Abstract
In the past several years, experiments using DNA microarrays have contributed to an increasingly refined molecular taxonomy of hematologic malignancies. In addition to the characterization of molecular profiles for known diagnostic classifications, studies have defined patterns of gene expression corresponding to specific molecular abnormalities, oncologic phenotypes, and clinical outcomes. Furthermore, novel subclasses with distinct molecular profiles and clinical behaviors have been identified. In some cases, specific cellular pathways have been highlighted that can be therapeutically targeted. The findings of microarray studies are beginning to enter clinical practice as novel diagnostic tests, and clinical trials are ongoing in which therapeutic agents are being used to target pathways that were identified by gene expression profiling. While the technology of DNA microarrays is becoming well established, genome-wide surveys of gene expression generate large data sets that can easily lead to spurious conclusions. Many challenges remain in the statistical interpretation of gene expression data and the biologic validation of findings. As data accumulate and analyses become more sophisticated, genomic technologies offer the potential to generate increasingly sophisticated insights into the complex molecular circuitry of hematologic malignancies. This review summarizes the current state of discovery and addresses key areas for future research.
Introduction
In the wake of the sequencing of the human genome, the field of genomics has grown to encompass a range of experimental approaches that take advantage of the vast quantity of genetic information that is now available. Large-scale, highly parallel surveys of gene expression, genetic polymorphisms, and protein abundance are among the analyses that have become both technically feasible and widely accessible. This review will focus on the utility and promise of the first wave of experimental use of nucleic acid microarrays to study hematologic malignancies.
The behavior of a transformed cell is determined by the state of activation of key cellular pathways, each with corresponding effects on gene expression. Chromosomal translocations and other oncogenic mutations alter the expression of sets of genes. Likewise, cellular differentiation is reflected in lineage-specific patterns of gene expression. Given that DNA microarrays quantitatively assay the abundance of mRNA transcripts on a genome-wide scale, this raises the possibility of using a single analytic platform to integrate information about myriad properties of a transformed hematopoietic cell.
Hematologic malignancies have been analyzed and classified on the basis of properties including morphology, cell surface markers, immunohistochemistry, and cytogenetic abnormalities. In principle, microarrays should capture much of the information assayed by these various techniques and broaden the scope of analysis to include pathways that are otherwise difficult to assess. Gene expression signatures might also help to refine the taxonomy of hematologic malignancies, predict response to therapy, and identify critical pathways that should be considered for therapeutic intervention.
The hematologic malignancies have been an attractive training ground for the development of genomic approaches to cancer. Importantly, biopsy specimens from hematologic malignancies can be sorted into purified cell populations based on the expression of cell surface markers. In addition, the extensive knowledge about the differentiation programs in hematopoiesis and the genetic abnormalities in hematologic malignancies have aided in the interpretation of complex gene expression data, facilitating the generation of testable biologic hypotheses that would be less tractable in solid tumors. This review discusses the basic experimental and analytic methodologies relevant to microarray approaches to hematologic malignancies.
While genomic technologies have tremendous potential, extracting the biologically meaningful structure from the large and often “noisy” data sets is challenging. In some cases, gene expression–based predictors of clinical outcome have been difficult to replicate in independent data sets. The validation of findings from microarray studies requires careful assessment of statistical significance, replication of findings in independent data sets, and experimental systems to test biologic hypotheses.
Methodologies in genomics
DNA microarrays
Multiple techniques have been developed to monitor expression of large numbers of genes including differential display,1 serial analysis of gene expression (SAGE),2 representational differential analysis,3 and DNA microarrays. Microarrays have emerged as the most commonly used method, generating quantitative information about the expression of thousands of genes with relative facility, rapidity, and reproducibility, as well as falling costs. Table 1 lists websites that contain protocols, data analysis software, and other information about microarray experiments.
Resources for microarray experiments
Website . | URL . | Resources . |
|---|---|---|
| National Human Genome Research Institute | http://research.nhgri.nih.gov/microarray/main.html | Protocols, web links |
| Broad Institute, Cancer Genomics Group | http://www.broad.mit.edu/cancer/ | Gene expression analysis software, web links |
| Stanford Genomics | http://genome-www.stanford.edu/ | Software, web links, cDNA microarray protocols |
| Jackson Laboratory, Statistical Genomics Group | http://www.jax.org/staff/churchill/labsite/ | Advice about the design of microarray experiments |
| Microarray Gene Expression Data (MGED) Society | http://www.mged.org/Workgroups/MIAME/miame_checklist.html | Minimal requirements for the publication of microarray data |
| Gene Expression Omnibus database | http://www.ncbi.nlm.nih.gov/geo/ | Repository of gene expression data |
| Array Express database | http://www.ebi.ac.uk/arrayexpress/ | Repository of gene expression data |
Website . | URL . | Resources . |
|---|---|---|
| National Human Genome Research Institute | http://research.nhgri.nih.gov/microarray/main.html | Protocols, web links |
| Broad Institute, Cancer Genomics Group | http://www.broad.mit.edu/cancer/ | Gene expression analysis software, web links |
| Stanford Genomics | http://genome-www.stanford.edu/ | Software, web links, cDNA microarray protocols |
| Jackson Laboratory, Statistical Genomics Group | http://www.jax.org/staff/churchill/labsite/ | Advice about the design of microarray experiments |
| Microarray Gene Expression Data (MGED) Society | http://www.mged.org/Workgroups/MIAME/miame_checklist.html | Minimal requirements for the publication of microarray data |
| Gene Expression Omnibus database | http://www.ncbi.nlm.nih.gov/geo/ | Repository of gene expression data |
| Array Express database | http://www.ebi.ac.uk/arrayexpress/ | Repository of gene expression data |
In DNA microarray experiments, DNA “probes” are arrayed on a platform such as a glass slide, nylon membrane, or silicon wafer. “Target,” cDNA or cRNA generated from sample RNA and labeled with a fluorescent dye or biotin, is hybridized to the microarray. A scanner then measures fluorescence at the site of each unique probe. Microarrays vary in the type of probe used, the manner in which probes are arrayed onto a solid support, and the method of target preparation. These differences can impact experimental design and interpretation, but it is becoming clear that robust and reproducible gene expression data can be generated on multiple platforms so the details of the microarrays themselves have become less critical.
The probes on a cDNA microarray are cDNA fragments, generated by polymerase chain reaction (PCR) amplification of cDNA clone inserts, that are robotically spotted onto a glass slide.4 For laboratories with access to the robotic equipment, this can be a relatively inexpensive microarray to produce. In practice, however, the chemistry for adhering DNA to glass is finicky, and the generation of high-quality cDNA arrays in individual research laboratories has been more problematic than one might hope. A technical consideration with cDNA microarrays is the variability in the amount of each probe that is robotically spotted on different arrays. To control for this inconsistency, sample RNA is often hybridized to the array in combination with a fixed amount of reference RNA, each labeled with a different fluor. cDNA arrays can also suffer from problems with cross-hybridization because cDNA probes often contain nonunique or repetitive sequences. Avoiding such nonunique regions is tedious when experiments are performed on a genome-wide scale. A significant advantage of cDNA arrays, however, is that they do not require prior sequence information. While this is no longer of significant value for human and mouse genomes, cDNA arrays are an attractive alternative for model organisms whose genomes are not yet sequenced. For other purposes, however, it is likely that cDNA arrays will give way to oligonucleotide arrays.
Oligonucleotide arrays are constructed with probes between 25 and 60 nucleotides in length that are either synthesized in situ on a silicon wafer or robotically spotted on glass slides.5 Unique oligonucleotide sequences can be selected by comparison to the entire genome and by the use of empiric rules governing hybridization properties. A challenge in all microarray experiments is that a single hybridization condition must be used for all probes. Short (25-mer) oligonucleotide arrays, such as those manufactured by Affymetrix (Santa Clara, CA), involve the in situ photolithographic synthesis of oligonucleotides. Because of the short probe length, hybridization specificity is controlled for by the inclusion of a second oligonucleotide probe that contains a sequence variant at the central (13th) nucleotide and by the selection of multiple (11-16) different probes representing each transcript. Due to the robustness of the manufacturing process, single-color hybridization is sufficient. In an alternative method, oligonucleotides are synthesized on microarrays in situ, a process that allows relative flexibility in the design of individual microarrays.6 The merits of various oligonucleotide microarray platforms are debated, but it is likely that the major determinant of experimental success relates to experimental design and the degree of biologic noise inherent in the experimental system. The extent to which increasingly sensitive and accurate microarrays will impact on biologic discovery remains to be determined.
Samples
Sources of RNA in studies of hematologic malignancies include peripheral blood, bone marrow, tissue biopsies, and cultured cells. Vagaries of sample collection and preparation may have dramatic effects on gene expression data. RNA production and degradation continue after a biopsy is performed, so samples should be processed or snap-frozen as expeditiously as possible. Remarkably, however, the fundamental biologic aspects of tissues appear to be preserved despite the heterogeneity of the sampling and labeling process.
A biopsy specimen, such as a bone marrow biopsy, is composed of a complex mix of malignant and nontransformed cells. Furthermore, the malignant cell population may be genetically diverse due to genetic instability. One approach to this cellular heterogeneity is to purify malignant cells by cell sorting or laser capture microdissection.7 This has the advantage of yielding a more homogeneous population of cells for study but has the disadvantage of increasing the processing time and extent of tissue manipulation. While nonmalignant components of tumors are often considered “contaminating,” it is likely that such nonmalignant cells carry important information regarding the pathogenesis of the malignancy in question. Cell sorting generally requires the use of fresh, prospectively collected specimens that are often not routinely available. Perhaps the greatest challenge in microarray analysis of clinical specimens is the availability of both properly stored tissues and appropriate clinical annotation including long-term follow-up. The lack of the latter has been particularly problematic, but banking efforts nationwide should yield improved resources in the years ahead.
Microarray experiments generally require approximately 5 μg of total RNA or approximately 5 × 105 cells. Rare cell populations or very small biopsy specimens may therefore have insufficient RNA for routine analysis. Techniques are being developed to amplify smaller quantities of RNA, such as performing 2 rounds of linear amplification by in vitro transcription.8 Through this procedure, as little as 10 ng of RNA or as few as 1000 cells can generate sufficient labeled cRNA to obtain reproducible microarray data. The fidelity of this amplification method likely does not match that of routine sample labeling, but for some applications it is the only alternative available. Whole-transcriptome PCR amplification approaches have been proposed, but these generally suffer from nonuniform amplification of transcripts across the genome. Formalin-fixed, paraffin-embedded tissues have been used for gene-specific reverse transcriptase–PCR (RT-PCR)–based detection of RNA.9 Methods for the application of such samples to microarray analysis have yet to be validated on a genome-wide scale and may be limited due to the fragmentation of RNA caused by formalin fixation.
Data analysis
The high dimensionality of microarray data presents new analytic challenges. The number of samples analyzed is often quite modest, but the number of genes can be enormous, often 20 000 or more. This situation is distinct from most clinical correlative studies in which a large number of samples are analyzed with respect to a limited number of variables. The statistical techniques for evaluating large gene expression data sets can be divided into 2 general categories: supervised learning and unsupervised learning. Supervised learning is used for class prediction, the identification of gene markers that correlate with known class distinctions, whereas unsupervised learning (often synonymous with clustering) is used for class discovery, the identification of a novel taxonomy based on underlying structure of the gene expression data.
Supervised learning uses known class labels to identify genes that correlate with classes such as cancer type or clinical outcome. Genes that correlate highly with a particular malignancy may be valuable diagnostic markers and may be candidates for further biologic investigation. There are 2 basic components to supervised learning–based classification: feature selection and classifier generation. There are multiple metrics that can be used for identifying the features (genes) correlated with a particular distinction of interest. These methods can generate slightly different lists of marker genes, but most methods are best suited for the identification of genes that are uniformly overexpressed in one class compared with the other. Better feature selection methods are therefore needed to identify genes with variable expression within a class, reflecting the complexity of biology. This may be particularly important, for example, in the identification of genes correlated with response to therapy in which treatment response may be governed by more than one biologic mechanism.
Marker genes that are differentially expressed between classes can be used to formulate a gene expression–based classifier. A large number of such classifiers have been reported, including weighted voting,10 k-nearest neighbors (k-NN),11 support vector machines,12,13 artificial neural networks,14 decision trees,15 and nearest shrunken centroid algorithms.16 For the most part, these different methods yield similar classification results, and it is yet to be determined which of these has the optimal properties for gene expression–based classification. In general, if only one algorithm is capable of generating accurate classification results, one should worry that a problem exists (such as a software bug). If biologically important structure exists in a dataset, it should in most cases be identifiable with multiple machine learning algorithms.
In the weighted voting classifier, each marker gene is given a vote weighted according to its value in discriminating between classes of interest. In k-NN analyses, the distance (in gene expression space) is calculated between a test sample and each of the samples in a training set. The class of the test sample is assigned to that of the neighboring (k) samples of known class. In the support vector machine algorithm, a hyperplane is defined that separates 2 classes in high-dimensional space. This method has been shown to have high accuracy but, like nearly all supervised learning methods, is prone to overfitting to an initial training set, resulting in classifiers that perform well on the data sets on which they were trained but perform less well when extended to other data sets. Thus with all methods, it is important to appropriately (and conservatively) estimate statistical significance in order to avoid overestimating accuracy due to overfitting.
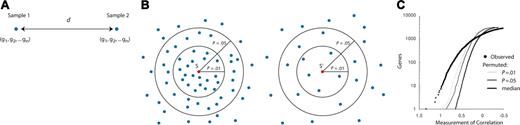
In some cases (eg, for biologic exploration), formal classifiers are not desired, but differentially expressed marker genes are sought between 2 experimental conditions. When comparing any 2 groups of samples, some degree of differential expression is always observed. The issue is therefore the extent to which the observed differential expression exceeds that expected by chance alone. This is generally based on some form of permutation testing such as that used in neighborhood analysis10 (Figure 1) or in calculation of the false discovery rate (FDR).17 In this manner, the class labels (eg, patient survival) are randomly permuted and gene expression correlates of these permuted labels are identified. The observed correlation can then be compared with the permuted values. This approach should be distinguished from simply randomizing the gene expression values themselves. Such a procedure would destroy the intrinsic (and extensive) correlation structure in gene expression data, thereby overestimating the significance of any observed gene-class correlations.
Neighborhood analysis. Panel A depicts 2 samples as vectors in gene expression space. The coordinates of the sample vector are composed of expression levels (g1, g2,..., gn) for each gene in the sample. The distance (d) can be calculated between 2 samples. Panel B is a schematic of a neighborhood analysis. A correlation is calculated between the expression of gene S and other genes with similar patterns of expression across different samples. On the right side of panel B, the coordinates of the genes have been randomly permuted, so the number of genes that correlate with S′ at a given level of significance is decreased. Panel C graphically illustrates a neighborhood analysis. The number of genes in a neighborhood increases as the measure of correlation decreases.
Neighborhood analysis. Panel A depicts 2 samples as vectors in gene expression space. The coordinates of the sample vector are composed of expression levels (g1, g2,..., gn) for each gene in the sample. The distance (d) can be calculated between 2 samples. Panel B is a schematic of a neighborhood analysis. A correlation is calculated between the expression of gene S and other genes with similar patterns of expression across different samples. On the right side of panel B, the coordinates of the genes have been randomly permuted, so the number of genes that correlate with S′ at a given level of significance is decreased. Panel C graphically illustrates a neighborhood analysis. The number of genes in a neighborhood increases as the measure of correlation decreases.
In unsupervised learning, samples are grouped together based solely on the gene expression data, without any a priori knowledge of the sample labels (eg, their biologic or clinical features). Hierarchical clustering, for example, creates a dendrogram based on pairwise similarities in gene expression within a set of samples.18 The length of the branches of the dendrogram reflects the similarity between genes or between samples. An alternative clustering algorithm, self-organizing maps (SOMs), groups samples (or genes) into a predefined number of clusters.19 The samples aggregate around “centroids,” which have been iteratively altered to fit the data. This algorithm is well suited for exploratory analyses of gene expression data and can generate a useful “executive summary” of the data. The method is limited, however, by the need to predefine the number of clusters, and in many cases the optimal number of clusters is not known. Other unsupervised learning methods include K-means clustering,20 principle component analysis (PCA),11 and nonnegative matrix factorization (NMF).21
The optimal statistical methodologies for analyzing microarray data continue to evolve, but in most cases these analytic approaches generally expose the same overall structure in a dataset. It is worth noting, however, that the most predominant patterns of coordinate gene expression identified by these methods are not necessarily the most biologically important. More subtle, yet biologically significant, structure may be buried beneath the dominant structure identified in a dataset and as such can be difficult to recover.
A challenge common to all of these methods is the difficulty in determining the biologic meaning of observed structure in a data set. In most cases, such biologic interpretation is highly subjective and thus fraught with potential for overinterpretation. It is expected that in the years ahead, as the functional annotation of the genome and of sets of coordinately regulated genes is elucidated, such biologic interpretation of genomic patterns will become less enigmatic.
Validation
Assessment of statistical significance and validation of findings are critical steps in the analysis of microarray data. Ideally, findings are replicated and confirmed with a separate set of samples. For this reason, investigators often divide their samples into a training set and a validation set. In this way, a classifier that is formulated using the training set can be assessed for its true accuracy using the validation set.
Often, insufficient samples are available to reserve a portion of the samples for an independent validation set. An alternative statistical technique for validating results is the “leave one out” cross-validation method. A classifier is created using the entire data set except one sample, and the classification of the remaining sample is predicted. The procedure is repeated leaving each sample out sequentially, and the percentage of samples correctly predicted is calculated. While this method is useful for estimating the robustness of a classifier, it tends to overestimate accuracy. True accuracy must thus await independent validation. Ultimately, scientific validation is achieved by repetition of experiments by other investigators at different institutions using independent data sets. Ideally, this would also be accomplished on alternative technology platforms (ie, types of microarrays). To facilitate this process, the international Microarray Gene Expression Data (MGED) group has created a standard for publication, the Minimal Information About a Microarray Experiment (MIAME) guidelines (Table 1), and submission of the raw microarray data at the time of publication is now expected. We believe that all published microarray data should be made publicly available in its entirety, without restriction. Repositories for such data have recently been established for this purpose (Table 1).
Molecular taxonomy of hematologic malignancies
Acute leukemia
Large-scale analyses of gene expression in leukemia have several experimental advantages relative to other malignancies. Compared with biopsies of solid tumors, bone marrow aspirates and peripheral blood samples are easier to obtain and are composed of a relatively uniform population of cells. Comparison of samples from patients with acute myeloid leukemia (AML) or acute lymphoblastic leukemia (ALL) by oligonucleotide microarray was an early test case for the potential of genomics-based classification.10 Of 6817 genes assayed in 38 patients, approximately 1100 correlated strongly with the AML-versus-ALL distinction. A class predictor was created by assigning a weighted vote to each of the 50 genes that correlated most highly with the AML-versus-ALL distinction, and accuracy of the predictor was confirmed both through cross-validation and using an independent data set. Using an unsupervised learning approach, the samples clustered correctly into the appropriate classification, AML or ALL, and further into 2 known subgroups, B-cell ALL and T-cell ALL. Gene expression data therefore independently revealed underlying biologic categories and predicted the diagnosis of unknown samples with high accuracy. Of course such distinctions were previously known. Nevertheless, this study demonstrated that biologic structure could be extracted from high-dimensional, biologic noisy data derived from patient samples.
More recently, studies of pediatric ALL have defined gene expression patterns associated with specific molecular abnormalities. In the largest such study, leukemic blasts from 360 cases of pediatric ALL were applied to DNA microarrays.22 By unsupervised clustering, 6 known clinical subtypes of ALL were identified (T-cell ALL, E2A-PBX1, BCR-ABL, TEL-AML1, MLL rearrangement, and hyperdiploid > 50 chromosomes), reflecting the major cytogenetic subclasses of the disease (Figure 2). A further subtype was observed in patients lacking specific cytogenetic abnormalities, but the biologic significance of this group still requires validation. Overall, the subgroups of pediatric ALL had gene expression profiles as divergent as those seen in different epithelial cancers. Consistent with these strong gene expression signatures, robust classifiers could be generated with accuracy exceeding 95%. Notably, this study was recently repeated on newer generation arrays (Affymetrix U133) yielding similar results.23 The implication of these studies is that cytogenetic classes can be accurately predicted based on gene expression alone. This is significant because ALL cytogenetics is notoriously difficult to perform, and only a small number of medical centers are able to generate reproducibly high-quality cytogenetic data from patient blasts. The availability of expression correlates of cytogenetic findings suggests that leukemia classification might be performed in a more robust, standardizable fashion using automatable gene expression profiling methods.
Multidimensional scaling plot of expression profiles from patients with ALL. Bone marrow samples from patients with ALL are each represented by a sphere. The color of the spheres corresponds to the indicated molecular abnormalities. The high dimensionality of the gene expression data has been reduced to the 3 dimensions that comprise the greatest variation across the dataset. In the top panel, the 3 component dimensions separate cases of T-ALL, E2A-PBX1, TEL-AML1, and hyperdiploid of more than 50 chromosomes from remaining ALL cases. In the bottom panel, 3 different component dimensions discriminate cases with the BCR-ABL translocation, MLL gene rearrangement, and a novel subgroup. Reprinted from Yeoh et al22 with permission.
Multidimensional scaling plot of expression profiles from patients with ALL. Bone marrow samples from patients with ALL are each represented by a sphere. The color of the spheres corresponds to the indicated molecular abnormalities. The high dimensionality of the gene expression data has been reduced to the 3 dimensions that comprise the greatest variation across the dataset. In the top panel, the 3 component dimensions separate cases of T-ALL, E2A-PBX1, TEL-AML1, and hyperdiploid of more than 50 chromosomes from remaining ALL cases. In the bottom panel, 3 different component dimensions discriminate cases with the BCR-ABL translocation, MLL gene rearrangement, and a novel subgroup. Reprinted from Yeoh et al22 with permission.
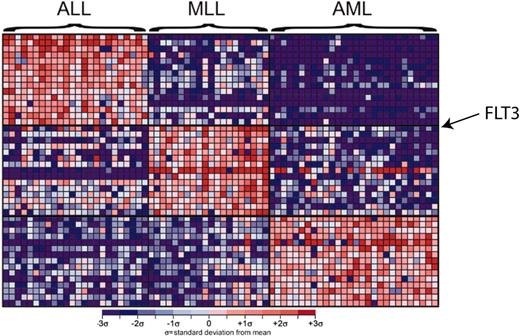
Other studies focused specifically on samples from patients with translocations involving the mixed-lineage leukemia (MLL) gene on chromosome 11q23.24-26 The expression profile of MLL samples was consistent with early lymphoid progenitor cells, suggesting a maturational arrest at an early stage in hematopoiesis. The MLL-specific gene expression profile included elevated expression of the FLT3 gene (Figure 3). Further analysis of the FLT3 gene revealed novel activating mutations in a subset of patients with MLL.25 FLT3 mutations have also been observed in patients with AML, further supporting the notion that FLT3 might be a functionally important target. To explore this possibility, the functional consequence of pharmacologic inhibition of FLT3 was explored in a xenograft model of MLL.25 PKC412, a small molecule inhibitor of FLT3, was administered to mice inoculated with the human MLL cell line, SEMK2-M1. Leukemic progression was dramatically abrogated in these mice, suggesting that FLT3 may in fact be a bona fide therapeutic target in MLL. These studies were further surprising in that they demonstrated that expression profiling primarily aimed at diagnostic classification can also yield important insights into the molecular pathophysiology of malignancy.
Gene expression profiles of bone marrow samples from patients with ALL, MLL, and AML. Each column represents a bone marrow sample and each row corresponds to a gene. The top marker genes for each diagnosis are shown. Shades of red indicate elevated expression while shades of blue indicate decreased expression. FLT3 is the gene that correlates most highly with the MLL leukemia subtype. Reprinted from Armstrong et al24 with permission.
Gene expression profiles of bone marrow samples from patients with ALL, MLL, and AML. Each column represents a bone marrow sample and each row corresponds to a gene. The top marker genes for each diagnosis are shown. Shades of red indicate elevated expression while shades of blue indicate decreased expression. FLT3 is the gene that correlates most highly with the MLL leukemia subtype. Reprinted from Armstrong et al24 with permission.
Similar progress has been made in the analysis of T-cell leukemia.27 Gene expression patterns revealed dysregulated expression of the key oncogenes HOX11, TAL1 (SCL), LYL1, LMO1, and LMO2 and indicated that these transcription factor–correlated gene expression programs were mutually exclusive. Importantly, overexpression of these key transcription factors was not limited to those cases in which the factors were rearranged by chromosomal rearrangement, consistent with the notion that aberrant expression can also occur through cis-acting mechanisms. Furthermore, the activation of transcription factor–associated gene expression patterns was linked to specific stages in normal thymocyte development, providing a biologic basis for divergent clinical behavior of T-cell ALL subgroups.
Two recent reports document the first large-scale efforts to catalog the diversity of gene expression profiles in AML.28,29 Bullinger et al28 used cDNA arrays to study 116 AML samples, whereas Valk et al29 used commercial oligonucleotide arrays to study 285 patients. Both studies clearly show that the well-recognized cytogenetic abnormalities seen in AML (eg, t(8;21) AML1/ETO, inv(16) CBFB/MHY11, t(15;17) PML/RARα) are associated with distinct gene expression signatures, suggesting that detection of these events in the future might be accomplished using gene expression–based techniques. Given the known association of cytogenetic abnormalities with clinical outcome in AML, it is of course not surprising that the gene expression correlates of these abnormalities are similarly predictive of survival.
Perhaps more interestingly, the studies also indicate that AMLs with “normal” karyotypes are largely distinguishable on the basis of gene expression from those with characteristic chromosomal translocation. Moreover, Bullinger et al28 argue that AMLs with normal karyotypes fall into 2 basic subgroups based on gene expression profiles that are associated with different overall survival. While this finding has potential clinical significance, it should be noted that the observation is based on a test set of only 22 patients, yielding a result of borderline significance (P = .046). Further validation is clearly needed. Nevertheless, the results are encouraging and suggest that the clinical diversity of AML may indeed be mapped onto distinct molecular programs that in the future might be translated into prognostic tests.
Diffuse large B-cell lymphoma
Diffuse large B-cell lymphoma (DLBCL) is a single diagnostic entity that encompasses lymphomas with a range of clinical behaviors and responses to therapy. Approximately 40% of patients are cured by anthracycline containing combination chemotherapy. Stratification of cases into subclasses with different clinical outcomes has not been possible with standard pathologic techniques. Gene expression profiling of DLBCL has led to important insights into the molecular heterogeneity of this important disease.
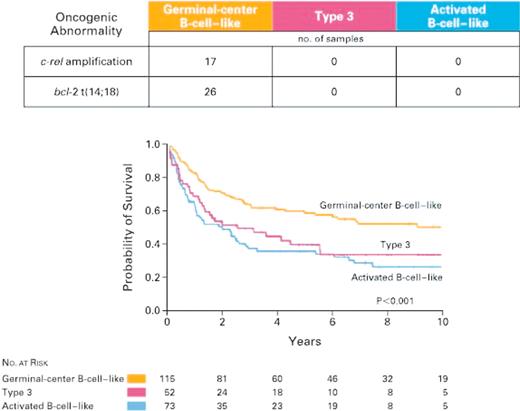
Alizadeh and colleagues30 were among the first to report heterogeneity of DLBCL from a global gene expression profiling perspective. A customized cDNA array (the “lymphochip”) was constructed containing 17 856 clones enriched in genes that are expressed in lymphoid cells or have been implicated in cancer biology. A broad spectrum of lymphoid malignancies, cell lines, and purified normal germinal center cells were profiled, and the data were subjected to unsupervised hierarchical clustering. This approach yielded 2 principal clusters of DLBCL: one that exhibited similarities to normal germinal center cells and one that exhibited similarities to in vitro–activated peripheral blood B cells. The t(14,18) translocation, involving the bcl-2 gene, was present in 26 cases, and amplification of the c-rel gene was found in 17 cases. Both cytogenetic abnormalities occurred exclusively in patients with germinal center B-cell–like DLBCL, evidence that the hierarchical clustering–defined DLBCL subclasses represented biologically meaningful classification.31 These results were interpreted to reflect differing cellular origins of DLBCL, and, importantly, these 2 groups were found to have significantly different clinical outcomes following standard therapy (Figure 4). Genes that predict clinical outcome clustered into previously defined gene expression signatures.30 Genes in the germinal center B cell, major histocompatibility complex (MHC) class II, and lymph node signatures predict a favorable outcome, while genes in the proliferation signature predict a poor outcome.31
Samples from 274 patients with DLBCL were assigned to one of 3 classes by hierarchical clustering according to the expression of 100 genes. Amplification of the c-rel locus and bcl-2 translocations were limited to the germinal center B-cell–like group. Kaplan-Meier estimates of overall survival after chemotherapy for 240 previously untreated patients are shown, demonstrating differences in clinical outcome between the gene expression subgroups. Reprinted from Rosenwald et al31 with permission.
Samples from 274 patients with DLBCL were assigned to one of 3 classes by hierarchical clustering according to the expression of 100 genes. Amplification of the c-rel locus and bcl-2 translocations were limited to the germinal center B-cell–like group. Kaplan-Meier estimates of overall survival after chemotherapy for 240 previously untreated patients are shown, demonstrating differences in clinical outcome between the gene expression subgroups. Reprinted from Rosenwald et al31 with permission.
This initial cell-of-origin classifier has undergone slight revision, but the basic observation of favorable prognosis of DLBCL harboring the germinal center gene expression pattern has been confirmed in 2 independent data sets31,32 and is independent of clinical prognostic factors (the International Prognostic Index). Whether or not the germinal center B-cell–like and activated B-cell–like signatures truly represent differing cells of origin of these tumors remains to be determined, but the prognostic value of the signatures appears clear. Interestingly, while unsupervised clustering was used for the discovery of the DLBCL cell-of-origin signatures, the genes represented on the array were enriched in those expressed in the germinal center, reflecting the notion that germinal center biology is important in the pathogenesis of DLBCL. As such, however, the analysis was not entirely unsupervised, and it remains to be determined whether the germinal center–like and activated B-cell–like subclasses represent the dominant structure in the molecular landscape of DLBCL.
An alternative approach to DLBCL outcome prediction was reported by Shipp and colleagues.33 Fifty-eight pretreatment samples with clinical follow-up were subjected to oligonucleotide microarray analysis followed by supervised learning–based outcome prediction. A 13-gene model was developed that assigned patients to 2 prognostic categories with 5-year overall survival of 70% versus 12%, but the accuracy of this exact model remains to be determined in independent data sets. Some of the genes in the outcome signature, however, were also correlated with outcome in the lymphochip study,30 including PKCβ, which was correlated with poor outcome in both studies. Nevertheless, the 13-gene outcome signature is largely nonoverlapping with the cell-of-origin signature, and it therefore will be important to determine to what extent these different studies are exposing different molecular aspects of DLBCL heterogeneity and to what extent the outcome prediction signatures withstand future clinical validation.
Another facet of DLBCL diagnostics is its distinction from other related diseases. Two recent papers demonstrated that the gene expression profile of mediastinal large B-cell lymphoma (MLBCL) is distinct from nonmediastinal DLBCL and DLBCL invading the mediastinum.34,35 Interestingly, MLBCL was found to have a molecular profile reminiscent of Hodgkin disease, with prominent expression of IL-13 receptor, STAT1, and TRAF1. While there has been a clinical suspicion that MLBCL represents a distinct molecular entity compared with DLBCL, this distinction is not always easy to make in clinical practice; the availability of molecular markers of the disease may prove useful. Whether MLBCL and Hodgkin disease share potential therapeutic targets remains to be determined.
Other hematologic malignancies
In addition to the key studies of acute leukemia and DLBCL, microarray technology has been applied to diverse hematologic malignancies including chronic lymphocytic leukemia (CLL),36,37 mantle cell lymphoma,38,39 mucosa-associated lymphoid tissue (MALT) lymphoma,40 Hodgkin disease,41,42 and other forms of leukemia and lymphoma.43-47 Smaller studies have started to address complex diagnostic entities such as myelodysplasia.
Different malignancies often share the same or related oncogenic events, and sets of genes associated with the outcome or behavior may apply to multiple malignancies. For example, a proliferation signature defined in germinal center B cells including, among other genes, elevated expression of G2/M phase regulators.48 The proliferation signature usefully predicts the proliferation rate and clinical outcome of mantle cell lymphoma38 and CLL.36 The characterization of other gene sets will facilitate the analysis of other malignancies.
Myelodysplasic syndromes (MDSs) are examples of more challenging malignancies to study by microarray. While the bone marrows of patients with leukemia are packed with relatively uniform blast cells, patients with MDS have marrows containing heterogeneous populations of cells along a continuum of normal and dysplastic differentiation. Furthermore, the molecular abnormalities and clinical classifications in MDS are less well characterized than, for example, the subtypes of leukemia defined by specific balanced translocations. No cell lines exist for in vitro examination of MDS. Initial microarray analyses of MDS have used unsorted bone marrow cells,49 purified CD34+ cells,50 and purified CD133+ cells.51 Further insights may be aided by larger data sets, focusing on patients with similar phenotypes or the same cytogenetic abnormalities, and sequential samples from patients with acute leukemia and antecedent MDS.
Experimental dissection of key pathways
In addition to their use in analyzing clinical specimens, DNA microarrays are valuable tools in more reductionist experimental paradigms. For example, the genetic targets of an oncogene may be identified by experimentally manipulating the expression of the oncogene and monitoring the effects by microarray. Gain-of-function experiments suffer from the fact that the appropriate cellular context for oncogene expression is often unknown. Similarly, loss-of-function experiments (whether by dominant-negative constructs, RNA interference, or pharmacologic treatment) suffer from potential lack of specificity. Such off-target effects may appear minimal when assaying only a limited number of analytes but can become hugely confounding when taking global views of the cell.52 In addition, dissecting the direct effects of a given perturbation from more indirect, downstream effects can be challenging. Careful time course studies can help with this, and cycloheximide can be employed as protein synthesis inhibitor to identify direct targets, though cycloheximide itself can produce significant global changes in gene expression. It is likely that the definitive elucidation of direct transcriptional targets will require both microarray-based expression profiling data and microarray-based genome-wide location analysis (eg, chromatin immunoprecipitation).
Microarrays have been used to study the targets of c-Myc, a transcription factor and cellular oncogene that is important in many malignancies including Burkitt lymphoma, in which the c-Myc is involved in chromosomal translocation. The genomic targets, including genes involved in cell growth, cell cycle, adhesion, and cytoskeletal organization, illustrate the myriad effects of c-Myc activation.53 Despite these studies, a complete understanding of the mechanisms of c-Myc action remains enigmatic. Similar experiments have highlighted the downstream targets for BCL-6, a transcriptional repressor that is translocated in many lymphomas,54 and Blimp-1, a transcriptional repressor involved in plasmacytic differentiation.55 The genetic targets of an oncogene may be determined computationally through patterns of coexpressed genes in gene expression databases. This strategy was successfully used and validated in the identification of CCAAT/enhancer-binding protein β (C/EBPβ) as a genetic target of cyclin D1 in cancer.56
The state of differentiation of a neoplastic cell is central to the phenotype of a malignancy. Microarray analyses of differentiation are largely descriptive, recapitulating what is already known about the differentiation process, but these experiments also generate novel findings and testable hypotheses. Gene expression signatures have been identified for multiple purified hematopoietic cell populations, including hematopoietic stem cells,57-61 plasma cells,62 and platelets.63 Using this type of gene expression data, AIDS-related primary effusion lymphoma was assigned to a plasmablastic cell of origin.64 The monitoring of differentiation of hematopoietic cell lines in vitro has revealed the complexity of genetic programs involved in hematopoietic differentiation. NB4 cells, derived from a patient with acute promyelocytic leukemia (APL), undergo neutrophilic differentiation in response to all-trans retinoic acid (ATRA). Microarray experiments revealed that the ubiquitin-activating enzyme E1–like (UBE1) gene is induced by ATRA in NB4 cells.19 Further experiments revealed that ATRA activates the UBE1 promoter, and overexpression of UBE1 triggers the degradation of PML/RARα and the apoptosis of APL cells.65
Molecular pharmacology
In principle, gene expression profiling offers the possibility of identifying specific pathways that are mutated in a patient's biopsy specimen and predicting the likelihood of response to a given therapeutic intervention. The ability to match targeted agents to appropriate tumors holds the promise of increased therapeutic efficacy and decreased toxicity. Despite this promising potential, the rate of clinically useful pharmacogenomic discovery has been slow. This is due in part to technical challenges (eg, noisy data) and also to the difficulty in obtaining sufficiently large numbers of uniformly treated patients with long-term clinical follow-up.
An early attempt to predict chemosensitivity was made in the gene expression profiling of established cancer cell lines. The NCI60, a set of 60 human cancer cell lines including 6 leukemia cell lines used by the National Cancer Institute for the testing of 70 000 potential chemotherapeutic agents, has been an experimental paradigm to assess whether gene expression profiles can predict response to a pharmacologic agent. One approach to analyzing this large dataset was to cluster the 60 cell lines in the space of all genes on the array.66 This largely recapitulated the organ of origin of the cell lines but did not provide direct insight into gene expression correlates of drug sensitivity. Supervised learning approaches were also applied to this dataset,67-69 and while there was some suggestion of predictability of chemosensitivity based on pretreatment expression profiles, for most compounds this was quite difficult. This was likely due in large measure to the great diversity of cell types within the NCI60 panel. For example, differences in chemosensitivity between leukemia cell lines and lung cell lines would be correlated with lineage-specific patterns of gene expression in addition to gene expression patterns actually governing chemosensitivity. The dissection of these 2 confounding factors is quite challenging, likely requiring larger data sets.
Applications in clinical diagnostics
It may be many years before we can obtain a gene expression profile for a patient's hematologic malignancy and use this information to formulate a prognosis and select from a large armamentarium of targeted therapies. Nevertheless, some applications of microarray-based research have the potential to find clinical utility much sooner. A challenging question is how to apply the information and technology from microarray-based experiments to clinical diagnostics in the near future.
The findings in genome-wide gene expression can be distilled into practical and useful clinical diagnostic tests in several ways (Table 2). A classifier based on a large number of genes would require a highly multiplexed detection method such as a customized or whole genome microarray. Bead or fiberoptic methods are also under development as alternative technologies for highly parallel measurement of gene expression.70,71 Although microarrays have not yet been introduced into clinical diagnostic laboratories, the cost and reproducibility would not be insurmountable. The gene expression–based distinction of ALL subtypes23 or the prediction of DLBCL clinical outcome31,33 would appear to be at present the closest to clinical development.
Clinical diagnostic tests developed from genome-wide gene expression data
Diagnostic methodology . | Advantages . | Disadvantages . | No. of features . |
|---|---|---|---|
| Whole genome microarray | Inclusive, universal platform for all applications | Expensive, complex data output | 1000s |
| Custom microarray | Allows assessment of many genes | Biased in only assessing selected genes | 10s to 100s |
| Other gene expression platforms: bead or fiberoptic based | Allows assessment of many genes | Requires further validation | 10s to 100s |
| Antibody microarray | Allows assessment of many proteins | Requires set of specific antibodies | 10s to 100s |
| Quantitative RT-PCR | Potential to analyze paraffin-embedded specimens | Challenging to multiplex | 10s |
| Flow cytometry | Routinely available | Limited to cell surface markers | 1 to several |
| Immunohistochemistry | Routinely available, spatial localization, assess morphology | Nonquantitative | 1 to several |
Diagnostic methodology . | Advantages . | Disadvantages . | No. of features . |
|---|---|---|---|
| Whole genome microarray | Inclusive, universal platform for all applications | Expensive, complex data output | 1000s |
| Custom microarray | Allows assessment of many genes | Biased in only assessing selected genes | 10s to 100s |
| Other gene expression platforms: bead or fiberoptic based | Allows assessment of many genes | Requires further validation | 10s to 100s |
| Antibody microarray | Allows assessment of many proteins | Requires set of specific antibodies | 10s to 100s |
| Quantitative RT-PCR | Potential to analyze paraffin-embedded specimens | Challenging to multiplex | 10s |
| Flow cytometry | Routinely available | Limited to cell surface markers | 1 to several |
| Immunohistochemistry | Routinely available, spatial localization, assess morphology | Nonquantitative | 1 to several |
A smaller number of genes could also be evaluated using alternative technologies such as multiplexed, quantitative RT-PCR. In other cases, microarray experiments may identify one or several markers that are useful by themselves. Such markers can be introduced into diagnostic laboratories immediately using traditional techniques such as flow cytometry and immunohistochemistry. For example, large-scale surveys of gene expression identified clusterin as a marker for anaplastic large-cell lymphoma,72 ZAP-70 as a marker of unmutated immunoglobulin locus in CLL,36,73 and CD58 as a marker of ALL cells.74
Conclusions
The collaboration of biologists, physicians, mathematicians, and many other scientists has created a fertile intellectual environment for the development of genomic approaches to questions of biologic and clinical relevance. Microarray technology is now widely accessible, and the evaluation of hematologic malignancies with gene expression profiling is burgeoning.
Genomic technologies offer a broad perspective into the molecular events in a transformed cell, but precision may be somewhat compromised in exchange for the large scope of the experiments. Validating the results of microarray experiments is one of the outstanding challenges for the future. Molecular signatures are being refined by developing larger data sets and integrating data across multiple platforms from multiple institutions. A primary aim will be to define gene expression–based classifiers and outcome predictors that are robust and reproducible, independent of subtleties of sample handling and methodology for detecting gene expression. Another major goal will be to develop high-throughput experimental systems to validate the numerous biologic hypotheses suggested by microarray data.
Increasingly, gene expression data are being correlated with information derived from other large-scale genomic technologies. High-throughput DNA sequencing, comparative genomic hybridization (CGH), and other DNA analyses facilitate the identification of genetic abnormalities in malignant cells. Patterns of methylation and chromatin structure can be surveyed on a genome-wide scale. Mass spectrometry and other proteomic technologies add information about the presence and activation of proteins.
Genomic technologies have the capacity to address the complexity of molecular networks in a transformed cell. Pathways that might be usefully targeted pharmacologically are highlighted and the process of drug development is facilitated.
Ultimately, genomic technologies will contribute to the central goals of parsing malignancies into diagnostic categories defined by specific molecular abnormalities and targeting the essential oncogenic pathways with specific therapies. As has been the case to date, this molecular reformation of oncology will likely be led by the hematologic malignancies. The challenge will be to bring this to fruition as quickly as possible, yet without compromising existing standards of care. The pace of such change, therefore, remains uncertain.
Prepublished online as Blood First Edition Paper, May 20, 2004; DOI 10.1182/blood-2004-01-0274.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal