Abstract
Kaposi sarcoma (KS) is the most common AIDS-associated malignancy and is characterized by angiogenesis and the presence of spindle cells. Kaposi sarcoma-associated herpesvirus (KSHV) is consistently associated with all clinical forms of KS, and in vitro infection of dermal microvascular endothelial cells (DMVECs) with KSHV recapitulates many of the features of KS, including transformation, spindle cell proliferation, and angiogenesis. To study the molecular mechanisms of KSHV pathogenesis, we compared the protein expression profiles of KSHV-infected and uninfected DMVECs. This comparison revealed that heme oxygenase-1 (HO-1), the inducible enzyme responsible for the rate-limiting step in heme catabolism, was up-regulated in infected endothelial cells. Recent evidence suggests that the products of heme catabolism have important roles in endothelial cell biology, including apoptosis and angiogenesis. Here we show that HO-1 mRNA and protein are up-regulated in KSHV-infected cultures. Comparison of oral and cutaneous AIDS-KS tissues with normal tissues revealed that HO-1 mRNA and protein were also up-regulated in vivo. Increased HO-1 enzymatic activity in vitro enhanced proliferation of KSHV-infected DMVECs in the presence of free heme. Treatment with the HO-1 inhibitor chromium mesoporphyrin IX abolished heme-induced proliferation. These data suggest that HO-1 is a potential therapeutic target for KS that warrants further study. (Blood. 2004;103: 3465-3473)
Introduction
Kaposi sarcoma-associated herpesvirus (KSHV; also human herpesvirus-8) is implicated in all clinical forms of KS1,2 as well as the lymphoproliferative disorders primary effusion lymphoma (PEL)3,4 and multicentric Castleman disease (MCD).5 Endothelial cells harbor the KSHV genome in vivo,1,6,7 are permissive for virus infection in vitro,8-11 and are thought to be the precursors of spindle cells.12-15 We and others have shown that in vitro infection of human dermal microvascular endothelial cells (DMVECs) with KSHV induces a spindle cell morphology and characteristics of a transformed phenotype, including loss of contact inhibition and growth in soft agar.8-11 Because explanted KS cells fail to maintain the KSHV genome following serial passage,16,17 in vitro infection of DMVECs has proved to be an invaluable tool for the study of KSHV pathogenic mechanisms.18,19
KSHV, like other herpesviruses, has a substantial coding capacity that includes viral genes unique to this human pathogen, genes that share homology with other members of the herpesviridae, as well as genes that appear to have originated in the host genome.20,21 Of note, at least 3 of these gene products, vFLIP (viral Fas-associating protein with death domain-like interleukin-1β [IL-1β]-converting enzyme [FLICE]-inhibitory protein), viral cyclin (vCYC), and latency-associated nuclear antigen (LANA), are believed to comprise the viral latency expression program and are consistently expressed in all virally infected cells in KS, PEL, and MCD.22-24 The gene product of open reading frame 71 (ORF 71), vFLIP, is thought to play an important role in prevention of apoptotic cell death.25 The activity of a homolog of cellular cyclin D, vCYC (ORF 72),26 together with the modulation of cellular transcription wrought by LANA (ORF 73)27 is thought to be involved in KSHV-induced host cell transformation. Additionally, LANA is indispensable for maintenance of the viral genome.28 Other gene products, such as viral G-protein-coupled receptor (vGCR)29,30 and vIL-6,31 are classified as lytic gene products and are expressed in only a minority of cells in vivo.32-34 Although reactivation of KSHV from the latent state and subsequent completion of the viral lytic expression cascade is incompatible with survival of a host cell, lytic gene products are thought to have paracrine influences on both latently infected and uninfected cells and are thus considered vital for lesion development.35
We have used our previously described in vitro KS model to dissect viral mechanisms of pathogenesis.18,19 The salient features of our model include recapitulation of KS cell physiology, including spindle cell formation, loss of contact inhibition, and anchorage-dependent growth restriction; long-term propagation of predominantly latently infected cells; and the ability to generate ageand passage-matched KSHV-infected and uninfected cultures.11 This system has been amenable to gene expression profiling by cDNA microarrays and has provided valuable information in this regard.18,19 In the present study, we employed a proteomics expression profiling approach to study alterations in host cell gene expression following in vitro infection of DMVECs with PEL-derived KSHV. Out of approximately 850 gene products screened by Western blotting, 52 genes exhibited significant up- or down-regulation in infected cells compared with uninfected controls. These altered genes have been implicated in diverse cellular processes, including apoptosis, cell division, metabolism, morphology, transcription, and tumorigenesis. One metabolic gene that showed significant up-regulation in KSHV-infected cells was heme oxygenase-1 (HO-1). Given recent evidence implicating HO-1 enzymatic activity in antiapoptotic responses,36,37 cell proliferation,38,39 and angiogenesis,38-41 we reasoned that up-regulation of this gene by KSHV infection may have important consequences for endothelial cell biology and KS pathophysiology.
HOs are responsible for the oxidative cleavage of the heme ring, the rate-limiting step in heme catabolism.42 Enzymatic degradation of heme releases carbon monoxide (CO), free iron, and biliverdin, which is subsequently converted to bilirubin by biliverdin reductase.42 To date, 3 isoforms have been identified: the stress-inducible HO-1 (HSP32) and the constitutive HO-2 and HO-3. These isozymes, products of distinct genes, differ in specific activity, inducibility, and tissue distribution. HO-2, an essentially uninducible isozyme, is present in high concentrations in tissues such as brain and testis.43 HO-3, also uninducible, is distinguished by a lesser degree of homology to the other isoforms and a markedly reduced specific activity; this isozyme is thought to function in heme transport within cells rather than heme catabolism.44,45 HO-1 is ubiquitously distributed in mammalian tissues and is strongly and rapidly up-regulated by noxious stimuli leading to oxidative stress such as transitional metals, glutathione-depleting agents, UV light, and heat shock.43 Heme, the natural substrate of HO-1, is itself a potent inducer of HO-1.43
In the present study we show that HO-1 mRNA and protein are up-regulated by KSHV infection of DMVECs in vitro as well as in KS biopsy tissue. Up-regulation of HO-1 enzymatic activity in vitro conferred a proliferative advantage to infected cells, suggesting that this enzyme may have an important role in KS pathophysiology.
Materials and methods
Reagents
Ethanolamine, pefabloc SC, leupeptin, glucose-6-dehydrogenase (type XV from baker's yeast), glucose-6-phosphate, and NADP+ (nicotinamide adenine dinucleotide) were from Sigma (St Louis, MO); pooled rat liver cytosol was from Cedra Corporation (Austin, TX); human skin total RNA was from Stratagene (La Jolla, CA); and Cu(II) mesoporphyrin IX (CuMP), Cr(III) mesoporphyrin IX (CrMP), and heme were from Porphyrin Products (Logan, UT). Stock solutions of methemalbumin (1.5 mM heme and 0.15 mM bovine serum albumin [BSA]), CuMP, and CrMP (1.0 mM) were prepared as described previously.46 Briefly, heme, CuMP, or CrMP was dissolved in 0.5 mL 10% (wt/vol) ethanolamine in deionized water. BSA was added to the heme solution in 2 mL deionized water. The volume was raised to 7 mL and adjusted to pH 7.4 with 1 N HCl and rapid stirring. The final volume was adjusted to 10 mL with deionized water. Heme and mesoporphyrin stock solutions were prepared in the dark and stored at -20°C for up to one month.
Derivation of KSHV-infected DMVECs
In vitro infection of DMVECs with KSHV was previously described in detail.11 Briefly, primary DMVECs (BioWhittaker, Walkersville, MD) were immortalized by retroviral-mediated expression of human papillomavirus (HPV) type 16 E6 and E7 genes and were subsequently infected with KSHV derived from tetradecanoyl phorbol acetate-treated body cavity-based lymphoma-1 (BCBL-1) cell culture supernatants. DMVECs were cultured in endothelial-serum-free medium (SFM) (GIBCO BRL, Gaithersburg, MD) supplemented with 10% human male type AB serum (HS; Sigma), penicillin (100 U/mL), streptomycin (100 μg/mL), glutamine (2 mM), endothelial cell growth supplement (50 μg/mL; Becton Dickinson, Bedford, MA), and G418 (200 μg/mL; GIBCO BRL). The BCBL-1 cell line was obtained through the AIDS Research and Reference Reagent Program (Division of AIDS, National Institute of Allergy and Infectious Disease, National Institutes of Health; contributed by Michael McGrath and Don Ganem) and was cultured in RPMI supplemented with 10% fetal calf serum, penicillin (100 U/mL), streptomycin (100 μg/mL), glutamine (2 mM), and 2-mercaptoethanol (5 × 10-5 M). KSHV infection of DMVECs was verified by polymerase chain reaction (PCR) amplification of the KS330 BamHI fragment of the ORF 26 gene, reverse transcription (RT)-PCR amplification of the spliced mRNA from the ORF 29 gene, and immunostaining for KSHV proteins. DMVECs were used for experimentation when more than 90% of cells expressed ORF 73 (LANA). Typically, 2% of infected cells in such LANA-positive cultures expressed the early lytic protein ORF 59 and less than 1% of ORF 59-positive cells expressed the late lytic glycoprotein ORF K8.1. Viral antigen expression was detected by immunofluorescent staining as previously described using antibodies generated and generously provided by Bala Chandran (The University of Kansas Medical Center, Kansas City, KS). For infection of primary DMVECs, the recombinant enhanced green fluorescent protein (EGFP)-expressing clone rKSHV.15247 was used.
BD PowerBlot
The western array screening service offered by BD Transduction Laboratories (Franklin Lakes, NJ) measures changes in protein expression by simultaneously probing control and experimental cell protein extracts with a panel of more than 850 monoclonal antibodies. KSHV-infected and uninfected DMVEC cultures were harvested according to BD Transduction Laboratories' specifications. Monolayers in T75 flasks were washed with phosphate-buffered saline (PBS) and lysed in 1 mL boiling lysis solution per flask (10 mM Tris [tris(hydroxymethyl)aminomethane], pH 7.4; 1 mM sodium ortho-vanadate; and 1% sodium dodecyl sulfate [SDS]). Lysates were scraped and transferred to 50 mL conical polypropylene tubes and subsequently heated in a microwave for 10 seconds uncapped. Cellular DNA was then sheared by passing lysates 10 times through a 25-gauge blunt needle. A small volume was removed and diluted 1:10 to reduce the SDS concentration to 0.1% and protein concentration was determined using the Bradford reagent according to manufacturer's instructions (Bio-Rad, Hercules, CA) with BSA as a standard. Samples were then shipped to BD Transduction Laboratories on dry ice where PowerBlot screening was conducted. Gradient SDS-polyacrylamide gel electrophoresis (SDS-PAGE; 4%-15%, 0.5-mm thick [Bio-Rad Criterion IPG well comb]) gels were loaded with 200 μg of protein in one continuous well across the entire width of the gel. This yields the equivalent of 10 μg protein per lane on a standard 10-well mini-gel. Gels were run 1.5 hours at 150 V. The gels were transferred to Immobilon-P membrane (Millipore, Bedford, MA) for 2 hours at 200 milliamps using a wet electrophoretic transfer TE Series apparatus (Hoefer Scientific Instruments, San Francisco, CA). Following transfer, membranes were air dried and subsequently rewet in methanol and blocked for 1 hour in blocking buffer (LI-COR, Lincoln, NE). Blocked membranes were then clamped into Western blotting manifolds that isolate 40 channels across each membrane. Each channel was then loaded with a complex antibody cocktail and incubated for 1 hour at 37°C. Blots were removed from manifolds, washed, and hybridized for 30 minutes at 37°C with secondary goat antimouse conjugated to Alexa680 fluorescent dye (Molecular Probes, Eugene, OR). Membranes were washed, dried, and scanned using the Odyssey Infrared Imaging System (LI-COR). Samples were run in triplicate and fold change was reported following analysis of blots using a 3 × 3 comparison method.
Western blotting
Uninfected and KSHV-infected cells were harvested by trypsinization and pelleted by centrifugation at 16 000 g for 30 minutes at 4°C and then resuspended in 100 μLof 1 × PBS. The resuspended pellet was frozen and then thawed 3 times to break open the cells. The total cell extract was then centrifuged at 16 000 g to spin out excess cellular debris. Total protein was measured with a Bradford assay kit (Bio-Rad). Samples for immunoblotting were prepared by taking 20 μg of total protein and combining with 3 × reducing sample buffer to make 30 μL total volume. The samples were boiled for 3 minutes and then centrifuged at 15 000 rpm for 1 minute to pellet insoluble material. Each sample (15 μL) was loaded on 2 identical 12% polyacrylamide gels and resolved by discontinuous electrophoresis (SDS-PAGE) as previously described.48 After electrophoresis, the gels were transferred to nitrocellulose and then subjected to immunoblot analysis using anti-HO-1 (Transduction Laboratories; catalog no. 610712) and anti-stress-activated protein kinase/c-Jun N-terminal kinase (anti-SAPK/JNK; Cell Signaling Technologies, Beverly, MA) antibodies as per the manufacturer's recommendations. The antigen-antibody complex was then incubated with antimouse immunoglobulin (Ig) horseradish peroxidase and antirabbit Ig horseradish peroxidase (Amersham Life Sciences, Arlington Heights, IL). The blot was developed by incubation with chemiluminescent substrate (Pierce, Rockford, IL) and exposed to Kodak BioMAX MR film.
Immunofluorescent staining
For detection of HO-1 protein, E6/E7-transformed DMVEC monolayers were rinsed 2 times with PBS containing 1% normal goat serum and 0.02% sodium azide (staining buffer), fixed in 2% paraformaldehyde, permeabilized with 0.05% triton, and stained with an anti-HO-1 monoclonal antibody (clone 23; BD Transduction Laboratories) and an anti-LANA rabbit polyclonal antibody followed by a goat antimouse fluorescein isothiocyanate (FITC)-labeled (Biosource International, Camarillo, CA) and goat antirabbit Alexa-labeled (Molecular Probes) secondary antibodies. For detection of HO-1 in nontransformed cells, primary DMVECs were infected at a low multiplicity of infection with the EGFP-expressing KSHV clone. Monolayers were fixed, permeabilized, and stained for HO-1 as for transformed cells. All antibodies were used at a 1:100 dilution in staining buffer and incubated with cell monolayers for 60 minutes at 37°C. Primary antibodies were omitted from duplicate monolayers to control for nonspecific binding of secondary antibody. Stained cells were mounted and examined on a Zeiss fluorescent microscope (Thornwood, NY).
Quantitative RT-PCR
RNA was isolated using an RNeasy Total RNA kit (QIAGEN, Valencia, CA). RNA samples were treated with RNase-free DNase I to remove any residual genomic DNA contamination (Ambion, Austin, TX). Quantification of RNA was performed by a 2-step method. First, cDNA was synthesized using superscript II (Invitrogen, Carlsbad, CA). Synthesized cDNA was diluted in H2O to a final concentration of 1 ng per reaction in TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA). Quantitative RT-PCR was performed on an ABI-PRISM 7700 Sequence Detection System (Applied Biosystems) under standard reaction conditions. Relative quantitation of gene expression, as outlined in the Applied Biosystems User Bulletin 2, was conducted to compare uninfected and KSHV-infected samples. Following primer efficiency validation on RNA from KSHV-infected DMVECs, the comparative cycle threshold (CT) method was performed as outlined in user bulletin 2. The comparative CT method compares differences in the threshold cycles between samples after normalization to an endogenous control, in this case glyceraldehyde phosphate dehydrogenase (GAPDH). CT values vary with target RNA concentration such that a higher target concentration yields an earlier threshold signal over background. Differences between samples during the exponential phase of PCR amplification were calculated by the following equation: ΔΔCT = (CT (HO-1) - CT (GAPDH)) infected - (CT (HO-1) - CT (GAPDH)) uninfected and converted to n-fold change units by the equation 2-ΔΔCT. The following primers were generated using primer express v1.1 (Advanced Biotechnologies, Columbia, MD): HO-1 forward, 5′-GCCCTTCAGCATCCTCAGTTC-3′, and reverse, 5′-GGTTTGAGACAGCTGCCACA-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGT-3′, and reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
Immunohistochemistry
Paraffin-embedded biopsy tissue from oral AIDS-KS lesions were sectioned and mounted on charged slides. The slides were deparaffinized, treated in citrate buffer (30 minutes, steamed), transferred to Tris-buffered saline (TBS)/Tween buffer, and placed on an automatic staining machine programmed for staining using the Vectastain Elite ABC kit with diaminobenzidine as the substrate (Vector Laboratories, Burlingame, CA). Polyclonal anti-HO-1 (Affinity Bioreagents, Golden, CO) was used at 1:40. Slides were counterstained with hematoxylin/eosin.
HO enzymatic activity
Crude endothelial cell protein extracts were prepared as previously described.49 Briefly, following overnight incubation in complete media alone or with 10 μM CuMP or CrMP, monolayers were rinsed with PBS and scraped directly into 300 μL sonication buffer (0.25 M sucrose, 20 mM Tris-HCl, 50 μg/mL Pefabloc SC, 4 μg/mL leupeptin; pH 7.4) sonicated on ice 2 times for 30 seconds and centrifuged for 20 minutes at 18 000g. The protein concentration of the resultant supernatant was determined using Bradford reagent as described above. HO activity was measured by the spectrophotometric determination of bilirubin production as described elsewhere.49 Final reaction concentrations were 1 mg endothelial cell protein extract, 50 μM heme, 2 mg/mL pooled rat liver cytosol, 1 mM MgCl2, 3 units glucose-6-dehydrogenase, 1 mM glucose-6-phosphate, and 2 mM NADP+ in 0.5 mL 0.1 M potassium phosphate buffer (pH 7.4) for 30 minutes at 37°C. The reaction was stopped by the addition of 2 volumes of chloroform. Bilirubin concentration in the chloroform extracts was determined using an Ultrospec 2100 Pro spectrophotometer (Amersham Pharmacia Biotech, Piscataway, NJ) to measure the difference in optical density at 464 and 530 nm and the equation A = c L ϵ assuming an extinction coefficient (ϵ) of 40 mM-1cm-1 for bilirubin in chloroform with a path length (L) of 1.0 cm. HO activity was reported as picomoles of bilirubin produced per milligram endothelial cell protein extract per hour.
Assessment of heme-induced proliferation
The effect of low-dose heme treatment on proliferation of KSHV-infected and uninfected DMVECs was assessed by direct cell counting. Cells were plated in 96-well plates at 1.25 × 104 cells per well and incubated overnight in complete medium. The next morning cells were refed with SFM without serum or ECGS in the presence or absence of CuMP or CrMP and incubated for 24 hours. The next morning (t = 0) cells were refed with SFM containing CuMP or CrMP in the presence or absence of 5 μM heme and incubated for an additional 48 hours (t = 48). Quadruplicate wells were trypsinized and counted by hemocytometer at t = 0 and 48. Trypan blue exclusion showed that cell viability was higher than 95% for all treatment conditions. To ensure that the ability of these cells to proliferate in this experimental setting was not nonspecifically limited, a separate experiment was conducted whereby the proliferative response of serum-starved cells was assessed following a 48-hour incubation in SFM containing 10% human serum in the presence or absence of endothelial cell growth supplement (50 μg/mL), a complex mixture containing known endothelial cell mitogens including vascular endothelial growth factor (VEGF) and basic fibroblast growth factor (bFGF).
Results
Western blot screening of KSHV-infected DMVECs
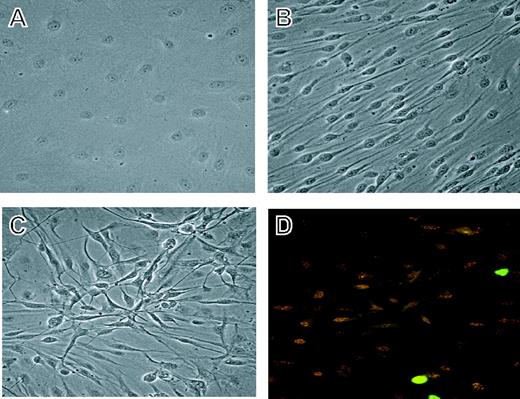
To screen changes in gene expression following experimental infection of DMVECs with KSHV, we raised age- and passage-matched cultures of KSHV-infected and uninfected cells (Figure 1) as previously described.11 Uninfected DMVECs retained the classical cobblestone morphology of normal endothelial cells and remained contact inhibited after serial passage (Figure 1A). KSHV-infected cultures, however, assumed a spindle cell morphology reminiscent of the neoplastic endothelial cell-derived spindle cells in KS lesions (Figure 1B). Cultures were harvested for expression profiling once infected cultures had become more than 90% spindled and ORF 73-positive (Figure 1C-D). Alterations in gene expression resulting from KSHV infection are most likely due to latent infection as staining for the lytic marker ORF 59 revealed less than 2% spontaneous lytic reactivation. We note, however, that a paracrine influence conferred by even sporadic reactivation could also effect gene expression.35 Total protein extracts were prepared according to BD Transduction Laboratories' specifications and analyzed by the PowerBlot method as described in “Materials and methods.”
Derivation of uninfected and KSHV-infected DMVECs. (A) Phase image of uninfected DMVECs showing the classical cobblestone appearance of normal endothelial cells. (B) Phase image of age- and passage-matched KSHV-infected DMVECs showing spindle cell morphology following viral infection. (C) Phase image of KSHV-infected DMVECs demonstrating loss of contact inhibition. Image shows approximately 3 layers of cells in a 3-dimensional focus. (D) Corresponding immunofluorescence microscopic image showing staining for the latent viral protein ORF 73/LANA (red) and the lytic viral protein ORF 59/progressivity factor-8 (green). Cells were used for experimentation at 3 weeks after infection when more than 90% of cells stained positively for ORF 73. Spontaneous lytic reactivation of KSHV, as assessed by ORF 59 expression, was generally restricted to less than 2% of cells. Original magnification, × 20.
Derivation of uninfected and KSHV-infected DMVECs. (A) Phase image of uninfected DMVECs showing the classical cobblestone appearance of normal endothelial cells. (B) Phase image of age- and passage-matched KSHV-infected DMVECs showing spindle cell morphology following viral infection. (C) Phase image of KSHV-infected DMVECs demonstrating loss of contact inhibition. Image shows approximately 3 layers of cells in a 3-dimensional focus. (D) Corresponding immunofluorescence microscopic image showing staining for the latent viral protein ORF 73/LANA (red) and the lytic viral protein ORF 59/progressivity factor-8 (green). Cells were used for experimentation at 3 weeks after infection when more than 90% of cells stained positively for ORF 73. Spontaneous lytic reactivation of KSHV, as assessed by ORF 59 expression, was generally restricted to less than 2% of cells. Original magnification, × 20.
Out of approximately 850 gene products screened by PowerBlot western array screening, 52 genes exhibited significant up- or down-regulation in infected cells compared with uninfected controls. Functional clustering of the data revealed several potentially important alterations in host cell physiology caused by KSHV infection (Table 1). Genes involved in cellular processes, including apoptosis, cell adhesion, morphology, proliferation, DNA damage repair, metabolism, protein sorting, transcription, and tumorigenesis, all showed significant changes in expression levels. We chose to focus further efforts on HO-1, a gene with potentially interesting and important implications in KS pathophysiology.
Genes modulated in DMVECs after KSHV infection
Unigene cluster . | Gene ID . | Symbol . | PowerBlot value . | Name . | Function . | Group . |
|---|---|---|---|---|---|---|
| Hs.82395 | A40036 | Fas | 8.8 | CD 95 | Apoptosis | A |
| Hs.89862 | Q15628 | TRADD | −1.5 | TNFR1-associated death domain protein | Apoptosis | A |
| Hs.381231 | NP_001219 | CASP8 | Absent | Caspase 8 | Apoptosis | A |
| Hs.287797 | P05556 | ITGB1 | 1.8 | Integrin beta 1 | Cell adhesion | CA |
| Hs.216381 | A45973 | Nexilin | −4.2 | Nexilin | Cell adhesion | CA |
| Hs.119537 | A38219 | KHDRBS1 | 3.0 | Sre-associated in mitosis, 68 kDa | Mitosis | CD |
| Hs.3849 | AB045981 | FKBP10 | 2.4 | FK506-binding protein 10, 65 kDa | Cell proliferation | CD |
| Hs.433496 | Q9UN86 | G3BP2 | 2.3 | Ras-GAP | Mitogenesis | CD |
| Hs. 104741 | NP_060962 | TOPK | −6.3 | T-LAK cell-originated protein kinase | MAPKK | CD |
| Hs.317432 | P54687 | BCAT1 | −3.5 | Branched chain aminotransferase 1 | Cell proliferation | CD |
| Hs.22868 | JN0805 | PTPN11 | −2.9 | Protein tyrosine phosphatase, nonreceptor type II | Protein-tyrosine phosphatase | CD |
| Hs.181046 | P51452 | DUSP3 | −2.8 | Dual specificity phosphatase 3 | Regulator of CDKs | CD |
| Hs.211593 | A45416 | PRKCQ | −2.5 | Protein kinase C theta | Cell proliferation | CD |
| Hs.75770 | NP_000312 | RBI | −1.9 | Retinoblastoma protein | Cell proliferation | CD |
| Hs. 169449 | P17252 | PRKCA | Absent | Protein kinase C alpha | Cell proliferation | CD |
| Hs.93183 | S51797 | VASP | 3.0 | Vasodilator-stimulated phosphoprotein | Cytoskeleton | CS |
| Hs. 13405 | Q9NQX3 | GPHN | 2.9 | Gephyrin | Cytoskeleton | CS |
| Hs.74034 | Q03135 | CAV1 | 2.6 | Caveolin 1 | Structural role in cavcolae | CS |
| Hs. 171374 | NP_055785 | KIFAP3 | 2.5 | Kinesin-associated protein 3 | Microtubule organization | CS |
| Hs.325474 | NP_149129 | CALD1 | −2.6 | L-Caldesmon | Cytoskeleton | CS |
| Hs. 146409 | A36382 | CDC42 | −2.4 | Cell division cycle 42 | Regulation of actin cytoskeleton | CS |
| Hs.377973 | NP_005563 | LMNA | −1.9 | Lamin A/C | Nuclear envelope formation | CS |
| Hs. 180141 | NP_068733 | CFL2 | −1.6 | Cofilin 2 | Actin-binding protein | CS |
| Hs.84981 | A32626 | Ku80 | 1.8 | Ku antigen 80 K chain | Double-stranded break repair | DR |
| Hs. 192803 | P23025 | XPA | Absent | Xeroderma pigmentosum, complementation group A | Nucleotide excision repair | DR |
| Hs.202833 | P09601 | HMOX1 | 4.2 | Heme oxygenase (decycling) 1 | Heme catabolism | M |
| Hs. 171280 | Q99714 | HADH2 | 2.6 | Hydroxyacyl-coenzyme A dehydrogenase, type II | Short-chain alcohol dehydrogenase | M |
| Hs. 170222 | A31311 | SLC9A1 | 2.3 | Solute carrier family 9, isoform 1 | pH homeostasis | M |
| Hs. 111039 | JC1343 | NMT1 | 2.1 | N-myristoyltransferase, type 1 | Myristylation | M |
| Hs.6906 | NP_005393 | RALA | 1.6 | RAS-like protein A | GTP-binding protein | M |
| Hs.1239 | A30325 | ANPEP | −3.5 | Aminopeptidase M | Enkephalin processing | M |
| Hs.122647 | NP_004799 | NMT-2 | −2.2 | N-myristoyltransferase, type 2 | Myristylation | M |
| Hs.9302 | Q13371 | PHLP | 2.4 | Phosducin-like protein | G-protein regulation | O |
| Hs.153924 | P53355 | DAP kinase | 1.9 | Death-associated protein kinase I | IFN-gamma-associated cell death | O |
| Hs.11866 | O14925 | TIMM23 | 2.7 | Translocase of inner mitochondrial membrane 23 | Mitochondrial protein translocation | PS |
| Hs.16258 | NP_570137 | RAB24 | 2.1 | RAB24 | Vesicle sorting | PS |
| Hs.84153 | Q13561 | DCTN2 | 1.9 | Dynactin 2 | Microtubule-based organelle transport | PS |
| Hs.300631 | NP_057610 | GS15 | 1.8 | Golgi SNARE, 15 kDa | Vesicle sorting | PS |
| Hs.75410 | P11021 | HSPA5 | 1.6 | Heat shock 70 kDa protein 5 | Prevents aggregation | PS |
| Hs.12797 | O60231 | DDX16 | 3.0 | DEAH-box protein 16 | Helicase | T |
| Hs.96055 | Q01094 | E2F1 | 1.8 | E2F transcription factor I | Transcription repressor | T |
| Hs.408442 | O14901 | TIEG2 | −5.7 | TGFB inducible early growth response 2 | Transcription repressor | T |
| Hs.21486 | P42224 | STAT1 | −2.6 | Signal transducer and activator of transcription 1 | Interferon-induced transcription | T |
| Hs.146355 | NP_009297 | ABL1 | 2.6 | Abelson murine leukemia viral oncogene homolog I | Oncogenesis | TG |
| Hs.41714 | NP_004314 | BAG-1 | 2.4 | Bcl-2-associated athanogene-1 | Oncogenesis | TG |
| Hs.1345 | A33166 | MCC | 2 | Mutated in colorectal cancer | Tumor suppressor | TG |
| Hs.343475 | ILYW_F | CTSD | 1.7 | Cathepsin D | Endopeptidase; estrogen inducible | TG |
| Hs.226795 | 14GS_B | GSTP1 | 1.5 | Glutathione S-transferase pi | Oncogenesis | TG |
| Hs.58169 | NP_006092 | HEC | −3.0 | Highly expressed in cancer | Oncogenesis | TG |
| Hs.82483 | Q15796 | Smad 2/3 | −2.1 | Mothers against decapentaplegic homolog 2 | Tumor suppressor | TG |
| Hs.77183 | P10398 | ARAF1 | Absent | A-Raf protooncogene | Oncogenesis | TG |
| Hs.118796 | NP_001146 | ANXA6 | −4.9 | Annexin A6 | Unknown | U |
Unigene cluster . | Gene ID . | Symbol . | PowerBlot value . | Name . | Function . | Group . |
|---|---|---|---|---|---|---|
| Hs.82395 | A40036 | Fas | 8.8 | CD 95 | Apoptosis | A |
| Hs.89862 | Q15628 | TRADD | −1.5 | TNFR1-associated death domain protein | Apoptosis | A |
| Hs.381231 | NP_001219 | CASP8 | Absent | Caspase 8 | Apoptosis | A |
| Hs.287797 | P05556 | ITGB1 | 1.8 | Integrin beta 1 | Cell adhesion | CA |
| Hs.216381 | A45973 | Nexilin | −4.2 | Nexilin | Cell adhesion | CA |
| Hs.119537 | A38219 | KHDRBS1 | 3.0 | Sre-associated in mitosis, 68 kDa | Mitosis | CD |
| Hs.3849 | AB045981 | FKBP10 | 2.4 | FK506-binding protein 10, 65 kDa | Cell proliferation | CD |
| Hs.433496 | Q9UN86 | G3BP2 | 2.3 | Ras-GAP | Mitogenesis | CD |
| Hs. 104741 | NP_060962 | TOPK | −6.3 | T-LAK cell-originated protein kinase | MAPKK | CD |
| Hs.317432 | P54687 | BCAT1 | −3.5 | Branched chain aminotransferase 1 | Cell proliferation | CD |
| Hs.22868 | JN0805 | PTPN11 | −2.9 | Protein tyrosine phosphatase, nonreceptor type II | Protein-tyrosine phosphatase | CD |
| Hs.181046 | P51452 | DUSP3 | −2.8 | Dual specificity phosphatase 3 | Regulator of CDKs | CD |
| Hs.211593 | A45416 | PRKCQ | −2.5 | Protein kinase C theta | Cell proliferation | CD |
| Hs.75770 | NP_000312 | RBI | −1.9 | Retinoblastoma protein | Cell proliferation | CD |
| Hs. 169449 | P17252 | PRKCA | Absent | Protein kinase C alpha | Cell proliferation | CD |
| Hs.93183 | S51797 | VASP | 3.0 | Vasodilator-stimulated phosphoprotein | Cytoskeleton | CS |
| Hs. 13405 | Q9NQX3 | GPHN | 2.9 | Gephyrin | Cytoskeleton | CS |
| Hs.74034 | Q03135 | CAV1 | 2.6 | Caveolin 1 | Structural role in cavcolae | CS |
| Hs. 171374 | NP_055785 | KIFAP3 | 2.5 | Kinesin-associated protein 3 | Microtubule organization | CS |
| Hs.325474 | NP_149129 | CALD1 | −2.6 | L-Caldesmon | Cytoskeleton | CS |
| Hs. 146409 | A36382 | CDC42 | −2.4 | Cell division cycle 42 | Regulation of actin cytoskeleton | CS |
| Hs.377973 | NP_005563 | LMNA | −1.9 | Lamin A/C | Nuclear envelope formation | CS |
| Hs. 180141 | NP_068733 | CFL2 | −1.6 | Cofilin 2 | Actin-binding protein | CS |
| Hs.84981 | A32626 | Ku80 | 1.8 | Ku antigen 80 K chain | Double-stranded break repair | DR |
| Hs. 192803 | P23025 | XPA | Absent | Xeroderma pigmentosum, complementation group A | Nucleotide excision repair | DR |
| Hs.202833 | P09601 | HMOX1 | 4.2 | Heme oxygenase (decycling) 1 | Heme catabolism | M |
| Hs. 171280 | Q99714 | HADH2 | 2.6 | Hydroxyacyl-coenzyme A dehydrogenase, type II | Short-chain alcohol dehydrogenase | M |
| Hs. 170222 | A31311 | SLC9A1 | 2.3 | Solute carrier family 9, isoform 1 | pH homeostasis | M |
| Hs. 111039 | JC1343 | NMT1 | 2.1 | N-myristoyltransferase, type 1 | Myristylation | M |
| Hs.6906 | NP_005393 | RALA | 1.6 | RAS-like protein A | GTP-binding protein | M |
| Hs.1239 | A30325 | ANPEP | −3.5 | Aminopeptidase M | Enkephalin processing | M |
| Hs.122647 | NP_004799 | NMT-2 | −2.2 | N-myristoyltransferase, type 2 | Myristylation | M |
| Hs.9302 | Q13371 | PHLP | 2.4 | Phosducin-like protein | G-protein regulation | O |
| Hs.153924 | P53355 | DAP kinase | 1.9 | Death-associated protein kinase I | IFN-gamma-associated cell death | O |
| Hs.11866 | O14925 | TIMM23 | 2.7 | Translocase of inner mitochondrial membrane 23 | Mitochondrial protein translocation | PS |
| Hs.16258 | NP_570137 | RAB24 | 2.1 | RAB24 | Vesicle sorting | PS |
| Hs.84153 | Q13561 | DCTN2 | 1.9 | Dynactin 2 | Microtubule-based organelle transport | PS |
| Hs.300631 | NP_057610 | GS15 | 1.8 | Golgi SNARE, 15 kDa | Vesicle sorting | PS |
| Hs.75410 | P11021 | HSPA5 | 1.6 | Heat shock 70 kDa protein 5 | Prevents aggregation | PS |
| Hs.12797 | O60231 | DDX16 | 3.0 | DEAH-box protein 16 | Helicase | T |
| Hs.96055 | Q01094 | E2F1 | 1.8 | E2F transcription factor I | Transcription repressor | T |
| Hs.408442 | O14901 | TIEG2 | −5.7 | TGFB inducible early growth response 2 | Transcription repressor | T |
| Hs.21486 | P42224 | STAT1 | −2.6 | Signal transducer and activator of transcription 1 | Interferon-induced transcription | T |
| Hs.146355 | NP_009297 | ABL1 | 2.6 | Abelson murine leukemia viral oncogene homolog I | Oncogenesis | TG |
| Hs.41714 | NP_004314 | BAG-1 | 2.4 | Bcl-2-associated athanogene-1 | Oncogenesis | TG |
| Hs.1345 | A33166 | MCC | 2 | Mutated in colorectal cancer | Tumor suppressor | TG |
| Hs.343475 | ILYW_F | CTSD | 1.7 | Cathepsin D | Endopeptidase; estrogen inducible | TG |
| Hs.226795 | 14GS_B | GSTP1 | 1.5 | Glutathione S-transferase pi | Oncogenesis | TG |
| Hs.58169 | NP_006092 | HEC | −3.0 | Highly expressed in cancer | Oncogenesis | TG |
| Hs.82483 | Q15796 | Smad 2/3 | −2.1 | Mothers against decapentaplegic homolog 2 | Tumor suppressor | TG |
| Hs.77183 | P10398 | ARAF1 | Absent | A-Raf protooncogene | Oncogenesis | TG |
| Hs.118796 | NP_001146 | ANXA6 | −4.9 | Annexin A6 | Unknown | U |
A indicates apoptosis; TNFR1, tumor necrosis factor receptor 1; absent, protein present in mock-infected cells but not in KSHV-infected cells (fold change cannot be called); CA, cell adhesion; CD, cell division; FK506, tacrolimus; GAP, GTPase-activating protein; T-LAK, lymphokine-activated killer cells with T-cell phenotype; MAPKK, mitogen-activated protein kinase kinase; CS, cell shape; DR, DNA repair; M, metabolism; GTP, guanosine 5′-triphosphate; O, other; IFN, interferon; PS, protein sorting; T, transcription; TGFB, transforming growth factor beta; TG, tumorigenesis; and U, unknown.
Experimentally infected DMVECs express increased levels of HO-1 mRNA and protein
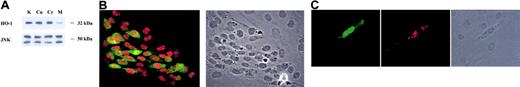
KSHV induction of HO-1 was of particular interest because virus up-regulation of this gene might confer a proliferative advantage to infected cells in the KS lesional environment. The PowerBlot data showing up-regulation of HO-1 in KSHV-infected DMVECs (Table 1) were independently confirmed in our laboratory by quantitative RT-PCR, Western blotting, and indirect immunofluorescence. KSHV-infected DMVECs expressed HO-1 mRNA at a level 3.48-fold higher than uninfected cells (Table 2). PowerBlot analysis revealed up-regulation of HO-1 by a factor of 4.24 and Western blot analysis comparing uninfected and KSHV-infected DMVECs confirmed significant viral induction of HO-1 (Figure 2A). As expected, modulation of HO-1 enzymatic activity by treatment with the heme analogs CrMP (inhibitory of HO-1 activity) and CuMP (noninhibitory) did not significantly alter the expression of HO-1 in KSHV-infected cells (Figure 2A).
Quantitative RT-PCR analysis of HO-1 expression
. | HO-1 expression . |
|---|---|
| Tissue culture compared with mock-infected cells | 3.48 |
| KS tissue compared with normal skin | 3.81 |
. | HO-1 expression . |
|---|---|
| Tissue culture compared with mock-infected cells | 3.48 |
| KS tissue compared with normal skin | 3.81 |
Up-regulation of HO-1 in KSHV-infected DMVECs. (A) Age- and passage-matched uninfected and KSHV-infected DMVECs were harvested and lysed with 3 freeze/thaw cycles. Clarified supernatants from whole-cell extracts were resolved on SDS-PAGE, transferred to nitrocellulose membranes, and probed with a monoclonal antibody (mAb) raised against the 32-kDa protein HO-1. HO-1 expression was increased in KSHV-infected extracts (lane K) relative to uninfected extract (lane M). Treatment of KSHV-infected cells with the heme analogs CuMP and CrMP (lanes Cu and Cr, respectively) did not significantly alter HO-1 expression. SAPK/JNK was used as a loading control. (B) Monolayers of KSHV-infected DMVECs were fixed with 2% paraformaldehyde, permeabilized with 0.05% Triton X, and stained with an anti-HO-1 mAb and an anti-ORF 73 polyclonal antibody followed by FITC-labeled goat antimouse and Texas Red-labeled goat antirabbit secondary antibodies. HO-1 staining (green) was found exclusively in cells expressing ORF 73 (red), but not in uninfected neighboring cells, which are visible in the phase image in the right panel. (C) The recombinant EGFP-expressing clone rKSHV.152 was used to infect primary DMVECs at a low multiplicity of infection. Monolayers were fixed and permeabilized as described in B and stained for HO-1 (red). EGFP-KSHV-positive, spindle-shaped DMVECs stained positive for HO-1 (red), whereas uninfected EGFP-negative cells, visible in the phase image in right panel, did not. Original magnification, × 20.
Up-regulation of HO-1 in KSHV-infected DMVECs. (A) Age- and passage-matched uninfected and KSHV-infected DMVECs were harvested and lysed with 3 freeze/thaw cycles. Clarified supernatants from whole-cell extracts were resolved on SDS-PAGE, transferred to nitrocellulose membranes, and probed with a monoclonal antibody (mAb) raised against the 32-kDa protein HO-1. HO-1 expression was increased in KSHV-infected extracts (lane K) relative to uninfected extract (lane M). Treatment of KSHV-infected cells with the heme analogs CuMP and CrMP (lanes Cu and Cr, respectively) did not significantly alter HO-1 expression. SAPK/JNK was used as a loading control. (B) Monolayers of KSHV-infected DMVECs were fixed with 2% paraformaldehyde, permeabilized with 0.05% Triton X, and stained with an anti-HO-1 mAb and an anti-ORF 73 polyclonal antibody followed by FITC-labeled goat antimouse and Texas Red-labeled goat antirabbit secondary antibodies. HO-1 staining (green) was found exclusively in cells expressing ORF 73 (red), but not in uninfected neighboring cells, which are visible in the phase image in the right panel. (C) The recombinant EGFP-expressing clone rKSHV.152 was used to infect primary DMVECs at a low multiplicity of infection. Monolayers were fixed and permeabilized as described in B and stained for HO-1 (red). EGFP-KSHV-positive, spindle-shaped DMVECs stained positive for HO-1 (red), whereas uninfected EGFP-negative cells, visible in the phase image in right panel, did not. Original magnification, × 20.
Double staining for KSHV LANA and HO-1 by indirect immunofluorescence (IF) showed that HO-1 was up-regulated in KSHV-infected, LANA-positive cells but not in uninfected neighboring cells (Figure 2B). A range of HO-1 staining intensities is apparent by IF in latently infected cells suggesting that in addition to viral components, cellular factors may also influence HO-1 expression in these cells. Parallel monolayers stained for the lytic marker ORF 59 showed that less than 1% of cells had undergone spontaneous lytic reactivation (data not shown), thus suggesting that latent infection with KSHV was sufficient for up-regulation of HO-1. HO-1 up-regulation by KSHV infection was confirmed in primary DMVECs infected with the recombinant EGFP-expressing clone rKSHV.15247 at a low multiplicity of infection (Figure 2C). Following one passage after infection, IF revealed HO-1 expression in EGFP-positive, spindle-shaped primary DMVECs but not neighboring EGFP-negative cells, suggesting a direct rather than paracrine effect of KSHV infection on up-regulation of HO-1.
HO-1 is up-regulated in KS tissue
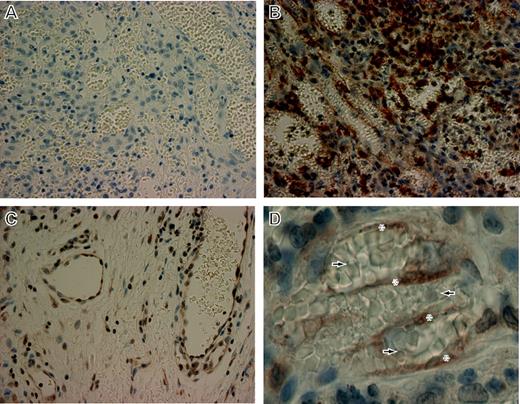
Quantitative RT-PCR on RNA extracted from a biopsy of cutaneous AIDS-KS revealed up-regulation of HO-1 message by a factor of 3.8 relative to normal skin (Table 2). HO-1 levels from both in vitro and in vivo sample sets were normalized to an internal control, GAPDH. HO-1 protein levels were assessed in biopsy tissue of oral AIDS-KS lesions by immunohistochemistry (IHC; Figure 3). Examination of normal tissue at the tumor margin showed HO-1 expression within the intact endothelial lining of normal vascular spaces as well as sporadic expression in nonvascular regions (Figure 3C). KS lesional tissue showed a higher degree of cellularity, numerous abnormal vascular spaces with extravasated erythrocytes, and substantially higher HO-1 reactivity compared with normal tissue (Figure 3B). Examination of the KS lesional tissue at higher magnification revealed numerous HO-1-expressing spindle-shaped cells traversing the abnormal vascular spaces (Figure 3D). The strong HO-1 reactivity observed in spindle-shaped cells within KS lesional tissue suggests that this protein may play an important role in the pathophysiology of KS.
HO-1 expression in oral AIDS-KS tissue.Paraffin-embedded biopsy tissues from oral AIDS-KS lesions were sectioned and mounted on charged slides. The slides were deparaffinized and stained with polyclonal anti-HO-1 and counterstained with hematoxylin/eosin. (A) Rabbit Ig was used as a control for secondary specificity in sections taken from identical tissue blocks. The panel shows low background staining in an area of tumor involvement. Original magnification, × 40. (B) A similar area of tumor involvement from a section derived from the same tissue block shows a high degree of cellularity, numerous abnormal vascular spaces with extravasated erythrocytes, and a marked level of HO-1 expression (dark brown). Original magnification, × 40. (C) HO-1 expression in normal tissue at the tumor margin of the same tissue section shown in panel B. HO-1 was found within the endothelial lining of normal vascular spaces and sporadically in nonvascular regions. Original magnification, × 40. (D) Higher magnification of an area of tumor involvement from the same tissue section shown in panel B showing HO-1 staining in spindle-shaped cells (*) traversing abnormal, erythrocyte-congested vascular spaces (arrows). Original magnification, × 63.
HO-1 expression in oral AIDS-KS tissue.Paraffin-embedded biopsy tissues from oral AIDS-KS lesions were sectioned and mounted on charged slides. The slides were deparaffinized and stained with polyclonal anti-HO-1 and counterstained with hematoxylin/eosin. (A) Rabbit Ig was used as a control for secondary specificity in sections taken from identical tissue blocks. The panel shows low background staining in an area of tumor involvement. Original magnification, × 40. (B) A similar area of tumor involvement from a section derived from the same tissue block shows a high degree of cellularity, numerous abnormal vascular spaces with extravasated erythrocytes, and a marked level of HO-1 expression (dark brown). Original magnification, × 40. (C) HO-1 expression in normal tissue at the tumor margin of the same tissue section shown in panel B. HO-1 was found within the endothelial lining of normal vascular spaces and sporadically in nonvascular regions. Original magnification, × 40. (D) Higher magnification of an area of tumor involvement from the same tissue section shown in panel B showing HO-1 staining in spindle-shaped cells (*) traversing abnormal, erythrocyte-congested vascular spaces (arrows). Original magnification, × 63.
HO-1 enzymatic activity is up-regulated in KSHV-infected cells compared with uninfected cells
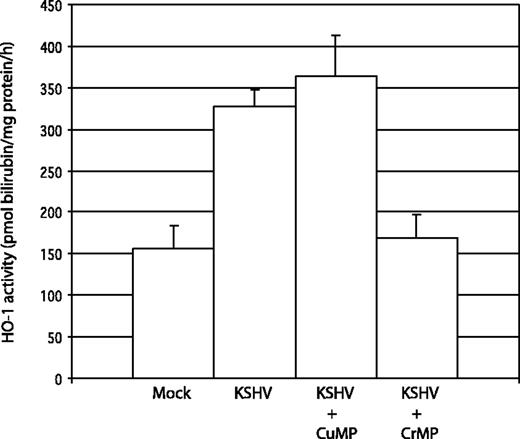
We verified that HO-1 induced by KSHV infection of DMVECs was enzymatically active by measuring the production of bilirubin by crude whole-cell extracts. HO-1 activity was increased 2.1-fold in KSHV-infected DMVECs over uninfected controls (Figure 4). Treatment of KSHV-infected cultures with an inhibitor of HO-1 activity, the heme analog CrMP (10 μM overnight), resulted in reduction of HO-1 activity to baseline levels, while treatment with the structurally related but noninhibitory CuMP had no net effect on HO-1 activity. Note that these analogs do not influence HO-1 expression levels (Figure 2A). Therefore, the HO-1 expressed in KSHV-infected DMVECs is enzymatically active and susceptible to pharmacologic inhibition.
Measurement of HO activity in DMVEC cell extracts. HO activity in uninfected and KSHV-infected DMVEC cell extracts was determined by spectrophotometric measurement of bilirubin production. Cell extracts were incubated with 50 μM heme for 30 minutes at 37°C. The reaction was stopped by the addition of 2 volumes of chloroform, and bilirubin concentration was determined by comparing the difference in optical density at 464 and 530 nm. HO activity was reported as picomoles of bilirubin produced per milligram endothelial cell protein extract per hour. KSHV-infected cells (KSHV) had 2.1-fold greater HO activity relative to uninfected cells (Mock). Treatment with the HO inhibitor CrMP reduced HO activity in infected cells to the level of uninfected cells (KSHV + CrMP), whereas treatment with the noninhibitory heme analog CuMP did not reduce HO activity (KSHV + CuMP). Results were determined in triplicate (error bars indicate ± SD).
Measurement of HO activity in DMVEC cell extracts. HO activity in uninfected and KSHV-infected DMVEC cell extracts was determined by spectrophotometric measurement of bilirubin production. Cell extracts were incubated with 50 μM heme for 30 minutes at 37°C. The reaction was stopped by the addition of 2 volumes of chloroform, and bilirubin concentration was determined by comparing the difference in optical density at 464 and 530 nm. HO activity was reported as picomoles of bilirubin produced per milligram endothelial cell protein extract per hour. KSHV-infected cells (KSHV) had 2.1-fold greater HO activity relative to uninfected cells (Mock). Treatment with the HO inhibitor CrMP reduced HO activity in infected cells to the level of uninfected cells (KSHV + CrMP), whereas treatment with the noninhibitory heme analog CuMP did not reduce HO activity (KSHV + CuMP). Results were determined in triplicate (error bars indicate ± SD).
HO-1 up-regulation increases proliferation of KSHV-infected DMVECs in the presence of free heme
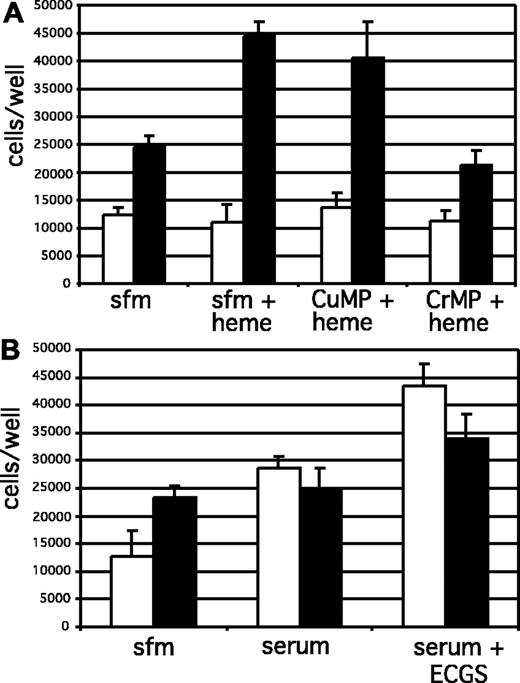
Because of the potential for increased exposure of KS cells to free heme and heme-containing proteins in KS tissue,50-54 we investigated the possibility that HO-1 up-regulation might affect the response of KSHV-infected cells to heme. Overexpression of HO-1 in endothelial cells by viral vector transduction has been shown to increase cell proliferation.38,39 To assess the effect of up-regulation of HO-1 on proliferation of DMVECs, uninfected and KSHV-infected cells were plated in 96-well plates and cultured for 24 hours in SFM with no growth supplements, followed by the addition of a nontoxic dose of heme (5 μM) for 48 hours. Selected cultures were additionally treated with the heme analogs CrMP (an inhibitor of HO-1 activity) and CuMP (noninhibitory analog). Proliferation of DMVECs in response to treatment was assessed by performing direct cell counts at 0 and 48 hours after heme treatment as described in “Materials and methods.” Importantly, treatment with heme had no effect on the proliferation of uninfected DMVECs but significantly stimulated the growth of KSHV-infected cells (Figure 5A). This increase in proliferation was due to HO-1 enzymatic activity, since inhibition of HO-1 activity by treatment with CrMP abolished this increase in cell division, whereas treatment with the noninhibitory heme analog CuMP did not. Uninfected cells did exhibit a robust proliferative response when serum-starved cells were incubated with 10% human serum in the presence or absence of endothelial cell growth supplement, a complex mixture containing known endothelial cell mitogens, indicating that the lack of proliferation of these cells in the presence of heme was due to an unresponsiveness to heme rather than a nonspecific limitation resulting from the experimental procedure employed (Figure 5B). Cell viability was higher than 95% by trypan blue exclusion for all treatment conditions (data not shown), indicating that the steady state of uninfected cell numbers under all treatment conditions was due to minimal proliferation in serum-free conditions rather than a cytotoxic effect of heme on these cells. Given the increased concentration of heme within the KS lesional microenvironment, up-regulation of HO-1 in KSHV-infected cells and the subsequent increase in cellular proliferation in the presence of the enzyme's substrate may provide a survival and growth advantage for spindle cells compared with uninfected cells. Thus, KSHV induction of HO-1 may represent a novel strategy for viral tumorigenesis.
Proliferation of DMVECs in response to free heme. The proliferative response of uninfected and KSHV-infected cells to heme was determined by direct cell counts. (A) Cells were plated in complete media overnight followed by culture in serum-free media (SFM) for 24 hours in the presence or absence of the heme analogs CrMP (to inhibit HO-1 activity) and CuMP (noninhibitory). Cells were then incubated with and without 5 μM heme in SFM and proliferation measured 48 hours later. Uninfected cells (□) did not proliferate in response to incubation with heme, whereas KSHV-infected cells (▪) proliferated to a greater extent following exposure to heme. Enhanced proliferation of KSHV-infected cells in the presence of heme was HO-dependent, since treatment with CrMP abolished the proliferative response. (B) The lack of proliferation of uninfected cells was due to an unresponsiveness to heme rather than a nonspecific limitation resulting from the experimental procedure employed. These cells did exhibit a robust proliferative response when serum-starved cells were incubated with 10% human serum in the presence or absence of endothelial cell growth supplement (ECGS; 50 μg/mL), a complex mixture containing known endothelial cell mitogens. Results were determined in quadruplicate (error bars indicate ± SD).
Proliferation of DMVECs in response to free heme. The proliferative response of uninfected and KSHV-infected cells to heme was determined by direct cell counts. (A) Cells were plated in complete media overnight followed by culture in serum-free media (SFM) for 24 hours in the presence or absence of the heme analogs CrMP (to inhibit HO-1 activity) and CuMP (noninhibitory). Cells were then incubated with and without 5 μM heme in SFM and proliferation measured 48 hours later. Uninfected cells (□) did not proliferate in response to incubation with heme, whereas KSHV-infected cells (▪) proliferated to a greater extent following exposure to heme. Enhanced proliferation of KSHV-infected cells in the presence of heme was HO-dependent, since treatment with CrMP abolished the proliferative response. (B) The lack of proliferation of uninfected cells was due to an unresponsiveness to heme rather than a nonspecific limitation resulting from the experimental procedure employed. These cells did exhibit a robust proliferative response when serum-starved cells were incubated with 10% human serum in the presence or absence of endothelial cell growth supplement (ECGS; 50 μg/mL), a complex mixture containing known endothelial cell mitogens. Results were determined in quadruplicate (error bars indicate ± SD).
Discussion
We report here the results of a proteomics screening approach undertaken to identify alterations in gene expression in DMVECs experimentally infected with KSHV. Of the 850 genes examined by Western blot array, 52 showed significant positive or negative regulation compared with uninfected DMVECs. In the present study we chose to focus our efforts on HO-1 because of the potential role this gene has in angiogenesis. Given the vascular nature of KS lesions, host genes involved in angiogenesis that are induced by KSHV infection may provide targets for pharmacologic intervention. Additionally, the availability of specific HO-1 inhibitors in clinical use for control of hyperbilirubinemia in neonates55,56 aided our initial dissection of the role of HO-1 up-regulation in KSHV-induced pathogenesis.
HO-1 is a heme-degrading enzyme that cleaves the porphyrin ring-releasing equimolar quantities of CO, free iron, and biliverdin. Free iron then stimulates the production of the iron-scavenging protein ferritin, while biliverdin is rapidly reduced to bilirubin by biliverdin reductase.42 The effects of these HO-1 metabolites on endothelial physiology suggest that HO-1 might play an important and interesting role in KS pathogenesis. For example, CO produced by HO-1 activity was shown to protect endothelial cells from both CD95/Fas- and tumor necrosis factor α (TNFα)-mediated apoptosis.36,37 In addition, generation of the cytoprotectant bilirubin, along with ferritin up-regulation following release of free iron, has been shown to protect endothelial cells from oxidative damage resultant from myriad noxious stimuli.57-59
In the present study we show that KSHV infection of DMVECs results in up-regulation of HO-1 at both the message and protein levels. In vitro, immunofluorescent analysis revealed up-regulation of HO-1 in infected DMVECs but not uninfected neighboring cells in both primary and E6/E7-transformed cultures. Recently it was shown by serial analysis of gene expression (SAGE) that HO-1 expression in AIDS-KS biopsy tissue is significantly greater than in control tissue.60 Here we independently confirm this finding with quantitative RT-PCR results on cutaneous AIDS-KS tissue that show elevated HO-1 mRNA compared with normal skin. Furthermore, immunohistochemistry on oral AIDS-KS lesional tissue revealed increased expression of HO-1 protein compared with normal tissue at the tumor margin. In vitro, up-regulated HO-1 was enzymatically active and susceptible to chemical inhibition with a heme analog, CrMP.
We show here that increased HO-1 activity in KSHV-infected endothelial cells results in increased cell proliferation following exposure to free heme, whereas uninfected cells grown under the same conditions did not exhibit a proliferative response. Because of the importance of angiogenesis in the pathophysiology of KS, heme-induced proliferation of KSHV-infected cells may be an important factor contributing to lesion development. There is evidence that KS lesions have locally elevated concentrations of hemoglobin and free heme and therefore contain ample substrate for HO-1. Intracellular junctions between neoplastic endothelial cells are frequently defective allowing the extravasation of large numbers of erythrocytes from vessel lumens into the surrounding tissue.50-53 These renegade cells become more numerous as the disease progresses and abnormal networks of slitlike vascular spaces expand. Erythrophagocytosis by KS cells is also a common feature.54 Many spindle cells contain membrane-bound intracytoplasmic erythrocytes (erythrophagosomes) surrounded by lysosomal granules. These erythrophagosomes show various stages of disintegration leading ultimately to extruded hemoglobin and empty ghost cells (dead red blood cells [RBCs]). Others have described the presence of hyaline globules in plaque- and nodular-stage KS, and histologic staining has revealed that these are the remnants of degenerated erythrocytes.51 In light of these histologic findings, it is conceivable that up-regulation of HO-1 by KSHV infection could confer a selective advantage to KS cells within the high heme lesional microenvironment compared with neighboring uninfected cells. The availability of potent inhibitors of HO-1 in clinical use elsewhere55,56 makes this enzyme a potentially important treatment target in KS.
Up-regulation of HO-1 has been recognized in other experimental pathogenesis systems. Infections by influenza virus,61 encephalomyocarditis virus,62 Listeria monocytogenes,63 and Rickettsia rickettsii64 all show significant up-regulation of HO-1, suggesting that this enzyme may participate in cellular responses to some intracellular pathogens. We have assessed the expression of HO-1 in our DMVEC system following infection with the KSHV-related rhesus rhadinovirus (RRV). RRV is associated with the development of lymphomas and retroperitoneal fibromatosis (RF) in rhesus macaques coinfected with simian immunodeficiency virus, an animal model similar to HIV and KSHV coinfection in humans (S. Wong, manuscript in preparation, May 2004). The expression profile of DMVECs infected with a recombinant EGFP-expressing clone of RRV was compared with that of uninfected cells by cDNA microarray (S.C.M., S.G.H., A.V.M., and S. Wong, manuscript in preparation, May 2004). HO-1 was found not to be induced in these cells by RRV infection (S. Wong, personal communication, October 2003). We verified this finding by taqman in our laboratory (data not shown). Alternately, another gene that we have reported as up-regulated by KSHV infection, cKit, was found to be up-regulated by RRV infection (data not shown), suggesting that there are similar yet distinct subsets of cellular genes induced by these closely related, endothelial-tropic viruses. It is interesting that RRV does not induce HO-1, a phenomenon that might be explained by the histologic differences that exist between RF and KS. One of the dominant histopathologic features of KS, the disorganized network of abnormal vascular spaces filled with extravasated erythrocytes, is notably absent in RF.65,66 It is conceivable that in the absence of a high heme microenvironment within RF tissue, RRV may not have experienced the selective pressure to up-regulate HO-1 because no proliferative advantage is to be derived by that enzyme's expression in the absence of substrate. Therefore, the specificity of HO-1 up-regulation in endothelial cells by KSHV infection may be a modulation of cellular gene expression resulting from the unique erythrocyte-rich microenvironment characteristic of KS.
Recent reports indicate that HO-1 expression in endothelial cells results in up-regulation of VEGF.40,67 Current studies in our laboratory are focused on defining the mechanism(s) by which KSHV induces HO-1 in DMVECs as well as determining the cellular pathways activated by HO-1-derived metabolites that result in enhanced proliferation of infected cells following exposure to heme.
Prepublished online as Blood First Edition Paper, January 15, 2004; DOI 10.1182/blood-2003-08-2781.
Supported in part by a sponsored research agreement from Virogenomics (A.V.M. and K.F.), NOT-HL-02-013 (A.V.M.), NIH-NIDCR PO1 DE-07496. The University of California, San Francisco (UCSF) AIDS Specimen Bank is supported by P30 A127763-12.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We acknowledge Dr Molly Kulesz-Martin and Dr Jon Hanifin (OHSU Department of Dermatology) for cutaneous AIDS-KS tissue from the Molecular Profiling Resource in Dermatology (partially supported by NIH CA31101 and OHSU Cancer Center grant CA69533); Dr Bala Chandran (University of Kansas Medical Center) for KSHV-specific antibodies; Dr Jeff Vieira (Department of Laboratory Medicine, University of Washington, and Program in Infectious Diseases, Fred Hutchinson Cancer Research Center) for the recombinant EGFP-expressing clone rKSHV.152; and Linda Jauron-Mills (Oregon Cancer Center) for help with IHC.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal