemia. We were able to test this hypothesis in Mll-AF9+/–/Hoxa9–/– mice. Mice expressing Mll-AF9 as a heterozygous knock-in mutation develop myeloid leukemia.13 The leukemia in these mice occurs with a latency period of about 6 months and is preceded by a preleukemic phase characterized by expansion of myeloid precursors.14,15 We compared the Mll-AF9–mediated myeloid expansion and leukemia development in the presence and absence of Hoxa9.
To identify other Hox genes that may be involved in leukemia caused by the Mll-AF9 fusion gene, we conducted gene expression profiling analysis. We studied gene expression profiles of bone marrow cells from wild-type, preleukemic, and leukemic mice and found dysregulation of several genes belonging to the Hox-a cluster in the preleukemic mice carrying the Mll-AF9 gene, with further progression of Hox abnormalities in mice with leukemia.
Materials and methods
Mice
Mll-AF9+/– (Mll) mice were obtained from Dr Terence H. Rabbitts (Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom), and Hoxa9–/– mice originally produced by Dr Mario R. Capecchi (Department of Human Genetics, Howard Hughes Medical Institute, University of Utah School of Medicine, Salt Lake City) were obtained from Dr H. Jeffrey Lawrence (Department of Medicine, Veterans Affairs Medical Center, University of California, San Francisco). These mice were bred to obtain Mll-AF9+/–/Hoxa9+/– mice, which were then backcrossed with Hoxa9–/– mice to obtain Mll-AF9+/–/Hoxa9–/– (Mll/Hox) mice. Genotyping of the mice was carried out using polymerase chain reaction (PCR) on genomic DNA at weaning and at humane killing. The primers and conditions for the PCRs were as described before.15,16
FACS analysis and culture of bone marrow cells
At the time of humane killing, femurs were flushed with serum containing media to obtain bone marrow cells. Single-cell suspensions were obtained by passing the cells through a 21-gauge needle, and red blood cell (RBC) lysis was performed by incubation in ACK (ammonium chloride lysing) buffer at room temperature for 5 minutes. After centrifugation, cells were resuspended in fluorescence-activated cell sorter (FACS) buffer (10% fetal bovine serum and 0.2% sodium azide in phosphate-buffered-saline). Antibodies used for FACS were directed against Gr-1, CD11b, and B220 (BD Pharmingen, San Diego, CA). Fluorescence was detected using a FACSCalibur flow cytometer and analyzed using the CellQuest-Pro software (BD Pharmingen).
For colony culture assays, cells were collected as above except that after RBC lysis cells were cultured in methylcellulose as described before.15 After 7 days in culture, colonies containing more than 50 cells were counted and classified as described.15 For replating analysis, cells from each colony were replated in methylcellulose medium. After 7 days in culture, colonies were counted and classified and the procedure was repeated 1 more time to result in a total of 3 generations of platings.
RNA extraction and microarray hybridization
We used 10 × 106 to 20 × 106 cells to extract total RNA using the RNeasy Mini Kit (Qiagen, Valencia, CA). Quality of the RNA was judged as being good if 2 distinct ribosomal RNA bands were seen on gel electrophoresis. Ten micrograms of RNA was then used for generation of labeled cRNA exactly as described according to the manufacturer's instructions (Affymetrix, Santa Clara, CA). Hybridization of the labeled cRNA fragments and washing, staining, and scanning of the arrays were carried out as per instructions of the manufacturer. Expression values were calculated using the MAS 5.0 software (Affymetrix).
Data analysis
We used the ratio of perfect-match to mismatch probes as expression values for each gene. Scanning for errors and linear normalization was carried out using the GeneData Expressionist Suite (GeneData, South San Francisco, CA). We used all the genes on the U74A array, ignoring the “absent” and “present” calls applied by MAS 5.0. Further details are provided as supplementary information on the Blood website (Table S1; see the Supplemental Table link at the top of the online article).
Results
Both the Mll-AF9+/– and the Mll-AF9+/–/Hoxa9–/– mice demonstrate myeloid expansion at 8 weeks
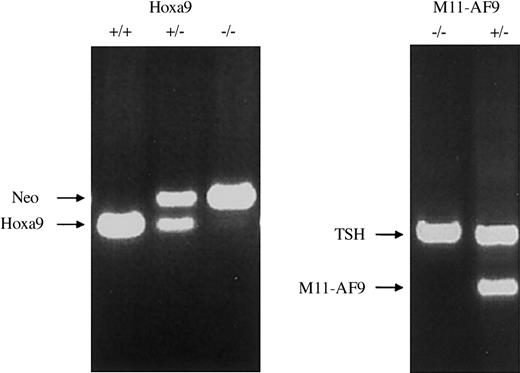
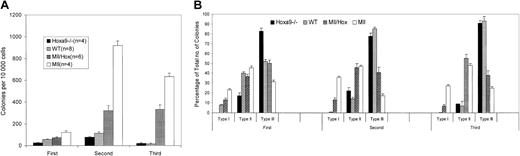
Because several studies have shown a strong association between the presence of an MLL fusion gene and high levels of expression of HOXA9, we used a genetic analysis to determine whether the Mll-AF9 fusion gene leukemia required the presence of Hoxa9. We obtained Mll-AF9+/–/Hoxa9–/– mice by crossbreeding Mll-AF9+/– mice with Hoxa9–/– mice. For the remainder of the text, the Mll-AF9+/– mice will be referred to as Mll mice and the Mll-AF9+/–/Hoxa9–/– mice as Mll/Hox mice. The genotypes of the mice were confirmed by PCR analysis of genomic tail DNA at weaning and again at humane killing (Figure 1). Leukemia development in Mll mice occurs in several steps. The earliest changes can be detected by colony-forming assays even in the prenatal stages.15 This assay allows us to quantify the self-renewal capacity of myeloid precursor cells in the bone marrow. We compared the bone marrow cells from 8-week-old Mll and Mll/Hox mice in a colony-forming assay. At this age, none of the mice show any signs of distress and a sufficient number of cells can be harvested from the bone marrow of a single mouse for colony assays. As shown in Figure 2A, the average numbers of colonies per 10 000 bone marrow cells from wild-type mice were 58 ± 3, 116 ± 11, and 16 ± 7 in the first, second, and third platings, respectively. The average number of colonies from Hoxa9–/– mice in the first plating was 26 ± 3, less than that for the wild-type mice. This difference was diminished by the second and third platings (Figure 2A). Colony numbers were significantly higher for 8-week-old Mll mice, with the average number of colonies being 121 ± 17, 919 ± 43, and 634 ± 31 for first, second, and third platings, respectively (Figure 2A). The Mll/Hox mice also showed a higher number of colonies compared with the wild-type and Hoxa9–/– mice but less than those of Mll mice, with the average number of colonies per 10 000 bone marrow cells being 73 ± 9, 323 ± 47, and 332 ± 42 for the 3 platings respectively (Figure 2A).
Generation of Mll and Mll/Hox mice. PCR results on mouse tail DNA showing wild-type, heterozygous, and homozygous Hoxa9 knock-out mice (left panel). Neo indicates neomycin-resistance gene. PCR for Mll-AF9 is shown on the right panel with the genotypes as indicated. TSH indicates β subunit of thyroidstimulating hormone used as an internal control for the PCR reaction.
Generation of Mll and Mll/Hox mice. PCR results on mouse tail DNA showing wild-type, heterozygous, and homozygous Hoxa9 knock-out mice (left panel). Neo indicates neomycin-resistance gene. PCR for Mll-AF9 is shown on the right panel with the genotypes as indicated. TSH indicates β subunit of thyroidstimulating hormone used as an internal control for the PCR reaction.
Myeloid colony-forming assays on bone marrow cells from 8-week-old mice. (A) A bar graph depicting total colony numbers formed in 3 generations of platings by cells from Mll (n = 4), Mll/Hox (n = 6), wild-type (WT) (n = 8), and Hoxa9–/– (n = 4) mice. Data represent means and standard errors. (B) Distribution of type I, type II, and type III colonies. Data represent means and standard errors of percentages of each colony type formed in each of the 3 generations of platings.
Myeloid colony-forming assays on bone marrow cells from 8-week-old mice. (A) A bar graph depicting total colony numbers formed in 3 generations of platings by cells from Mll (n = 4), Mll/Hox (n = 6), wild-type (WT) (n = 8), and Hoxa9–/– (n = 4) mice. Data represent means and standard errors. (B) Distribution of type I, type II, and type III colonies. Data represent means and standard errors of percentages of each colony type formed in each of the 3 generations of platings.
When cultured under myeloid growth conditions, murine bone marrow cells expressing the Mll-AF9 gene form 3 types of morphologically distinct colonies.15 Type I colonies are compact, type II colonies have a compact center and a halo of loose cells, and type III colonies have dispersed cells but no center. Type I colonies are composed mostly of immature myeloid precursors, while the cells in type II and type III colonies are progressively more differentiated. Compared with wild-type mice, bone marrow cells from both the Mll and Mll/Hox mice resulted in a greater proportion of type I colonies (Figure 2B). Type I colonies are rarely seen after replating cells from the wild-type mice, while no type I colonies are seen in the case of Hoxa9–/– mice in any of the platings. In contrast, type I colonies persisted in the second and third platings of cells from both the Mll and Mll/Hox mice. However, the average proportions of type I colonies in the Mll/Hox mice were smaller compared with those of Mll mice (Figure 2B). Taken together, these data suggest that 8-week-old Mll/Hox mice display an enhanced expansion of myeloid precursors, although at lower levels than the Mll mice.
Mll/Hox mice develop leukemia with the same incidence and latency as the Mll mice
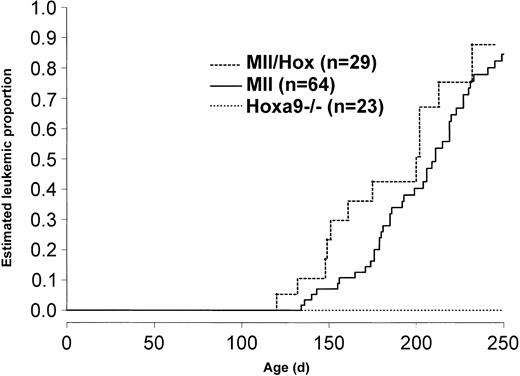
Mice were observed and killed when they showed signs of distress. Postmortem examination revealed that both the Mll/Hox and the Mll mice showed features of leukemia, including splenomegaly, cervical lymphadenopathy, hepatomegaly, and pale femurs. The Mll/Hox and Mll mice developed signs and symptoms and postmortem features of leukemia at the same median time and with the same incidence (Figure 3). For comparison, we also studied age-matched wild-type and Hoxa9–/– mice, and as shown in Figure 3, none of the Hoxa9–/– or wild-type (not shown) mice have developed leukemia.
Incidence of leukemia in Mll and Mll/Hox mice. Kaplan-Meier curves for risk of death from leukemia in Mll and Mll/Hox mice compared with that in Hoxa9–/– mice.
Incidence of leukemia in Mll and Mll/Hox mice. Kaplan-Meier curves for risk of death from leukemia in Mll and Mll/Hox mice compared with that in Hoxa9–/– mice.
Leukemia from the Mll/Hox mice showed a greater block in myeloid differentiation than leukemia from the Mll mice
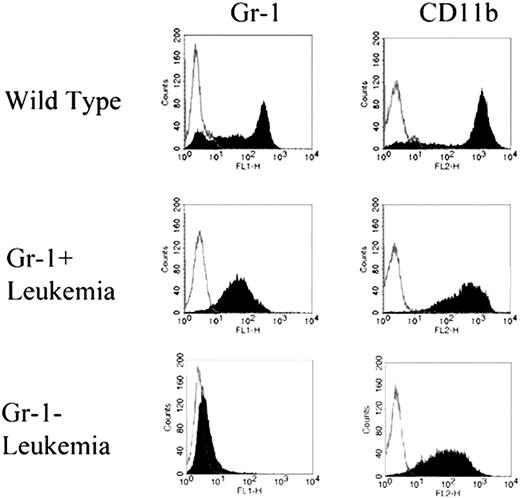
We next sought to determine if the type of leukemia differed between the Mll and the Mll/Hox mice. To analyze the leukemias, we examined the composition of cells from spleen and bone marrow of leukemic mice. Immunophenotyping analysis of bone marrow cells was particularly revealing. As shown in Figure 4, two types of staining patterns were observed for Gr-1 in the bone marrow cells from leukemic mice. One was a broad, low-intensity fluorescence profile, with the average percentages of Gr-1–expressing cells in the leukemic mice being higher than those in the Hoxa9–/– and the wild-type mice. We termed this type of leukemia “Gr-1+,” with most of the cells expressing Gr-1 and also CD11b (Mac-1α). Some of the leukemic mice had only a few Gr-1–positive cells with a much weaker staining, while CD11b was widely expressed. We termed this type of leukemia “Gr-1–,” reflecting a greater block in myeloid differentiation than that seen in the Gr-1+ leukemias. As expected and as shown in Figure 4, both Gr-1+ and Gr-1– leukemias differed significantly from the wild-type marrow in their Gr-1 and CD11b staining patterns. Both the Gr-1+ and Gr-1– leukemias displayed a monocytic component, with more than 20% of the leukocytes in peripheral blood being monocytes. On the other hand, only the Gr-1+ leukemias displayed a neutrophilic component characterized by peripheral neutrophilia.
FACS staining profiles of Gr-1+ and Gr-1–. Immunophenotyping using anti–Gr-1 and anti-CD11b antibodies on bone marrow cells from wild-type and leukemic mice. Based on the Gr-1 staining patterns, leukemias were classified as Gr-1+ or Gr-1–. Both Gr-1+ and Gr-1–leukemias displayed identical staining profiles for CD11b.
FACS staining profiles of Gr-1+ and Gr-1–. Immunophenotyping using anti–Gr-1 and anti-CD11b antibodies on bone marrow cells from wild-type and leukemic mice. Based on the Gr-1 staining patterns, leukemias were classified as Gr-1+ or Gr-1–. Both Gr-1+ and Gr-1–leukemias displayed identical staining profiles for CD11b.
Gr-1+ and Gr-1– leukemias were seen in both the Mll and the Mll/Hox mice. However, a higher proportion of Mll/Hox mice (6 of 10) developed the Gr-1– type of leukemia compared with Mll mice (4 of 28) (P = .004, χ2 test). More detailed results on Gr-1+ and Gr-1– leukemia are shown in Table 1.
Bone marrow composition of leukemic (MII and MII/Box) and nonleukemic (wild-type and HoxA9-/-) mice
Mouse type/cell type . | MII . | MII/Hox . | Wild-type . | Hoxa9-/- . |
|---|---|---|---|---|
| Flow cytometry | ||||
| % Gr-1 | 49 ± 6 | 66 ± 6 | ||
| Gr-1+ | 72 ± 3*† | 83 ± 9*‡ | — | — |
| Gr-1- | 20 ± 3*‡ | 31 ± 4*§ | — | — |
| % CD11b | 85 ± 3* | 96 ± 1* | 60 ± 5 | 77 ± 5 |
| % B220 | 9 ± 2* | 11 ± 5* | 23 ± 2 | 28 ± 4 |
| Morphology | ||||
| % blasts | 17 ± 2* | 14 ± 3* | 2 ± 0.4 | 3 ± 0.6 |
| % RBC precursors | 12 ± 1* | 12 ± 4* | 39 ± 3 | 42 ± 5 |
Mouse type/cell type . | MII . | MII/Hox . | Wild-type . | Hoxa9-/- . |
|---|---|---|---|---|
| Flow cytometry | ||||
| % Gr-1 | 49 ± 6 | 66 ± 6 | ||
| Gr-1+ | 72 ± 3*† | 83 ± 9*‡ | — | — |
| Gr-1- | 20 ± 3*‡ | 31 ± 4*§ | — | — |
| % CD11b | 85 ± 3* | 96 ± 1* | 60 ± 5 | 77 ± 5 |
| % B220 | 9 ± 2* | 11 ± 5* | 23 ± 2 | 28 ± 4 |
| Morphology | ||||
| % blasts | 17 ± 2* | 14 ± 3* | 2 ± 0.4 | 3 ± 0.6 |
| % RBC precursors | 12 ± 1* | 12 ± 4* | 39 ± 3 | 42 ± 5 |
Data represent mean ± standard error. MII, n = 28; MII/Hox, n = 10; wild-type, n = 17; and Hoxa9-/-, n = 6.
Statistically significant difference between leukemic and nonleukemic mice (P < .05).
n = 24.
n = 4.
n = 6.
Additional results of phenotyping shown in Table 1 demonstrate expected differences between wild-type and leukemic mice but no significant differences between the Gr-1+ and Gr-1– leukemias using other markers. We also evaluated bone marrow smears of the mice in our study to compare the morphologic composition of the bone marrow cells. We defined myeloblasts morphologically by their large size and high nucleus-cytoplasm ratio. Using these criteria, the average percentages of blasts in the leukemic mice were higher than those in the wild-type and Hoxa9–/– mice (Table 1; P < .001). Morphologically, the myeloblasts in either the Gr-1+ or Gr-1– leukemias or the Mll or Mll/Hox mice could not be differentiated. The leukemic mice also displayed significant infiltration of their spleens with the cellular compositions of spleens reflecting those of the bone marrow (data not shown).
Hoxgene dysregulation in Mll preleukemic and leukemic mice
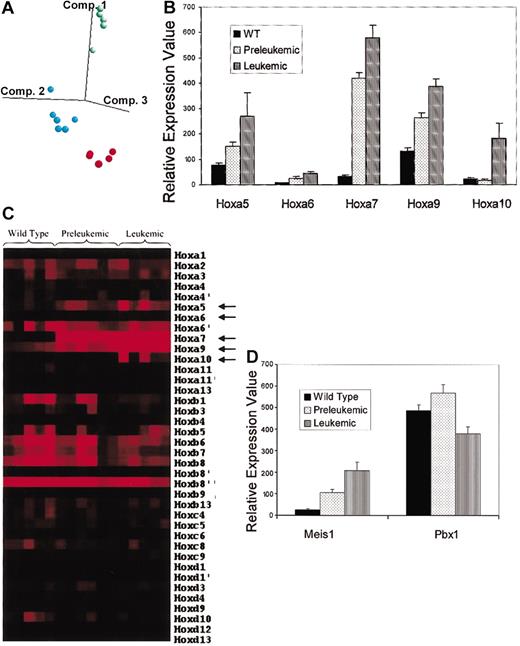
Our results demonstrating that Hoxa9 influenced the phenotype but not the incidence of Mll-AF9 leukemia encouraged us to look further at other Hox genes and non-Hox genes that may be important in Mll fusion gene leukemias. Also, we have previously published cellular data suggesting that a preleukemic stage precedes the leukemic stage in the development of overt leukemia in mice expressing Mll-AF9 as a germ-line knock-in mutation.15 In this study we sought to determine whether gene expression profiling could provide additional support for this hypothesis. Using Affymetrix oligonucleotide microarrays, we analyzed bone marrow RNA from leukemic Mll mice, 8-week-old preleukemic Mll mice, and wild-type mice. To select genes that were differentially expressed consistently among the 3 groups, we applied an analysis of variance (ANOVA) (P < .005) to the data from about 12 000 genes on the murine array. This filtered set contained about 500 genes (Table S1). Hierarchic clustering analysis using these genes correctly grouped the mice into their respective classes (data not shown). In a principle components analysis carried out with these 500 genes, the samples were again segregated according to their class label (Figure 5A). In this study we focused on the Hox family of genes to evaluate the role of Hox dysregulation in leukemogenesis. Included in the list of 500 differentially expressed genes were 5 members of the Hox-a cluster: Hoxa5, Hoxa6, Hoxa7, Hoxa9, and Hoxa10. As shown in Figure 5B, the expression levels of Hoxa5, Hoxa6, and Hoxa9 all increased 2-fold to 3-fold (average) in preleukemic Mll mice compared with those in the wild-type mice and increased further in the leukemic Mll mice. Expression levels of Hoxa7, on the other, hand increased an average 12-fold in the preleukemic mice compared with wild-type mice and increased further in the leukemic mice. The levels of Hoxa10 did not differ among the wild-type and preleukemic mice but were an average 7-fold higher in the leukemic mice. Although there were minor differences between mice within each group in the degree of overexpression for a given gene, they followed the same overall trend.
Microarray analysis of bone marrow cells from Mll and wild-type mice. (A) Principal components analysis using the genes identified in the ANOVA. Each of the colored dots represents a mouse sample: red indicates WT; blue, preleukemic; and green, leukemic. The first 3 eigenvectors are used to map the samples in 3-dimensional space. (B) Relative expression values of Hoxa5, Hoxa6, Hoxa7, Hoxa9, and Hoxa10 in wild-type (WT), preleukemic, and leukemic mice. Data represent means and standard errors. (C) Hot-map of Hox gene expression. Each column represents a mouse sample while each row represents a Hox gene transcript. Increasing expression levels are represented by progressively brighter shades of red while darker shades represent relatively lower expression. Arrows point to genes showing significant changes in expression between wild-type, preleukemic, and leukemic mice. (D) Relative expression values of the TALE protein genes Meis1 and Pbx1 in wild-type (WT), preleukemic, and leukemic mice. Data represent means and standard errors.
Microarray analysis of bone marrow cells from Mll and wild-type mice. (A) Principal components analysis using the genes identified in the ANOVA. Each of the colored dots represents a mouse sample: red indicates WT; blue, preleukemic; and green, leukemic. The first 3 eigenvectors are used to map the samples in 3-dimensional space. (B) Relative expression values of Hoxa5, Hoxa6, Hoxa7, Hoxa9, and Hoxa10 in wild-type (WT), preleukemic, and leukemic mice. Data represent means and standard errors. (C) Hot-map of Hox gene expression. Each column represents a mouse sample while each row represents a Hox gene transcript. Increasing expression levels are represented by progressively brighter shades of red while darker shades represent relatively lower expression. Arrows point to genes showing significant changes in expression between wild-type, preleukemic, and leukemic mice. (D) Relative expression values of the TALE protein genes Meis1 and Pbx1 in wild-type (WT), preleukemic, and leukemic mice. Data represent means and standard errors.
Overall, we evaluated expression levels of 32 Hox genes represented on the murine microarray (Affymetrix U74A). As shown in Figure 5C, none of the Hox genes other than Hoxa5, Hoxa6, Hoxa7, Hoxa9, and Hoxa10 demonstrated significant changes in the levels of their expression between the 3 groups.
TALE (three–amino acid loop extension) proteins are known to collaborate with HOX proteins in DNA binding and have also been implicated in causing leukemia.12 We compared expression levels of TALE protein genes in wild-type, preleukemic Mll, and leukemic Mll mice. As shown in Figure 5D, expression levels of Meis1 increased an average 4-fold in the bone marrow cells of preleukemic mice and further increased in the leukemic mice when compared with those of wild-type mice. On the other hand, such an increase was not seen in expression levels of Pbx1, and these levels in fact decreased by 33% in the leukemic mice compared with the preleukemic mice.
Collectively, these results suggest a direct effect of Mll-AF9 on certain genes in the Hox-a cluster, even before the development of frank leukemia. The parallel changes in expression levels of Meis1 and genes of the Hox-a cluster suggest that they share regulatory elements.
Discussion
Our results demonstrate that Mll-AF9 results in the expansion of myeloid precursors, Hox gene dysregulation, and the eventual development of myeloid leukemia. The colony-forming assay demonstrated that preleukemic Mll/Hox mice displayed an increased self-renewal capacity of myeloid progenitors, similar to Mll mice. However, this increase was at lower levels than that found in Mll mice. The relatively smaller number of colonies noted in the Mll/Hox mice suggests a reduced clonogenic capacity of myeloid precursors resulting from the absence of Hoxa9.
Most Mll mice developed Gr-1+ leukemia, while most Mll/Hox mice developed Gr-1– leukemia. The differences between the Mll and Mll/Hox mice suggest that specific Hox genes, particularly Hoxa9, are important in determining the leukemic phenotype. Previous studies have shown that in leukemia generated by enforced coexpression of a Hox gene (Hoxb3 or Hoxa9) and of the Hox cofactor Meis1, the phenotype of the leukemia is determined by the specific Hox gene.17 In their study, Thorsteinsdottir et al reported a relatively immature myeloid leukemia with a block in neutrophil maturation associated with enforced expression of Hoxa9. In contrast, enforced expression of Hoxb3 was associated with a relatively mature leukemia. A direct comparison of these results and ours is not possible because of differences in both the leukemia models and the methods of analysis. However, both suggest that Hox gene expression is important in determination of the phenotype of the leukemia. Overall, the results from the 2 models suggest that Hoxa9 does play an important role in the definition of leukemic phenotype but not in the incidence of leukemia.
Our gene expression profiling data show that, in Mll mice, 5 Hox genes were overexpressed: Hoxa5, Hoxa6, Hoxa7, Hoxa9, and Hoxa10. This suggests that Mll-AF9 induces dysregulation of several Hox genes and that multiple genes play a role in the myeloid expansion and leukemia development seen in the Mll mice. Interestingly, all the Hox genes affected by Mll-AF9 belong to the Hox-a cluster that is highly conserved in evolution.18 There is evidence indicating that interactions between neighboring Hox genes in the same cluster may be important in defining cell and tissue identity.19,20 Thus, the difference in leukemic phenotypes noted between the Mll and Mll/Hox mice might be due to interactions of different Hox proteins. There may also be functional redundancy within members of the same Hox cluster. Consequently, in the absence of Hoxa9, overexpression of any of the other genes in the Hox-a cluster may be sufficient for the development of leukemia. Alternatively, studies of Hox genes in mice suggest that paralogous Hox genes may overlap in their functions.21,22 Thus, Hoxb9 or Hoxd9 might substitute for Hoxa9 in the Mll/Hox mice, resulting in leukemia. However, this is unlikely because no changes in expression of any of the Hox-b or Hox-d genes were observed in the Mll mice.
We have also shown in this study that the preleukemic and leukemic Mll mice show differences in their bone marrow gene expression profiles. The increase in expression of Hoxa5, Hoxa6, Hoxa7, and Hoxa9 occurred in the preleukemic mice and can be attributed to the presence of the Mll-AF9 gene. However, the increase in expression of Hoxa10 did not occur until the mice developed leukemia. Additional events that cause further dysregulation of Hox gene expression such as overexpression of Hoxa10 result in the eventual development of leukemia. A caveat for interpreting these results is that the apparent overexpression of certain genes in the leukemic mice compared with the preleukemic mice might be caused by a selective expansion of cells that express these genes.
The analogous changes in expression of a few Hox genes and the Hox cofactor Meis1 in the Mll mice suggest that these genes share regulatory elements and that this coactivation might be important in the development of leukemia. Previous studies have shown that the leukemia caused by overexpression of Hoxa9 is accelerated with the coexpression of Meis1.8 Similarly, coactivation of Meis1 and Hoxa7 or Hoxa9 by proviral integration leads to myeloid leukemia in BXH-2 mice.12
The Mll/Hox mice showed preleukemic myeloid expansion and eventually developed myeloid leukemia. The incidence and time course of the leukemia was identical in the Mll and Mll/Hox mice. These findings directly demonstrate that Hoxa9 is not required for the development of leukemia in a germ-line murine model of Mll-AF9 leukemia. A recent report by Ayton and Cleary concluded that Hoxa9 was required for induction of leukemia in a murine retroviral transduction/transplantation model of MLL-ENL.23 This contrasting conclusion may be due to differences in the method of introduction of the fusion gene or functional differences resulting from the fusion gene itself. In their study, Ayton and Cleary used retroviral transduction to introduce the MLL-ENL fusion gene into primary myeloid progenitor cells from wild-type and Hoxa9–/– mice. Similar to our observations with Mll-AF9, the Hoxa9–/– cells yielded fewer type I colonies than the wild-type cells when transduced with MLL-ENL. When transplanted into recipients, however, the MLL-ENL–transduced Hoxa9–/– cells did not result in leukemia. A control experiment in which Hoxa9 was reintroduced into Hoxa9–/– cells resulted in partial rescue of the leukemic phenotype with about 50% leukemia incidence. The explanation for this latter observation is not clear. Our model differs from the MLL-ENL model in that all hematopoietic tissues contain the Mll-AF9 fusion gene whereas the retroviral MLL-ENL model contains the fusion gene only in a small subset of hematopoietic cells. Given the broad expression of Mll-AF9 in hematopoietic cells of knock-in mice, it is possible that the transformed cell type may differ between the 2 model systems. Complete comparative analysis of the cellular phenotype in the 2 models should be helpful in this regard. The report by Forster et al demonstrating that mice carrying the Mll-ENL fusion gene as a conditional knock-in mutation develop myeloid leukemia with a phenotype similar to that found in the Mll-AF9 knock-in mice is noteworthy.24
There are other potentially important differences between the 2 models. One is that the level of expression of the introduced genes is likely to differ. In the knock-in model the fusion gene is under the control of the endogenous promoter, whereas in the retroviral model it is controlled by exogenous promoters. Also, haploinsufficiency of MLL that is present in human leukemias is also present in the knock-in model but not in the retroviral model. Finally, differences between the biologic effects of MLL-ENL and Mll-AF9 on Hoxa9 cannot be ruled out from the current models. Additional comparative studies will be necessary to further evaluate this possibility.
The overexpression of Hoxa9, Hoxa10, and Meis1 in the leukemic Mll mice is similar to the pattern of HOX gene deregulation observed in human MLL leukemias.3 These results suggest that the murine model is suitable for further studies of this disease process. These results are also in agreement with those reported by Ayton and Cleary with one possible exception: Viral transfection of murine myeloid progenitors with MLL-ENL did not result in an increase in expression of Hoxa6, whereas we saw a 2-fold increase.23 There are several possible explanations for this difference, including different Hox profiles of cells that are transformed by MLL-ENL and Mll-AF9. Differential expression of Hox genes in hematopoietic cells is a function of the stage of differentiation.25 As stated above, it is possible that the transformed cell might be different in the knock-in Mll-AF9 and retroviral MLL-ENL models. Additional studies will be needed to determine whether there are significant differences between the Hox profiles of the cells transformed by these 2 fusion genes.
Our results suggest a paradigm that may be applicable not only to leukemias with MLL-AF9 rearrangements but to any leukemia with dysregulated HOX gene expression. We propose that the nature of the transformed cell, including the phenotype and malignant potential, is defined not by the expression of a single HOX gene but by the expression pattern of several HOX genes— the “HOX code.” The “HOX code” has been extensively studied in vertebrate development and describes the process by which clustered HOX genes function in a colinear manner resulting in precise temporal sequencing to produce embryonic axis patterning.26 The colinear ordering of the HOX-A cluster has been highly conserved throughout long periods of evolution in mammals.
As a practical matter, if more than one HOX gene is critical for leukemogenesis, targeting of these genes for therapy will probably require inactivation of more than one HOX gene.
Prepublished online as Blood First Edition Paper, November 13, 2003; DOI 10.1182/blood-2003-07-2582.
Supported in part by a grant to J.H.K. from the National Institutes of Health (R01 87053). A.R.K. is the William Kennedy research fellow of the National Childhood Cancer Foundation.
The online version of the article contains a data supplement.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Terence H. Rabbitts, Dr H. Jeffrey Lawrence, and Dr Mario R. Capecchi for providing us with the transgenic mice; Suzanne Grindle for analysis of the microarray data; Robin Bliss for statistical analysis; and the staff of the Informatics Core and the Flow Cytometry Core facilities at the University of Minnesota Cancer Center.
Supplemental data
The data obtained from scanned images of the microarrays were uploaded onto an Oracle-based data server using GeneData Expressionist Suite (GeneData, South San Francisco, CA). Individual chips were assessed for quality of hybridization and other errors. The expression values were then transformed to logarithmic scale (base 2), and a linear scaling factor was applied to data from each chip. The normalized data from the chips were then compared in an ANOVA test (P < .005) to select genes that were significantly different between wild-type, preleukemic, and leukemic mice. When the genes selected by the ANOVA were used in a hierarchical clustering analysis, the samples were correctly grouped into their respective classes. A list of the genes selected by the ANOVA, along with their expression values, is included. The raw data and the ANOVA-selected list were mined for Hox genes since we were particularly interested in the changes in expression levels of this group of genes. The list selected by the ANOVA included 5 of the Hox genes: Hoxa5, Hoxa6, Hoxa7, Hoxa9, and Hoxa10. The expression values of these Hox genes in each group of mice are represented as means and standard errors in Fig. 5B. The expression values of all the Hox genes represented on the Affymetrix U74Av2 array are represented in Fig. 5C as a “hotmap” created using Treeview (http://rana.lbl.gov/EisenSoftware.htm).

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal