Acute myelogenous leukemia 1 (AML1; runt-related transcription factor 1 [Runx1]) is a member of Runx transcription factors and is essential for definitive hematopoiesis. Although AML1 possesses several subdomains of defined biochemical functions, the physiologic relevance of each subdomain to hematopoietic development has been poorly understood. Recently, the consequence of carboxy-terminal truncation in AML1 was analyzed by the hematopoietic rescue assay of AML1-deficient mouse embryonic stem cells using the gene knock-in approach. Nonetheless, a role for specific internal domains, as well as for mutations found in a human disease, of AML1 remains to be elucidated. In this study, we established an experimental system to efficiently evaluate the hematopoietic potential of AML1 using a coculture system of the murine embryonic para-aortic splanchnopleural (P-Sp) region with a stromal cell line, OP9. In this system, the hematopoietic defect of AML1-deficient P-Sp can be rescued by expressing AML1 with retroviral infection. By analysis of AML1 mutants, we demonstrated that the hematopoietic potential of AML1 was closely related to its transcriptional activity. Furthermore, we showed that other Runx transcription factors, Runx2/AML3 or Runx3/AML2, could rescue the hematopoietic defect of AML1-deficient P-Sp. Thus, this experimental system will become a valuable tool to analyze the physiologic function and domain contribution of Runx proteins in hematopoiesis.
Introduction
Acute myelogenous leukemia 1 (AML1)/runt-related transcription factor 1 (Runx1) belongs to a family of transcriptional regulators called Runx, which contain a conserved 128–amino acid Runt domain responsible for sequence-specific DNA binding.1 Runx proteins make heterodimeric complexes with a partner protein, CBFβ/PEBP2β (core-binding factor β/polyomavirus enhancer–binding protein 2β),2-4 and this association is essential for its biologic activity.5-7 There are 3 known mammalian Runx family members: AML1/Runx1, Runx2/AML3, and Runx3/AML2. Typically, Runx functions as a transcriptional activator of target gene expression. Under some conditions, however, it can repress the transcription of specific genes.
AML1 was originally identified on chromosome 21 as the gene that is disrupted in the (8;21)(q22;q22) translocation, which is one of the most frequent chromosome abnormalities associated with human AML.8,9 Subsequently, AML1 was shown to be one of the most frequent targets of leukemia-associated gene aberrations.10,11 Moreover, somatic point mutations of the AML1 gene were also demonstrated in patients with AML and myelodysplastic syndrome (MDS).12-14 In addition to a role in leukemic transformation, gene-targeting studies in mice have demonstrated that AML1 is essential for early development of definitive hematopoiesis. AML1-deficient embryos develop through the yolk sac stage but die around 12 to 13 days of gestation following complete block of fetal liver hematopoiesis.15,16
AML1 includes at least 3 alternative splicing forms: AML1a, AML1b, and AML1c.17 In AML1b and AML1c, the carboxy (C)–terminal to the Runt domain lies in a region that contains sequences of defined biochemical functions, which are absent in AML1a. Several functional domains have been identified in the C-terminal half, such as trans-activation domain,18,19 trans-repression domain,20 and VWRPY motif.21-23
During vertebrate embryogenesis, hematopoietic development consists of 2 distinct waves of discrete cellular components known as primitive and definitive hematopoiesis.24 In mice, the first wave of primitive hematopoiesis, which consists predominantly of a large and nucleated erythroid cell, emerges in the yolk sac at 7.5 embryonic days after coitus (dpc). Then, primitive hematopoiesis begins to be replaced around 9.5 dpc by definitive hematopoiesis, generally described as the second wave. Progenitors for definitive hematopoiesis originate from para-aortic splanchnopleural (P-Sp) region at 7.5 to 9.5 dpc,25,26 and long-term repopulating hematopoietic stem cells (LTR-HSCs) that can reconstitute adult mice appear in the aorta-gonad-mesonephros (AGM) at 10.5 to 11.5 dpc.27,28 These cells subsequently colonize the fetal liver, where they expand and differentiate. Active sites for definitive hematopoiesis are transferred to bone marrow and spleen prior to birth and function throughout life within these organs. Recent studies have shown that AML1 is expressed in the hematopoietic cell clusters within the P-Sp/AGM region and AML1-deficient embryos are devoid of these hematopoietic clusters.29-32
The hematopoietic defect of AML1-deficient mice could be replicated in vitro by several culture systems, including the P-Sp/AGM culture33,34 and the embryonic stem (ES) cell culture.35 In these systems, hematopoietic cells are generated in wild-type cultures but not in AML1-deficient cultures. Recently, a gene knock-in approach was used to demonstrate rescue in vivo of hematopoiesis from the AML1-deficient ES cells.35,36 This hematopoietic rescue requires the trans-activation domain of AML1 but not the C-terminal trans-repression subdomain. However, no report has elucidated roles for specific internal domains or disease-related mutations of AML1 in hematopoiesis.
In the present study, we used a coculture system of cells derived from the P-Sp region with a layer of a stromal cell line, OP9, in which hematopoietic cell development of various lineages is efficiently induced.37 The cultured P-Sp–derived cells show a significant colony-forming activity in semisolid culture with appropriate cytokines, as well as distinct surface expression of hematopoietic markers. In this culture system, AML1-deficient P-Sp–derived cells failed to show any hematopoietic activity. This defect was efficiently rescued by reactivating AML1 by retroviral-mediated expression. Using this system, we then examined a hematopoietic potential of a series of AML1 mutants and demonstrated that the hematopoietic rescue of AML1-deficient P-Sp regions require transcriptionally active forms of AML1. We also showed that enforced expression of other Runx transcription factors, Runx2/AML3 or Runx3/AML2, could rescue the hematopoietic defect of AML1-deficient P-Sp regions. These results provide evidence that transcriptional activity of AML1 is essential for hematopoietic development from P-Sp regions. In addition, this coculture system makes a useful method to determine functional consequences of AML1 on its hematopoietic potential.
Materials and methods
Mice and embryos
AML1-deficient mice were generated as described previously38 and were crossed onto the C57BL/6 background. To generate embryos, timed matings were set up between AML1+/– males and AML1+/– females. The time at midday (12:00) was taken to be 0.5 dpc for the plugged mice.
In vitro P-Sp culture
P-Sp culture was performed as described previously39 with a minor modification. In brief, isolated P-Sp regions of 9.5 dpc embryos were dissociated by incubation with 250 U/mL dispase (Godo Shusei, Tokyo, Japan) for 20 minutes and cell dissociation buffer (Gibco BRL, Carlsbad, CA) for 20 minutes at 37°C, washed once in phosphate-buffered saline (PBS), followed by vigorous pipetting. Approximately 5 × 104 P-Sp–derived cells were suspended in 300 μL serum-free StemPro media (Life Technologies, Gaithersburg, MD) supplemented with 50 ng/mL stem cell factor (SCF), 5 ng/mL interleukin (IL3; gifts from Kirin Brewery, Takasaki, Japan), and 10 ng/mL murine oncostatin M (R&D Systems, Minneapolis, MN). Single-cell suspensions were seeded on preplated OP-9 stromal cells in the 24-well plate, followed by incubation at 37°C.
Plasmid construction
The cDNAs of human AML1a, AML1b, and various AML1 mutants were subcloned as EcoRI-EcoRI fragments into the retrovirus vector pMY/internal ribosomal entry site–enhanced green fluorescent protein (IRES-EGFP; pMY/IG).40 C-terminal deletion mutants of AML1b, AML1b-R139G, AML1b-S249/266A, and AML1b-K24/43R were constructed as described previously.13,41-43 For construction of AML1bΔ(205-332), we deleted the PvuII-BstPI fragment from AML1b, filled the resultant plasmid with a Klenow fragment, and religated it. AML1bΔ(181-210) was created by polymerase chain reaction (PCR) with the insertion of a BglII restriction site to join the fragments. Flag-tagged human Runx2/AML3 cDNA was inserted into the BamHI and the EcoRI restriction sites of pMYs/IRES-EGFP (pMYs/IG).40 Flag-tagged human Runx3/AML2 cDNA was inserted into the SacII and the XhoI sites of the same vector.
Retroviral transduction
Plat-E packaging cells44 (2 × 106) were transiently transfected with 3 μgof AML1, Runx2/AML3, Runx3/AML2, or AML1 mutants; mixed with 9 μL of FuGENE6 (Roche Molecular Biochemicals, Indianapolis, IN); followed by incubation at 37°C. Supernatant containing retrovirus was collected 48 hours after transfection and used immediately for infection. Retroviral transduction to the cells derived from AML1-deficient P-Sp regions was performed as described previously with minor modification.45 In brief, the viral supernatant was added to the P-Sp culture together with 10 μg/mL Polybrene (Sigma, St Louis, MO). After 72 hours of incubation, virus-containing medium was replaced by standard culture medium. The cells were incubated for another 10 days and processed for analysis. To confirm the expression of Runx proteins, NIH3T3 cells were also infected with the same viral supernatants. The number of retrovirus-infected cells was evaluated by the expression of green fluorescent protein (GFP).
Colony-forming cell (CFC) assay
The nonadherent or semiadherent cells rescued from AML1-deficient P-Sp regions were used for CFC assay. Cells (6 × 104) were plated into MethoCult3434 medium (StemCell Technologies, Vancouver, BC, Canada) and cultured in a 5% CO2 incubator at 37°C. Colony types were determined at day 7 by morphologic appearance and by Wright-Giemsa staining of each colony.
Flow cytometry analysis
Flow cytometry analysis was performed in a FACScalibur with the Cellquest program (Becton Dickinson, San Jose, CA) after addition of propidium iodide to exclude dead cells. For surface staining, cell suspensions collected from the P-Sp cultures were incubated on ice for 30 minutes in the presence of various mixtures of labeled monoclonal antibodies. The monoclonal antibodies used were phycoerythrin (PE)–conjugated anti–granulocyte 1 (anti-Gr1), anti–macrophage antigen 1 (anti-Mac1), anti–stem cell antigen 1 (anti-Sca1), allophycocyanin (APC)–conjugated anti-CD45, anti–c-Kit, and biotin-conjugated anti-CD34. Biotinylated antibodies were then counterstained with PE- or APC-conjugated streptavidin. Isotype-matched antibodies conjugated with the appropriate fluorochrome were used as negative controls.
Western blot analysis
Retrovirus-infected NIH3T3 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer.41 Whole-cell lysates containing 100 μg of proteins were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (Immobilon; Millipore, Bedford, MA). The membrane was blocked with 10% skim milk, treated with anti-AML1 (PC284L; Oncogene, Cambridge, MA) or anti-Flag (M2; Sigma), washed, and reacted with the rabbit anti–immunoglobulin G (anti-IgG) antibody coupled to horseradish peroxidase. The blot was visualized using the enhanced chemiluminescence (ECL) system (Amersham Pharmacia Biotech, Piscataway, NJ).
Results
Retroviral expression of AML1 rescues hematopoiesis by AML1-deficient P-Sp region
AML1-deficient mice die in midgestation as a result of a complete block in fetal liver hematopoiesis, indicating the strict in vivo requirement of AML1 in definitive hematopoiesis. Consistently, the primary culture system of the P-Sp region has demonstrated that the failure of hematopoiesis in the fetal liver is preceded by a hematopoietic defect in the P-Sp region, from which the development of hematopoietic cells was never detected in AML1-deficient embryos.34,45 When the cells isolated from wild-type P-Sp regions at 9.5 dpc were cocultured with OP9 stromal cells, small and round-shaped nonadherent cells were produced in 5 days (Figure 1A). These cells were thought to represent a hematopoietic cell population of various lineages because they expressed hematopoietic cell surface markers and generated hematopoietic cell colonies when plated into a semisolid culture (Figure 2; data not shown). In contrast, the cells from AML1-deficient P-Sp regions failed to develop any hematopoietic cells (Figure 1B), which coincides with the notion that AML1 is a prerequisite for hematopoietic cell production in the P-Sp region. Thus, AML1-dependent hematopoiesis could be recapitulated in vitro, and we went on further to examine whether reactivation of a transcriptionally active form of AML1 can rescue this hematopoietic defect. First, by packaging the pMY/IG-AML1b in Plat-E cells, we generated the AML1b-IRES-GFP retrovirus that expresses AML1b and GFP. Then we infected the cells derived from the AML1-deficient P-Sp region with the AML1b-IRES-GFP retrovirus and cultured for an additional 10 days. Interestingly, AML1b-infected cultures generated numerous small and round cells with a nonadherent property (Figure 1C). In contrast, cultures infected with the empty vector (control) produced no such cells (Figure 1D). These nonadherent cells were morphologically indistinguishable from the hematopoietic cells generated from the wild-type P-Sp cells and proliferated continuously for more than 30 days. These results suggest that lack of P-Sp hematopoiesis can be complemented by retrovirus-mediated reactivation of AML1 in this culture system.
Retroviral expression of AML1b rescues hematopoiesis from AML1-deficient P-Sp regions. Photographs were taken with a Nikon Eclipse TE2000-U (Nikon Sankei, Tokyo, Japan) at a magnification of × 100 after 5 days of culture (A-B) and 14 days of culture (C-D). (A) Hematopoietic cells emerged at day 5 from wild-type P-Sp regions. (B) No hematopoietic cells were observed in the culture of AML1-deficient P-Sp regions, showing only the background of OP9 cells. (C) AML1b-transduced P-Sp regions from an AML1-deficient embryo generated numerous round, nonadherent, or semiadherent cells. (D) A control culture infected with mock virus failed to generate any hematopoietic cells.
Retroviral expression of AML1b rescues hematopoiesis from AML1-deficient P-Sp regions. Photographs were taken with a Nikon Eclipse TE2000-U (Nikon Sankei, Tokyo, Japan) at a magnification of × 100 after 5 days of culture (A-B) and 14 days of culture (C-D). (A) Hematopoietic cells emerged at day 5 from wild-type P-Sp regions. (B) No hematopoietic cells were observed in the culture of AML1-deficient P-Sp regions, showing only the background of OP9 cells. (C) AML1b-transduced P-Sp regions from an AML1-deficient embryo generated numerous round, nonadherent, or semiadherent cells. (D) A control culture infected with mock virus failed to generate any hematopoietic cells.
Expression of the hematopoietic markers on the rescued cells from AML1-deficient P-Sp culture. Flow cytometric profiles of the cells stained with antibodies against Gr1/Mac1, CD45, CD34, Sca1, and c-Kit. Note that the rescued cells from AML1-deficient P-Sp regions are GFP-positive and express various hematopoietic cell surface markers. Flow cytometric profiles of the rescued cells are similar to those of hematopoietic cells in wild-type P-Sp culture. GFP intensity (marking retrovirally transduced cells) is plotted on the x-axis and intensity of counterstaining of hematopoietic surface markers is plotted on the y-axis. Isotype-matched control staining of the hematopoietic cells from wild-type P-Sp regions is also shown.
Expression of the hematopoietic markers on the rescued cells from AML1-deficient P-Sp culture. Flow cytometric profiles of the cells stained with antibodies against Gr1/Mac1, CD45, CD34, Sca1, and c-Kit. Note that the rescued cells from AML1-deficient P-Sp regions are GFP-positive and express various hematopoietic cell surface markers. Flow cytometric profiles of the rescued cells are similar to those of hematopoietic cells in wild-type P-Sp culture. GFP intensity (marking retrovirally transduced cells) is plotted on the x-axis and intensity of counterstaining of hematopoietic surface markers is plotted on the y-axis. Isotype-matched control staining of the hematopoietic cells from wild-type P-Sp regions is also shown.
Rescued cells retain the features of hematopoietic cells
In the previous study, Mukouyama et al45 described that the retroviral transfer of AML1 into the AML1-deficient P-Sp region gave rise to the production of small and round cells in the culture with an appropriate combination of cytokines. However, under their experimental condition in which no stromal cell layer was employed, the recovered cells showed neither CFC activity nor expression of hematopoietic cell surface markers, such as CD45 and c-Kit, which indicates that the rescue of the hematopoietic defect is incomplete, if it occurs at all. To determine whether the nonadherent cells recovered under our experimental condition retain the features of hematopoietic cells, we examined CFC activity and surface markers of those cells, both of which are distinctly detected in wild-type P-Sp–derived cells in our coculture system. On the 10th day of culture, the nonadherent cells were collected and seeded into a semisolid medium. As shown in Figure 3, these cells generated a number of mixed, granulocyte/macrophage, and erythroid colonies, indicating that the recovered cells should contain various types of CFCs, possibly including definitive lineages. In addition, the flow cytometric analysis revealed that the rescued cells expressed hematopoietic cell surface markers, such as a marker of hematopoietic cells, CD45; myeloid markers Gr1/Mac1; and markers of hematopoietic progenitors c-Kit, Sca1, and CD34 (Figure 2). Their expression profiles were nearly identical to those of hematopoietic cells generated from the wild-type P-Sp cells (Figure 2).
Colony formation of the rescued cells from AML1-deficient P-Sp. The hematopoietic defect of AML1-deficient P-Sp regions was rescued by retroviral expression of AML1b, and the rescued cells were plated into MethoCult3434 medium. The rescued cells generated various types of hematopoietic colonies including definitive origins. Representative hematopoietic colonies by 7 days of culture are shown. (A-C) Morphology of the colonies. (D-F) Cytospin preparation of corresponding cell populations. Cytospins were stained with Wright-Giemsa. Mix indicates mixed colony; GM, granulocyte/macrophage colony; and E, erythroid colony. Photographs were taken with a Nikon Eclipse TE2000-U (Nikon Sankei) at a magnification of × 100.
Colony formation of the rescued cells from AML1-deficient P-Sp. The hematopoietic defect of AML1-deficient P-Sp regions was rescued by retroviral expression of AML1b, and the rescued cells were plated into MethoCult3434 medium. The rescued cells generated various types of hematopoietic colonies including definitive origins. Representative hematopoietic colonies by 7 days of culture are shown. (A-C) Morphology of the colonies. (D-F) Cytospin preparation of corresponding cell populations. Cytospins were stained with Wright-Giemsa. Mix indicates mixed colony; GM, granulocyte/macrophage colony; and E, erythroid colony. Photographs were taken with a Nikon Eclipse TE2000-U (Nikon Sankei) at a magnification of × 100.
Thus, in contrast to the previous report,45 we found that the nonadherent cells derived from AML1-deficient P-Sp regions in our culture system retained the features of hematopoietic cells that are indistinguishable from those of wild-type P-Sp–derived cells. It is, thus, likely that seeding onto OP9 stromal cells may provide a more favorable environment for production of hematopoietic cells from P-Sp regions and/or the expansion of P-Sp–derived hematopoietic cells.
Hematopoietic potential of AML1 mutants
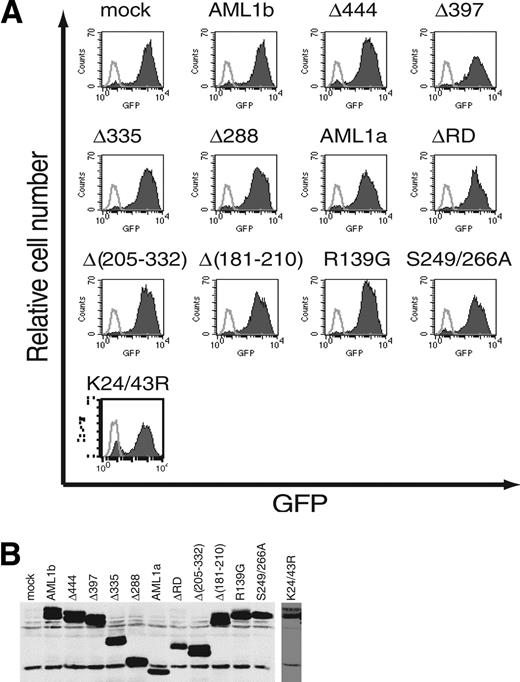
Using this experimental system, we then analyzed the hematopoietic potential of various AML1 mutants (Figure 4). We generated retroviruses that express a variety of AML1 mutants, including serial C-terminal truncation, deletion of functional domains, and substitution of specific residues. NIH3T3 cells were infected with these viruses and the titer of the retroviruses was evaluated by flow cytometric measurement of GFP-positive NIH3T3 cells (Figure 5A). Coincidently, retrovirus-mediated expression of each mutant in infected cells was confirmed by Western blotting (Figure 5B).
Structures of AML1 and its mutants. The structures of various AML1 mutants are presented schematically. Runt indicates the Runt domain; AD, trans-activation domain; ID, inhibitory domain; VWRPY, VWRPY motif; m, a binding region for mSin3A. R→G means a missense mutation at codon 139, which lead to a change of amino acid (R139G; single-letter amino acid code). S→A means a missense mutation at codon 249 and 266, which lead to changes of amino acids (S249A and S266A). K→R means a missense mutation at codon 24 and 43, which lead to changes of amino acid (K24R and K43R).
Structures of AML1 and its mutants. The structures of various AML1 mutants are presented schematically. Runt indicates the Runt domain; AD, trans-activation domain; ID, inhibitory domain; VWRPY, VWRPY motif; m, a binding region for mSin3A. R→G means a missense mutation at codon 139, which lead to a change of amino acid (R139G; single-letter amino acid code). S→A means a missense mutation at codon 249 and 266, which lead to changes of amino acids (S249A and S266A). K→R means a missense mutation at codon 24 and 43, which lead to changes of amino acid (K24R and K43R).
Infection efficiencies of retroviruses expressing AML1 or its mutants. (A) The efficiency of retrovirus-mediated gene transfer of AML1 or its mutants was estimated by infecting NIH3T3 cells. Retrovirus-infected cells were evaluated by the expression of GFP (shaded histograms). Also shown are the noninfected NIH3T3 cells (open histograms). All the retroviruses infected more than 80% of NIH3T3 cells. (B) Expression of AML1 or its mutant proteins in infected NIH3T3 cells. The expression is monitored by immunoblotting of whole-cell lysates with anti-AML1.
Infection efficiencies of retroviruses expressing AML1 or its mutants. (A) The efficiency of retrovirus-mediated gene transfer of AML1 or its mutants was estimated by infecting NIH3T3 cells. Retrovirus-infected cells were evaluated by the expression of GFP (shaded histograms). Also shown are the noninfected NIH3T3 cells (open histograms). All the retroviruses infected more than 80% of NIH3T3 cells. (B) Expression of AML1 or its mutant proteins in infected NIH3T3 cells. The expression is monitored by immunoblotting of whole-cell lysates with anti-AML1.
Among those mutants, we first used a series of C-terminal deletion mutants including AML1a and examined their hematopoietic potential by delivering them into the AML1-deficient P-Sp region in our coculture system. AML1bΔ444 and AML1bΔ397, which possess the trans-activation subdomain, retained the ability to rescue the hematopoietic defect of the AML1-deficient P-Sp region (Figure 6A-B). In contrast, AML1bΔ335, AML1bΔ288, and AML1a, which lack the trans-activation domain, failed to produce any hematopoietic cells (Figure 6C-E). In addition, AML1bΔ(205-332), which retains the C-terminal region but lacks a half of the activation domain, has also lost the hematopoietic potential (Figure 6G). Thus, consistent with the observation in the previous report,35 in vitro hematopoietic rescue requires the trans-activation domain of AML1, whereas the C-terminal repression domain including VWRPY motif is dispensable for this function.
Hematopoietic potential of the AML1 mutants. Cells isolated from AML1-deficient P-Sp regions were infected with retrovirus containing the AML1 mutants. Each retrovirus contained (A) AML1bΔ444, (B) AML1bΔ397, (C) AML1bΔ335, (D) AML1bΔ288, (E) AML1a, (F) AML1bΔRD, (G) AML1bΔ(205-332), (H) AML1bΔ(181-210), (I) AML1b-R139G, (J) AML1b-S249/266A, or (K) AML1b-K24/43R. AML1bΔ444 (A), AML1bΔ397 (B), AML1bΔ(181-210) (H), AML1b-S249/266A (J), and AML1b-K24/43R (K) retain the ability to rescue the hematopoietic defect of AML1-deficient P-Sp regions, whereas other mutants do not. Shown are phase-contrast microscopic views of these cultures at 14 days. Photographs were taken with a Nikon Eclipse TE2000-U (Nikon Sankei) at a magnification of × 100.
Hematopoietic potential of the AML1 mutants. Cells isolated from AML1-deficient P-Sp regions were infected with retrovirus containing the AML1 mutants. Each retrovirus contained (A) AML1bΔ444, (B) AML1bΔ397, (C) AML1bΔ335, (D) AML1bΔ288, (E) AML1a, (F) AML1bΔRD, (G) AML1bΔ(205-332), (H) AML1bΔ(181-210), (I) AML1b-R139G, (J) AML1b-S249/266A, or (K) AML1b-K24/43R. AML1bΔ444 (A), AML1bΔ397 (B), AML1bΔ(181-210) (H), AML1b-S249/266A (J), and AML1b-K24/43R (K) retain the ability to rescue the hematopoietic defect of AML1-deficient P-Sp regions, whereas other mutants do not. Shown are phase-contrast microscopic views of these cultures at 14 days. Photographs were taken with a Nikon Eclipse TE2000-U (Nikon Sankei) at a magnification of × 100.
The Runt domain of AML1 is essential for both DNA binding and heterodimerization with CBFβ, but its role in hematopoietic development has not yet been directly investigated. Therefore, we next examined the hematopoietic potential of AML1bΔRD, a deletion mutant that lacks the Runt domain and is defective for both DNA binding and heterodimerization with CBFβ. As shown in Figure 6F, AML1bΔRD could not rescue hematopoiesis from AML1-deficient P-Sp regions, indicating an essential role for the Runt domain in the hematopoietic potential of AML1. To elucidate more explicitly a role of DNA binding of AML1, we used AML1b-R139G, a mutant isolated from a patient with MDS, which harbors point mutation causing substitution of Arg139 in the Runt domain with Gly.13 The DNA-binding ability is severely impaired in AML1b-R139G, whereas heterodimerization with CBFβ is spared. As shown in Figure 6I, AML1b-R139G also failed to show any hematopoietic potential. These results indicate that DNA binding of AML1 through the Runt domain is also indispensable for in vitro hematopoietic rescue of the AML1-deficient P-Sp region.
Among corepressors that are recruited by AML1 is mSin3A, which may contribute to AML1-mediated repression of gene transcription, as well as to intracellular stability of AML1.20,46 Indeed, the AML1 mutant that cannot interact with mSin3A [AML1bΔ(181-210)] is defective for repression of the p21 promoter in the in vitro transcription response assay. Posttranscriptional modification is also one of the important mechanisms that regulate AML1 function.42,43 For example, transcriptional activity of AML1 is enhanced by extracellular signal-regulated kinase (ERK)–dependent phosphorylation on Ser249 and Ser266, whereas p300-mediated acetylation on Lys24 and Lys43 augments DNA binding of AML1. To clarify roles of these regulatory mechanisms in the hematopoietic potential of AML1, we used 3 types of AML1 mutants: AML1bΔ(181-210), AML1b-S249/266A, and AML1b-K24/43R. AML1bΔ(181-210) is an internal deletion mutant that lacks the binding domain for the mSin3A.20 In AML1b-S249/266A, the 2 target serines for ERK-mediated phosphorylation were replaced with alanines, which results in lack of ERK-induced enhancement of the transcriptional activity. Finally, AML1b-K24/43R is an acetylation-defective mutant, in which the 2 lysine residues were substituted with arginines. Remarkably, all of these mutants retained the ability to rescue the hematopoietic defect in contrast to the mutants of the Runt domain (Figure 6H,J,K). The cells rescued by these AML1 mutants contained CFCs, expressed hematopoietic cell surface markers, and were morphologically indistinguishable from the rescued cells by wild-type AML1b (data not shown). From these findings, we concluded that the hematopoietic potential of AML1 does not require the interaction with mSin3A, ERK-dependent phosphorylation, or p300-mediated acetylation. Some posttranslational modifications, as well as repressor activities, of AML1 may not necessarily be required for early hematopoietic development. Given that all of these mutants retain a basal activity of gene transcription,20,42,43 however, these results again argue a close correlation between the transcriptional activity of AML1 and its hematopoietic potential.
Runx2/AML3 and Runx3/AML2 have the capacity to rescue the hematopoietic defect of AML1-deficient P-Sp regions
In addition to AML1, there are 2 other known mammalian Runx transcription factors, Runx2/AML3 and Runx3/AML2. To determine whether these Runx proteins have the capacity to substitute for AML1 in hematopoiesis, we infected AML1-deficient P-Sp with retroviruses carrying Runx2/AML3 or Runx3/AML2. The infection efficiency and protein expression were assessed by the same method used for AML1 mutants (Figure 7A-B). Interestingly, enforced expression of either Runx2 or Runx3 in AML1-deficient P-Sp resulted in the generation of numerous hematopoietic cells (Figure 7C). There is no difference among the rescued hematopoietic cells by all 3 Runx proteins in terms of morphology, expression of surface markers, and CFC activity (data not shown). These results suggest redundant roles among Runx proteins in early hematopoietic development.
Runx2/AML3 and Runx3/AML2 have the capacity to rescue the hematopoietic defect of AML1-deficient P-Sp regions. (A) The efficiency of retrovirus-mediated gene transfer of Runx2/AML3 or Runx3/AML2 was estimated by infecting NIH3T3 cells. Retrovirus-infected cells were evaluated by the expression of GFP (shaded histograms). Also shown are the noninfected NIH3T3 cells (open histograms). (B) Expression of 3 Runx proteins (AML1, Runx2/AML3, and Runx3/AML2) in infected NIH3T3 cells. The expression is monitored by immunoblotting of whole-cell lysates with anti-Flag. (C) Both Runx2/AML3 and Runx3/AML2 have the capacity to rescue the hematopoietic defect of AML1-deficient P-Sp regions. Shown are phase-contrast microscopic views of these cultures at 14 days visualized using a Nikon Eclipse TE2000-U (Nikon Sankei) at a magnification of × 100.
Runx2/AML3 and Runx3/AML2 have the capacity to rescue the hematopoietic defect of AML1-deficient P-Sp regions. (A) The efficiency of retrovirus-mediated gene transfer of Runx2/AML3 or Runx3/AML2 was estimated by infecting NIH3T3 cells. Retrovirus-infected cells were evaluated by the expression of GFP (shaded histograms). Also shown are the noninfected NIH3T3 cells (open histograms). (B) Expression of 3 Runx proteins (AML1, Runx2/AML3, and Runx3/AML2) in infected NIH3T3 cells. The expression is monitored by immunoblotting of whole-cell lysates with anti-Flag. (C) Both Runx2/AML3 and Runx3/AML2 have the capacity to rescue the hematopoietic defect of AML1-deficient P-Sp regions. Shown are phase-contrast microscopic views of these cultures at 14 days visualized using a Nikon Eclipse TE2000-U (Nikon Sankei) at a magnification of × 100.
Discussion
The striking phenotype of AML1-deficient mice has demonstrated an essential role for AML1 in the formation of definitive hematopoiesis during development. However, domain contribution of AML1 in early hematopoietic development has not yet been fully elucidated. Here we described an assay for AML1 function based on the ability to rescue hematopoiesis from the AML1-deficient P-Sp regions. Using this system, we found that the hematopoietic potential of AML1 was closely related to its transcriptional activity. Among those mutants used in this study, AML1bΔ444, AML1bΔ397, and AML1bΔ(181-210) are transcriptionally active in a luciferase assay (Kurokawa et al41 ; data not shown). AML1b-S249/266A and AML1b-K24/43R also retain a basal activity of gene transcription. All of these transcriptionally active mutants of AML1 could confer hematopoietic activity on AML1-deficient P-Sp regions. On the contrary, other mutants that lose the transcriptional activation ability (Lutterbach et al,20 Kurokawa et al41 ; data not shown) did not rescue the hematopoietic defect. Previously, the C-terminal deletion mutants of AML1b were analyzed with the ES cell culture system. Among them, the mutants containing trans-activation subdomains retained the hematopoietic potential.35 In the present study, we extended these analyses by examining various AML1 mutants and clearly demonstrated that the transcriptional activity of AML1 is essential for in vitro hematopoietic rescue of AML1-deficient P-Sp regions.
AML1 can also function as a transcriptional repressor depending on the target gene and the cellular context by recruiting corepressors, such as transduction-like enhancer of split (TLE) and mSin3A. Of these, TLE interacts with AML1 by recognizing its C-terminal VWRPY motif,21-23 and mSin3A interacts mainly through the region between amino acids 181 and 210.20 As shown in the current study, the deletion mutants of AML1 that do not interact with these corepressors retained the hematopoietic potential. Therefore, repressor activity of AML1 appears dispensable in early hematopoietic development. Recently, investigations using T-cell–specific AML1-deficient mice demonstrated that AML1 has critical functions during thymocyte development.47 In addition, the trans-repression activity of AML1 was suggested to play a role in early thymocyte development.36 Therefore, the function of AML1 as a transcriptional repressor should be important for appropriate T-cell differentiation. Because the culture system described here lacks the ability to support T-cell development, we are currently establishing another in vitro culture system to investigate the domain contribution of AML1 in T-cell development.
Although a genetic mutation of AML1 has been found in patients with hematologic malignancies,12-14 the precise mechanisms of leukemogenesis caused by these mutations remain uncertain. Significant in this regard is our observation that AML1b-R139G, the AML1 mutant found in a MDS patient,13 has lost the hematopoietic potential. This is the first direct evidence that the point mutation in the AML1 gene, which was found in a patient with hematologic malignancy, leads to loss of its biologic activity in hematopoiesis. Taken together, this experimental system should contribute to further clarification of the molecular basis for leukemogenesis mediated by subtle mutations in the AML1 gene.
Our study clearly demonstrated that both Runx2/AML3 and Runx3/AML2 have the capacity to rescue the hematopoietic defect of AML1-deficient P-Sp regions. Moreover, we also showed that “human” Runx proteins could substitute for the “murine” AML1 in early hematopoietic development because we used human cDNAs in this study. These results are consistent with the fact that the Runt and the trans-activation domains, which are essential for hematopoiesis, are highly conserved among members of mammalian Runx family. Thus, Runx-mediated hematopoietic activity depends on the evolutionarily conserved domains in Runx proteins.
In summary, we established an experimental culture system to efficiently examine the hematopoietic potential of Runx transcription factors. By analysis of the mutants, we precisely mapped the region responsible for the hematopoietic potential of AML1 and demonstrated that the transcriptional activity of AML1 is essential for early hematopoietic development. Furthermore, our results suggest a functional redundancy of mammalian Runx proteins in hematopoiesis.
Prepublished online as Blood First Edition Paper, July 22, 2004; DOI 10.1182/blood-2004-04-1535.
Supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science and by Health and Labour Sciences Research grants from the Ministry of Health, Labour, and Welfare.
An Inside Blood analysis of this article appears in the front of this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank M. Ohki for the gift of the human AML1 cDNA, Y. Ito for the human Runx2/AML3 and Runx3/AML2 cDNAs, T. Kitamura for the Plat-E packaging cells and pMY/IRES-EGFP retrovirus vector, T. Nakano for the OP9 stromal cells, and Kirin Brewery Pharmaceutical Research Laboratory for cytokines.







This feature is available to Subscribers Only
Sign In or Create an Account Close Modal